Abstract
Megakaryopoiesis is a complex process that involves major cellular and nuclear changes and relies on controlled coordination of cellular proliferation and differentiation. These mechanisms are orchestrated in part by transcriptional regulators. The key hematopoietic transcription factor stem cell leukemia (SCL)/TAL1 is required in early hematopoietic progenitors for specification of the megakaryocytic lineage. These early functions have, so far, prevented full investigation of its role in megakaryocyte development in loss-of-function studies. Here, we report that SCL critically controls terminal megakaryocyte maturation. In vivo deletion of Scl specifically in the megakaryocytic lineage affects all key attributes of megakaryocyte progenitors (MkPs), namely, proliferation, ploidization, cytoplasmic maturation, and platelet release. Genome-wide expression analysis reveals increased expression of the cell-cycle regulator p21 in Scl-deleted MkPs. Importantly, p21 knockdown-mediated rescue of Scl-mutant MkPs shows full restoration of cell-cycle progression and partial rescue of the nuclear and cytoplasmic maturation defects. Therefore, SCL-mediated transcriptional control of p21 is essential for terminal maturation of MkPs. Our study provides a mechanistic link between a major hematopoietic transcriptional regulator, cell-cycle progression, and megakaryocytic differentiation.
Introduction
Megakaryocytes (MKs) are specialized blood cells that release platelets, the effectors of coagulation processes. During megakaryopoiesis, megakaryocyte progenitors (MkPs) coordinately proliferate and differentiate to develop into mature MKs that, ultimately, shed platelets. This complex biologic process requires profound cellular and nuclear changes (cytoplasm remodeling, cell size increase, nuclear polyploidization, and cytoskeletal dynamics) relying on an exquisite coordination of key cellular and molecular mechanisms.1
At an early stage in their development, MKs enter endomitosis (or abortive mitosis), during which DNA replicates without cell division. This process results in nuclear polyploidization, increase in cell size, and is linked to platelet formation.2,3 Formation of the demarcation membrane system (DMS, a reservoir for proplatelet membranes) and secretory granules reflects some of the specific characteristics of the cytoplasmic maturation of MKs.4,5
Signaling by thrombopoietin (TPO), the major megakaryocytic cytokine, induces PI3K activity, downstream of which lies the mammalian target of rapamycin pathway.6 Mammalian target of rapamycin is a kinase that controls cell size and cell-cycle progression in mammals and Drosophila.7,8 It also regulates key MK attributes (proliferation, cell size, cytoskeleton organization, and platelet formation) in part through control of G1/S cell-cycle progression.6,9 This control is mediated by sequential action of the cell-cycle regulators cyclin D3 (CCND3) and P21 (cyclin-dependent cell-cycle inhibitor, CDKN1a).9 In mouse MKs, cyclin D3 expression is high during endomitosis, leading to polyploidization10 ; in contrast, P21 expression is high in mature high ploidy MKs leading to G1/S progression blockade, thereby inducing cell-cycle arrest and terminal maturation.9
Numerous nuclear regulators play important roles in the transcriptional control of megakaryopoiesis.11,12 Several lines of evidence suggest that the basic helix-loop-helix protein stem cell leukemia (SCL)/TAL1, critical for hematopoietic specification13-16 and erythroid maturation17 (and references therein), is also functionally involved in megakaryopoiesis.
Enforced expression of SCL in human hematopoietic progenitors or mouse bone marrow cells enhances megakaryopoiesis.18-20 Loss-of-function studies have shown a requirement for SCL in megakaryopoiesis both during embryonic development and at the adult stage. Indeed, SCL is necessary for development of fetal liver megakaryopoiesis.21 In adult mice, analysis of short-term defects in Scl-deleted bone marrow cells revealed a block in MK development and loss of MkPs in vitro.22,23 This block was in part the result of perturbation of early committed hematopoietic progenitors and therefore precluded full analyses of the function of SCL in MKs. Characterization of long-term defects in the same mouse model revealed a late defect in MK differentiation under stress conditions leading to defective platelet production.24 In steady state, Scl-deleted mice displayed normal numbers of MKs with normal polyploidization. This overall mild phenotype probably resulted from the development of compensatory mechanisms. Taken together, these loss-of-function analyses strongly suggest that full investigation of the functions of SCL in MK differentiation requires a lineage-specific deletion approach.
SCL exerts its activity as part of large multiprotein complexes. Initial studies in erythroid cells have characterized its interaction with other transcription factors (E47, its heterodimerization partner, the LIM-domain binding protein LDB1, and the LIM-only domain protein LMO2) to form the SCL “core” complex.25,26 SCL acquires activating or repressing functions on recruitment of cofactors. ETO2 is a critical corepressor; its recruitment to the SCL complex determines the onset of red blood cell27 and MK terminal differentiation.28 SCL exerts its transcriptional control over a range of target genes either directly on binding to its recognition DNA motif, an E-box (CANNTG), or independently of its DNA binding activity.17,29 Interaction with the hematopoietic transcription factor GATA1 leads to formation of a pentameric complex composed of SCL, E47, LMO2, LDB1, and GATA1,30 that selectively activates target genes on binding to E-box/GATA composite DNA motifs in red cells.29,31,32 When acting independently of its DNA binding site, SCL is mostly recruited through GATA motifs.29,32 Similar SCL-containing multiprotein complexes are observed in MKs.25 Their function and genomic targets remain unknown.
So, despite evidence supporting the function and transcriptional activity of Scl in megakaryopoiesis, its exact role in this lineage remained to be fully characterized. To achieve this, we selectively inactivated SCL in MKs by crossing Scl-floxed mice (Sclfl/fl) to a recently described MK-specific Pf4-Cre recombinase mouse strain.33-35 We show that SCL is critically required for key aspects of megakaryocytic development. This function is mediated in part through the transcriptional control of p21 expression and cell-cycle progression.
Methods
Ploidy
Bone marrow cells derived from 5-FU–treated mice were plated in TPO-containing StemPro-34 medium (Invitrogen). After 4 days, cells were stained with CD41-FITC or IgG1-FITC antibodies (BD Biosciences PharMingen) and fixed in 70% ethanol for 3 hours at 4°C. Cells were incubated in 500 μL of 0.1% Triton X-100 (Sigma-Aldrich) containing 50 μg/mL propidium iodide (Sigma-Aldrich) and 200 μg/mL RNAse A (Sigma-Aldrich). Ploidy of CD41+ cells was analyzed on a Cyan analyzer (DakoCytomation).
Day 4 liquid cultures of MKs derived from sorted MkPs were stained with CD41-FITC or IgG1-FITC antibodies (BD Biosciences PharMingen). Cells were incubated in Iscove modified Dulbecco medium/10% FCS containing 0.01mM Hoechst 33342 (Sigma-Aldrich) at 37°C for 2 hours. Ploidy was analyzed as described earlier in “Ploidy.”
Proliferation assay
Purified MkPs were grown in IMDM/10% FCS supplemented with TPO (1% conditioned medium), erythropoietin (2 U/mL), IL-3 (10 ng/mL), IL-11 (5 ng/mL), Kit ligand (50 ng/mL), FMS-like tyrosine kinase-3 ligand (10 ng/mL), and granulocyte macrophage colony-stimulating factor (2 ng/mL). At day 4, 10μM 5-bromo-2′-deoxyuridine (BrdU) was added for 4 hours, cells harvested, stained with CD41-FITC or IgG antibodies, and cell-cycle analysis performed using BrdU Flow Kit (BD Biosciences PharMingen).
Microarray analysis
Expression profiling was performed using Sentrix Mouse-6 Expression BeadChip arrays from Illumina on 3 independent Cre:Sclwt/wt and Cre;Sclfl/fl MkP populations. Experiments were performed as described.29 Expression array data have been submitted to Gene Expression Omnibus database (accession number GSE24969).
Lentivirus production
Antisense shRNA oligonucleotides were designed in the coding region of the murine p21 gene, p21 scramble shRNA sequences were designed as control (supplemental Methods, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). The oligonucleotides were introduced into pSuper plasmid (BVTech) downstream of PolIII H1 promoter. H1-shRNA DNA fragments were transferred into the lentiviral vector pTRIP/δU3EF1α-green fluorescent protein (GFP).36,37 Lentiviral supernatant was produced as described.36,37
MkP transduction
A total of 1 to 3 × 104 purified Cre;Sclfl/fl MkPs were plated in 300 μL of IMDM/10% FCS containing cytokines (see “Proliferation assay”). Cells were transduced with viruses expressing p21 shRNA or control sequences at a multiplicity of infection of 50 to 100. At 48 ours later, cells were FACS-sorted and GFP+ cells used in colony-forming cell assays, or replated in liquid culture and assessed for ploidy and cell cycle, or cytospun for acetylcholinesterase (AChE) staining.
ChIP assays
A total of 107 MKD1 cells (ES cell–derived MK cell line bearing floxed Scl alleles, H.C., P.V., C.P. manuscript in preparation) or 0.5 to 1 × 106 primary MKs at day 4 of the TPO culture were used for each chromatin immunoprecipitation (ChIP) assay. Experimental conditions and antibodies were as described.17,25,28
Luciferase assay
Luciferase expression was driven by sequences of the mouse p21 proximal promoter and first intron (−90 to 647 relative to transcriptional start site) or mutated versions of them (produced by PCR-based mutagenesis) using the pGL4 reporter plasmid (Promega). MKD1 cells were transfected with Amaxa cell line nucleofector kit V (VCA-1003, Lonza Group) following the manufacturer's recommendations. After 24 hours, luciferase and β-galactosidase (to normalize for transfection efficiency) activities were measured using standard procedures. To excise Scl floxed alleles, MKD1 cells were cotransfected with EF1α-GFP-Cre plasmid or EF1α-GFP plasmid (control) along with p21-736 reporter. After 24 hours, GFP+ cells were sorted and luciferase and β-galactosidase assayed.
Results
In vivo generation of SclΔ/Δ MKs
A conditional knockout approach was used to specifically inactivate SCL in vivo in MKs. Sequences coding for the core domain of SCL,38 the basic helix-loop-helix region, were targeted in murine ES cells by homologous recombination; a LoxP-flanked Neo-IRES-TK selection cassette was introduced into intron 6 of Scl and another LoxP site into the 3′ untranslated region sequences (supplemental Figure 1A). Neomycin-resistant ES cell clones were identified and characterized by Southern blotting (data not shown). Clones with Cre-mediated excision of the Neo-TK cassette (Sclwt/fl) were obtained from 2 independently targeted ES cell clones (Sclwt/fl-Neo). Two independent Sclwt/fl clones were injected into C57Bl/6 blastocysts, and mouse lines heterozygous for the Scl floxed allele were generated. Supplemental Figure 1B-C shows a representative genomic analysis of F1 mice. Heterozygous Sclwt/fl mice were bred to homozygosity and Sclfl/fl mice presented at a Mendelian ratio.
To selectively abolish the function of SCL in adult megakaryopoiesis, Sclfl/wt mice were bred to a transgenic strain expressing Cre recombinase driven by the regulatory sequences of platelet factor 4 (Pf4), a MK-specific CXC chemokine.34 Newborn mice (thereafter referred to as Cre;Sclfl/fl, Cre;Sclwt/fl, and Cre;Sclwt,wt) were obtained at the expected Mendelian ratio. Homozygous mice lived to adulthood.
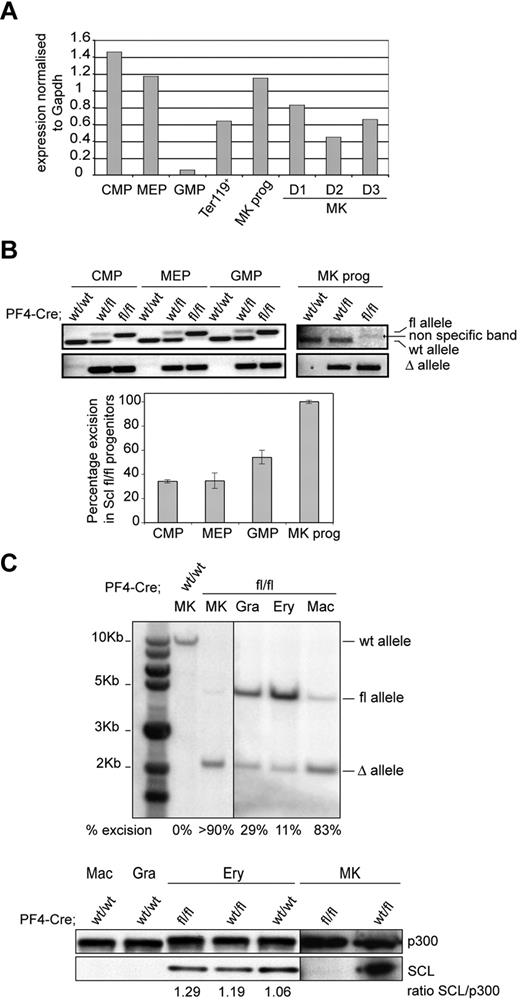
Because of Scl's broad expression in bone marrow hematopoietic progenitors and differentiated cells (Scl is highly expressed in common myeloid progenitor [CMP], bipotent erythroid/megakaryocytic progenitor [MEP], MkPs, erythroid [Ter119+] cells, and in in vitro-cultured MKs [MK, D1-D3], Figure 1A), we evaluated the level of Cre-mediated excision of the Sclfl allele in CMP, MEP, granulocyte-macrophage progenitor (GMP), and MkP populations isolated from Cre;Sclwt/wt, Cre;Sclwt/fl, and Cre;Sclfl/fl mice. Following the PCR strategy detailed in supplemental Figure 1C (top panel), both excised (Δ) and floxed (fl) alleles were amplified from CMP, MEP, and GMP derived from Cre;Sclwt/fl and Cre;Sclfl/fl mice (Figure 1B top panel). The data suggested partial excision of the Sclfl allele in these populations. Indeed, semiquantitative PCR analysis performed on the deleted (Δ) allele (Figure 1B bottom panel) showed 34% excision in Cre;Sclfl/fl-derived CMP and MEP and 54% in Cre;Sclfl/fl-derived GMP. In contrast, full excision was observed in Cre;Sclfl/fl MkPs, as shown by the complete absence of the amplified floxed allele with 100% excision (Figure 1B top and bottom panels). Importantly, we did not observe significant differences in the percentages of each progenitor compartment in Cre;Sclfl/fl-derived bone marrow compared with controls (supplemental Figures 2-3).
Complete absence of SCL protein in Cre;Sclfl/fl MKs. (A) Analysis of Scl mRNA levels in hematopoietic cells. RNA was isolated from wild-type C57Bl/6 bone marrow progenitor populations (CMP, MEP, GMP, and MkP) as well as from mature hematopoietic cells (Ter119+ splenocytes; bone marrow MKs cultured in vitro for the indicated number of days D1-D3). Quantitative reverse transcription PCR was performed using Scl-specific primers. The y-axis represents the enrichment in cDNA sequences normalized to Gapdh gene control sequences. (B) Analysis of PF4-Cre–mediated excision of Scl floxed alleles in hematopoietic progenitor compartments. (Top) PCR reactions showing amplification of floxed, wild-type (fl and wt, top panel), and excised (Δ, bottom panel) alleles from material isolated from CMP, MEP, GMP, and MkPs purified from Cre;Sclwt/wt, Cre;Sclwt/fl, and Cre;Sclfl/fl bone marrows, as indicated. (Bottom) Semiquantitative PCR reaction measuring the percentage of excision of Scl floxed alleles in Cre;Sclfl/fl progenitors (supplemental Methods). Data are the mean ± SD of 2 independent experiments. (C) SCL protein is not expressed in Cre;Sclfl/fl MKs. (Top) Southern blot analysis of genomic DNA isolated from purified Cre;Sclfl/fl hematopoietic cells. Gra indicates granulocytes; Ery, erythroid cells; and Mac, macrophages. Percentages of excision of the Scl floxed alleles are indicated. MKs from Cre;Sclwt/wt mice served as controls. (Bottom) Nuclear extracts from hematopoietic cells derived from Cre;Sclwt/wt, Cre;Sclwt/fl, and Cre;Sclfl/fl were analyzed by Western blotting for SCL expression. P300 was used as loading control. Vertical lines have been inserted to indicate repositioned gel lanes.
Complete absence of SCL protein in Cre;Sclfl/fl MKs. (A) Analysis of Scl mRNA levels in hematopoietic cells. RNA was isolated from wild-type C57Bl/6 bone marrow progenitor populations (CMP, MEP, GMP, and MkP) as well as from mature hematopoietic cells (Ter119+ splenocytes; bone marrow MKs cultured in vitro for the indicated number of days D1-D3). Quantitative reverse transcription PCR was performed using Scl-specific primers. The y-axis represents the enrichment in cDNA sequences normalized to Gapdh gene control sequences. (B) Analysis of PF4-Cre–mediated excision of Scl floxed alleles in hematopoietic progenitor compartments. (Top) PCR reactions showing amplification of floxed, wild-type (fl and wt, top panel), and excised (Δ, bottom panel) alleles from material isolated from CMP, MEP, GMP, and MkPs purified from Cre;Sclwt/wt, Cre;Sclwt/fl, and Cre;Sclfl/fl bone marrows, as indicated. (Bottom) Semiquantitative PCR reaction measuring the percentage of excision of Scl floxed alleles in Cre;Sclfl/fl progenitors (supplemental Methods). Data are the mean ± SD of 2 independent experiments. (C) SCL protein is not expressed in Cre;Sclfl/fl MKs. (Top) Southern blot analysis of genomic DNA isolated from purified Cre;Sclfl/fl hematopoietic cells. Gra indicates granulocytes; Ery, erythroid cells; and Mac, macrophages. Percentages of excision of the Scl floxed alleles are indicated. MKs from Cre;Sclwt/wt mice served as controls. (Bottom) Nuclear extracts from hematopoietic cells derived from Cre;Sclwt/wt, Cre;Sclwt/fl, and Cre;Sclfl/fl were analyzed by Western blotting for SCL expression. P300 was used as loading control. Vertical lines have been inserted to indicate repositioned gel lanes.
Next, we purified mature hematopoietic cells from bone marrow (MKs and macrophages) and spleen (erythroid cells and granulocytes) derived from Cre;Sclfl/fl mice and assessed the level of excision of the Sclfl allele by Southern blot (Figure 1C top). Greater than 90% excision was observed in the Cre;Sclfl/fl MK sample. This correlated with a complete absence of the SCL protein (Figure 1C bottom). Low-level excision in Cre;Sclfl/fl Ter119+ erythroid cells (11%, Figure 1C top) did not affect protein levels (Figure 1C bottom). Finally, the Scl sequences were also excised in granulocytes and macrophages (29% and 83%, respectively). However, because SCL is not expressed in these 2 lineages (Figure 1C bottom), genomic excision in these cells probably does not affect hematopoiesis.
In conclusion, we successfully generated a conditional knockout model with complete ablation of SCL expression specifically in the megakaryocytic lineage.
Cre;Sclfl/fl mutant mice are thrombocytopenic
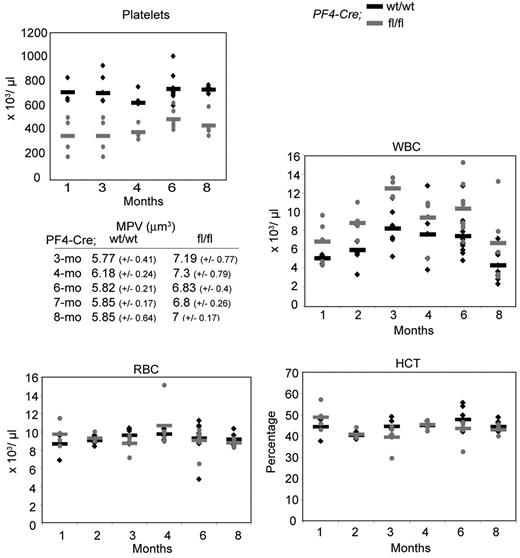
Hematologic parameters were determined over an 8-month period (Figure 2). Platelet counts in mutant Cre;Sclfl/fl mice were decreased on average by 1.5- to 2-fold, with increased mean platelet volume compared with controls, suggesting perturbed terminal megakaryopoiesis and platelet release. Mutant mice also showed an average of 1.5- to 2-fold increase in white blood cell numbers. Red blood cell counts and hematocrit were similar, suggesting no gross perturbation in the mutant erythroid lineage.
Cre;Sclfl/fl mice are thrombocytopenic. Peripheral blood parameters were analyzed in mice over an 8-month period. (♦) represents Cre;Sclwt/wt mice; and  , Cre;Sclfl/fl mice. Each diamond or dot represents an individual mouse. Horizontal bars represent the mean. Platelets, P = 1.8 × 10−12; MPV, mean platelet volume, P = 1.2 × 10−5; WBC, white blood cells, P = 3.7 × 10−5; RBC, red blood cells, P > .1; and HCT, hematocrit, P > .1. For each time point, the analysis was performed on 5 to 8 Cre;Sclfl/fl mice or 3 to 11 Cre;Sclwt/wt mice.
, Cre;Sclfl/fl mice. Each diamond or dot represents an individual mouse. Horizontal bars represent the mean. Platelets, P = 1.8 × 10−12; MPV, mean platelet volume, P = 1.2 × 10−5; WBC, white blood cells, P = 3.7 × 10−5; RBC, red blood cells, P > .1; and HCT, hematocrit, P > .1. For each time point, the analysis was performed on 5 to 8 Cre;Sclfl/fl mice or 3 to 11 Cre;Sclwt/wt mice.
Cre;Sclfl/fl mice are thrombocytopenic. Peripheral blood parameters were analyzed in mice over an 8-month period. (♦) represents Cre;Sclwt/wt mice; and  , Cre;Sclfl/fl mice. Each diamond or dot represents an individual mouse. Horizontal bars represent the mean. Platelets, P = 1.8 × 10−12; MPV, mean platelet volume, P = 1.2 × 10−5; WBC, white blood cells, P = 3.7 × 10−5; RBC, red blood cells, P > .1; and HCT, hematocrit, P > .1. For each time point, the analysis was performed on 5 to 8 Cre;Sclfl/fl mice or 3 to 11 Cre;Sclwt/wt mice.
, Cre;Sclfl/fl mice. Each diamond or dot represents an individual mouse. Horizontal bars represent the mean. Platelets, P = 1.8 × 10−12; MPV, mean platelet volume, P = 1.2 × 10−5; WBC, white blood cells, P = 3.7 × 10−5; RBC, red blood cells, P > .1; and HCT, hematocrit, P > .1. For each time point, the analysis was performed on 5 to 8 Cre;Sclfl/fl mice or 3 to 11 Cre;Sclwt/wt mice.
FACS analysis of bone marrow hematopoietic lineages showed no significant difference in the numbers of Ter119+, Mac1+, and Gr1+ cells between Cre;Sclfl/fl and control mice (supplemental Figure 4), suggesting that the levels of excision observed in the CMP, MEP, and GMP compartments (Figure 1B) did not perturb the differentiation of downstream lineages. We did, however, observe an increase in the percentage of B220+ cells in Cre;Sclfl/fl mutant mice.
Abnormal in vitro growth of SclΔ/Δ MkPs
The in vitro growth capacity of bone marrow CMP, MEP, and MkPs was analyzed in colony assays. Mutant Cre;Sclfl/fl CMPs and MEPs gave rise to numbers of erythroid, megakaryocytic, and mixed colonies similar to that generated from wild-type controls, and to increased numbers of myeloid colonies (Figure 3A-B). Morphologically, the colonies appeared normal (not shown).
Impaired megakaryocytic colony formation from Cre;Sclfl/fl MkPs. Bone marrow progenitors (CMP [A], MEP [B], MkP [C]) purified from Cre;Sclwt/wt wild-type (black bars), Cre;Sclwt/fl heterozygous (open bars), and Cre;Sclfl/fl homozygous (gray bars) mice were plated in methylcellulose. Colony-forming units-erythroid (CFUE), burst-forming units-erythroid (BFUE), MK, mixed, and myeloid colonies were scored at day 7. Data are the mean ± SD of 2 independent experiments performed in triplicate. *P < .01. (Bottom) Representative MK colonies grown from Cre;Sclwt/wt and Cre;Sclfl/fl MkPs. The photographs were taken using an inverted IX51 Olympus microscope with a Jenoptik C14 camera. Openlab, Version 3 software (Improvision) was used for image acquisition, and images were exported into Adobe Photoshop, Version CS2 (Adobe Systems). Scale bars represent 20 μm. Note the difference in the morphology and size of the colonies. A control MK shading platelet (arrow) is shown. Scale bar represents 50 μm.
Impaired megakaryocytic colony formation from Cre;Sclfl/fl MkPs. Bone marrow progenitors (CMP [A], MEP [B], MkP [C]) purified from Cre;Sclwt/wt wild-type (black bars), Cre;Sclwt/fl heterozygous (open bars), and Cre;Sclfl/fl homozygous (gray bars) mice were plated in methylcellulose. Colony-forming units-erythroid (CFUE), burst-forming units-erythroid (BFUE), MK, mixed, and myeloid colonies were scored at day 7. Data are the mean ± SD of 2 independent experiments performed in triplicate. *P < .01. (Bottom) Representative MK colonies grown from Cre;Sclwt/wt and Cre;Sclfl/fl MkPs. The photographs were taken using an inverted IX51 Olympus microscope with a Jenoptik C14 camera. Openlab, Version 3 software (Improvision) was used for image acquisition, and images were exported into Adobe Photoshop, Version CS2 (Adobe Systems). Scale bars represent 20 μm. Note the difference in the morphology and size of the colonies. A control MK shading platelet (arrow) is shown. Scale bar represents 50 μm.
In the megakaryocytic compartment, Cre;Sclfl/fl MkPs were not significantly affected in the production of mixed colonies, erythroid colonies were detected at a very low frequency, and there was an increase in the numbers of myeloid colonies (Figure 3C). Of note, this myeloid expansion, detected from all mutant progenitors, is in agreement with previous observations from our SCL DNA-binding mutant mouse model17 and probably reflects the role of SCL in fate determination.
Specific defects in the production of MK colonies were noticed (Figure 3C). Strikingly, the number of MK colonies derived from Scl-deleted MkPs was decreased 5- to 7-fold compared with controls. Furthermore, the size of mutant MK colonies was reduced to 2 or 3 cells/colony compared with 10 to 15 cells/colony in control plates. Finally, proplatelet-forming MKs were detected in control samples but absent in Cre;Sclfl/fl cultures. Collectively, these data suggested impaired cell survival, proliferation, and/or maturation of MkPs. To address this, we analyzed several aspects of megakaryopoiesis in freshly isolated bone marrow MKs.
SclΔ/Δ MKs display defects in proliferation, polyploidization, and cytoplasmic maturation
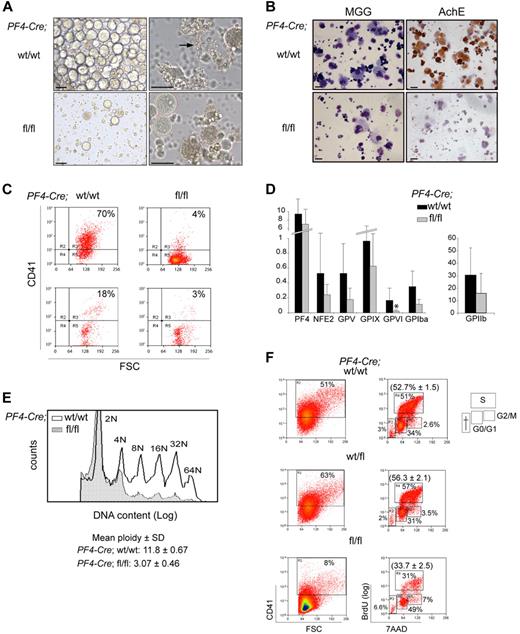
To generate sufficient numbers of MKs, mice were treated with 5-FU and in vitro–cultured bone marrow MKs were harvested. Cells isolated from Cre;Sclfl/fl mice gave rise to sparse cultures compared with control samples that consisted of large, round cells (Figure 4A left panels). Proplatelet-forming MKs were detected in control samples but not in Scl-deleted samples (Figure 4A right panels, arrow). Several assays were used to further characterize MK cultures. First, May-Grunwald-Giemsa staining revealed polylobulated nuclear morphology and abundant cytoplasm of large, mature MKs, and confirmed the low number of MKs in mutant Cre;Sclfl/fl populations (Figure 4B left panels). Second, AChE activity, specific to immature and mature MKs, was detected in control cultures but was absent from mutant cultures (Figure 4B right panels), suggesting an early block in megakaryocytic differentiation in SclΔ/Δ cells. Very rare AChE-positive MKs could be seen in mutant cultures derived from sorted MkPs (supplemental Figure 5). Third, analysis of CD41 (MK cell surface marker) expression showed that 70% of the cells obtained from Cre;Sclwt/wt bone marrow were CD41+ as opposed to 4% in Cre;Sclfl/fl cultures (Figure 4C top). Analysis of blood cells from control and mutant mice confirmed this difference (Figure 4C bottom panels).
Cre;Sclfl/fl MKs display several maturation defects. (A) Bone marrow cells derived from 5-FU–treated mice (Cre;Sclwt/wt and Cre;Sclfl/fl, as indicated) were cultured in TPO-containing medium for 3 days. The morphology of cells at day 3 of culture is shown. The arrow points to a wild-type MK shading platelets. Images of MK culture were taken using an inverted IX51 Olympus microscope with a Jenoptik C14 camera. Openlab, Version 3 software (Improvision) was used for image acquisition, and images were exported into Adobe Photoshop, Version CS2 (Adobe Systems). Scale bars represent 50 μm. (B) The cultures shown in panel A are stained with May-Grunwald-Giemsa (left) and for AChE activity (right). The photographs were taken using an Olympus BX60 microscope with a Qimaging camera. Openlab, Version 3 software (Improvision) was used for image acquisition, and images were exported into Adobe Photoshop, Version CS2 (Adobe Systems). Scale bars represent 20 μm. (C) CD41 expression was analyzed by flow cytometry in MKs derived from 5-FU–treated mice (top) and peripheral blood (bottom). Genotypes are as indicated. (D) Real-time quantitative PCR expression analysis of MK-specific genes. MKs were derived from Cre;Sclwt/wt (black bars) and Cre;SCLfl/fl (gray bars) 5-FU–treated mice. The y-axis represents enrichment in cDNA sequences normalized to Gapdh gene control sequences. Data are the mean ± SD of 7 independent experiments. *P < .01. (E) Analysis of the ploidy of Cre;Sclwt/wt and Cre;Sclfl/fl MKs. Bone marrow cells derived from 5-FU–treated mice were stained using propidium iodide and ploidy analyzed by flow cytometry on the CD41+ population. Peaks representing each ploidy class are labeled. Hatched histogram represents Cre;Sclfl/fl; and empty histogram, Cre;Sclwt/wt. One representative experiment of 3 is shown; the mean ploidy ± SD of all 3 experiments is shown. P < .01. (F) Cell-cycle analysis of CD41+ cells. Cre;Sclw/wt, Cre;Sclwt/fl, and Cre;Sclfl/fl bone marrow MkPs were purified, cultured for 4 days, and cell-cycle phases determined by analysis of BrdU incorporation and 7-amino-actinomycin D staining. The percentage of CD41+ cells in each culture is shown on the left. The percentage of cells in G0/G1, S, and G2M phases is shown on the right. One representative experiment of 3 is shown. The percentage (mean ± SD) of the cells in S phase in the 3 experiments is in parentheses. P = .006.
Cre;Sclfl/fl MKs display several maturation defects. (A) Bone marrow cells derived from 5-FU–treated mice (Cre;Sclwt/wt and Cre;Sclfl/fl, as indicated) were cultured in TPO-containing medium for 3 days. The morphology of cells at day 3 of culture is shown. The arrow points to a wild-type MK shading platelets. Images of MK culture were taken using an inverted IX51 Olympus microscope with a Jenoptik C14 camera. Openlab, Version 3 software (Improvision) was used for image acquisition, and images were exported into Adobe Photoshop, Version CS2 (Adobe Systems). Scale bars represent 50 μm. (B) The cultures shown in panel A are stained with May-Grunwald-Giemsa (left) and for AChE activity (right). The photographs were taken using an Olympus BX60 microscope with a Qimaging camera. Openlab, Version 3 software (Improvision) was used for image acquisition, and images were exported into Adobe Photoshop, Version CS2 (Adobe Systems). Scale bars represent 20 μm. (C) CD41 expression was analyzed by flow cytometry in MKs derived from 5-FU–treated mice (top) and peripheral blood (bottom). Genotypes are as indicated. (D) Real-time quantitative PCR expression analysis of MK-specific genes. MKs were derived from Cre;Sclwt/wt (black bars) and Cre;SCLfl/fl (gray bars) 5-FU–treated mice. The y-axis represents enrichment in cDNA sequences normalized to Gapdh gene control sequences. Data are the mean ± SD of 7 independent experiments. *P < .01. (E) Analysis of the ploidy of Cre;Sclwt/wt and Cre;Sclfl/fl MKs. Bone marrow cells derived from 5-FU–treated mice were stained using propidium iodide and ploidy analyzed by flow cytometry on the CD41+ population. Peaks representing each ploidy class are labeled. Hatched histogram represents Cre;Sclfl/fl; and empty histogram, Cre;Sclwt/wt. One representative experiment of 3 is shown; the mean ploidy ± SD of all 3 experiments is shown. P < .01. (F) Cell-cycle analysis of CD41+ cells. Cre;Sclw/wt, Cre;Sclwt/fl, and Cre;Sclfl/fl bone marrow MkPs were purified, cultured for 4 days, and cell-cycle phases determined by analysis of BrdU incorporation and 7-amino-actinomycin D staining. The percentage of CD41+ cells in each culture is shown on the left. The percentage of cells in G0/G1, S, and G2M phases is shown on the right. One representative experiment of 3 is shown. The percentage (mean ± SD) of the cells in S phase in the 3 experiments is in parentheses. P = .006.
Next, we examined mRNA expression of selected MK-specific markers by real-time PCR (Figure 4D). Of all the markers tested, only GpV1 expression was markedly down-regulated (7-fold reduction) in Scl-null MKs compared with control cells. Expression of Pf4, Nf-e2, GpV, GpIX, GpIα, and IIb was not significantly altered in mutant cells.
A typical feature of MK maturation is polyploidization, where MKs switch from a classic mitotic division process to an endomitotic process to increase their ploidy. To assess whether this process was affected in Scl-deleted MKs, we measured the DNA content of bone marrow–derived CD41+ cells by flow cytometry. A markedly lower ploidy level was observed from Cre;Sclfl/fl cells (mean ploidy 3N) compared with control Cre;Sclwt/wt cells (mean ploidy 12N) (Figure 4E).
Next, we determined the effect of absence of SCL on cell cycle. Cre;Sclwt/wt, Cre;Sclwt/fl, and Cre;Sclfl/fl MkPs were purified and the cell cycle analyzed by BrdU incorporation. Similar to findings obtained from cultured MKs (Figure 4C), homozygous mutant MkPs gave rise to less CD41+ cells than the control cells (Figure 4F left panels). Analysis of BrdU incorporation in CD41+ cells revealed that Cre;Sclfl/fl MKs had a lower proliferation rate with only 31% of BrdU+ cells compared with 51% and 57% in SCL-expressing MKs (Cre;Sclwt/wt and Cre;Sclwt/fl, respectively), leading to an accumulation of cells in G0/G1 (Figure 4F right panels).
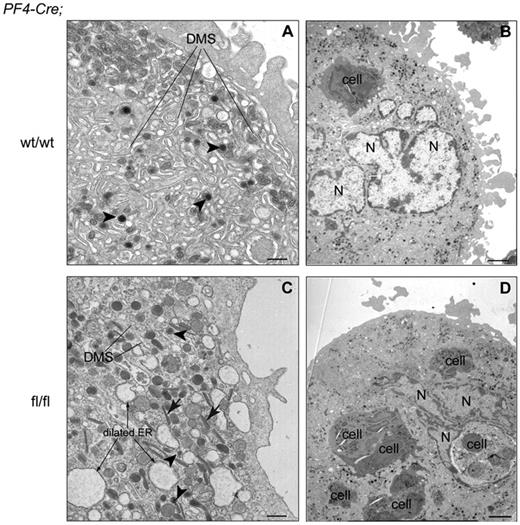
Finally, ultrastructural studies showed rarefaction and disorganization of the DMS accompanied by dilated rough endoplasmic reticulum in Cre;Sclfl/fl MKs, confirming a perturbed cytoplasmic maturation (compare Figure 5A,C). In addition, unusual elongated organelles containing crystalline structure were noticed in the mutant (Figure 5C arrow). Emperipolesis (presence of cells within the MKs) was markedly increased in mutant MKs (compare Figure 5B,D). To assess the characteristics of platelets produced in vivo from SCL-deficient MKs, platelet aggregation and dense granule secretion were monitored after stimulation with low concentrations of platelet agonists. These properties appeared relatively unchanged in the absence of SCL (supplemental Figure 6).
SclΔ/Δ MKs show ultrastructural defects.Cre;Sclwt/wt (A-B) and Cre;Sclfl/fl (C-D) bone marrow MKs were analyzed by electron microscopy. Note the absence of organized DMS in mutant MKs. Note the presence of 6 cells within the mutant MK (emperipolesis). N indicates polylobulated nucleus; and ER, endoplasmic reticulum. Arrowheads indicate α-granules; and arrows, crystalline structure. (A,C) Scale bars represent 500 nm. (B,D) Scale bars represent 2 μm.
SclΔ/Δ MKs show ultrastructural defects.Cre;Sclwt/wt (A-B) and Cre;Sclfl/fl (C-D) bone marrow MKs were analyzed by electron microscopy. Note the absence of organized DMS in mutant MKs. Note the presence of 6 cells within the mutant MK (emperipolesis). N indicates polylobulated nucleus; and ER, endoplasmic reticulum. Arrowheads indicate α-granules; and arrows, crystalline structure. (A,C) Scale bars represent 500 nm. (B,D) Scale bars represent 2 μm.
Collectively, these data indicate that SCL regulates critical aspects of megakaryopoiesis, namely, proliferation, polyploidization, and cytoplasmic maturation.
Numerous biologic processes are affected in SclΔ/Δ MkPs
To identify the molecular mechanisms underlying the early cellular defects observed in Cre;Sclfl/fl MkPs, we performed whole genome expression analyses. A total of 145 genes grouped into 8 biologic functions were significantly differentially expressed in the mutant populations compared with controls (from 1.4- to 7-fold, Table 1).
Differentially expressed genes in SclΔ/Δ MkPs
| Gene symbol . | Fold change* . | Accession no. . |
|---|---|---|
| Adhesion/migration | ||
| Col18a1† | 7.77 | NM_009929.2 |
| Megf10 | 3.63 | NM_001001979 |
| Emilin2 | 3.29 | NM_145158 |
| Cd72 | 2.73 | NM_007654.1 |
| Serpine2† | 2.22 | AK045954 |
| Map17 | 2.12 | NM_026018.1 |
| Cd63† | 1.77 | NM_007653.1 |
| Emid1 | 1.75 | NM_080595.1 |
| Cxcr4† | 1.75 | NM_009911.2 |
| Ltbp1 | 1.61 | NM_019919.2 |
| Mfge8 | 1.45 | NM_008594 |
| Tens1 | 0.66 | XM_109868 |
| Elmo1 | 0.6 | NM_080288.1 |
| Esam1 | 0.55 | NM_027102.1 |
| Selp† | 0.4 | NM_011347.1 |
| Cell cycle/apoptosis | ||
| Cdkn1a† | 3.26 | NM_007669.2 |
| Tnfrsf18 | 1.97 | NM_009400.1 |
| Rassf2 | 1.64 | NM_175445.3 |
| Bin1 | 1.62 | AK041729.1 |
| Bcl2† | 1.59 | NM_177410.1 |
| Ndg2 | 1.5 | NM_175329.3 |
| Tmbim4 | 1.5 | NM_026617.1 |
| Cryab | 1.37 | NM_009964.1 |
| Nalp6 | 0.65 | NM_133946.1 |
| Catnal1 | 0.47 | NM_018761.2 |
| Transcription | ||
| Mta3 | 2.2 | NM_054082.1 |
| Carhsp1 | 2.04 | NM_025821.2 |
| Lyl1 | 1.77 | NM_008535.1 |
| Rcor1 | 1.66 | NM_054048.1 |
| Gata2 | 1.57 | NM_008090.3 |
| Hes6 | 1.52 | NM_019479.2 |
| Foxp1 | 1.48 | NM_053202.1 |
| Mta2 | 1.48 | NM_011842.2 |
| Chd7 | 0.71 | XM_149413.3 |
| Fhl1 | 0.69 | NM_010211.1 |
| Ccndbp1 | 0.68 | NM_010761.1 |
| Hoxb5 | 0.67 | NM_008268.1 |
| Mysm1 | 0.65 | NM_177239.1 |
| Fos | 0.61 | NM_010234.2 |
| Ctdspl2 | 0.59 | XM_283758.1 |
| Tle1 | 0.52 | NM_011599.2 |
| Ear10 | 0.35 | NM_053112.1 |
| Ear2 | 0.34 | NM_007895 |
| Tal1 | 0.2 | NM_011527.1 |
| Immune response | ||
| Igh-6 | 6.66 | XM_354710.1 |
| Cst7 | 2.57 | NM_009977.1 |
| Clecsf8 | 2.44 | NM_010819.1 |
| Cd59a | 0.54 | NM_007652.2 |
| Signal transduction | ||
| Fcer1g† | 6 | NM_010185.2 |
| P2ry14 | 2.84 | NM_133200.2 |
| Wbscr5 | 2.51 | NM_022964.2 |
| Ppp1r1c | 2.13 | NM_033264.1 |
| Cd69 | 1.98 | XM_132882.1 |
| Mrvi1† | 1.97 | NM_194464.1 |
| Itgb3bp | 1.88 | NM_026348.2 |
| Il15 | 1.87 | NM_008357.1 |
| Centd1† | 1.81 | XM_132099.5 |
| Rpel1 | 1.79 | NM_198419.2 |
| Dok4 | 1.75 | NM_053246.1 |
| Tas1r1 | 1.7 | NM_031867.1 |
| Fgf3 | 1.69 | NM_008007.1 |
| Irak3 | 1.69 | NM_028679.2 |
| Dusp4 | 1.69 | NM_176933 |
| Cysltr1 | 1.63 | NM_021476.2 |
| Diras2 | 1.68 | NM_001024474 |
| Lat† | 1.54 | NM_010689.2 |
| Plekhg5 | 1.5 | NM_001004156 |
| Agtrap | 1.48 | NM_009642.3 |
| Rab27a | 1.48 | NM_023635.2 |
| Ly6e | 1.46 | NM_008529 |
| Il18r1 | 1.45 | NM_008365.1 |
| Cd244 | 1.36 | NM_018729.1 |
| Dusp1 | 0.69 | NM_013642.1 |
| Grb10 | 0.67 | AK012646 |
| Gucy1a3 | 0.62 | NM_021896.3 |
| Rab11a | 0.62 | NM_017382 |
| Fcgr2b | 0.6 | NM_010187.1 |
| Pdgfb | 0.57 | NM_011057.2 |
| Drctnnb1a | 0.56 | NM_053090 |
| Il6ra | 0.48 | AK020663 |
| Gnaz† | 0.45 | NM_010311.2 |
| Pde10a | 0.37 | NM_011866.1 |
| Transport | ||
| Alox5 | 5.53 | XM_132832.3 |
| Tnni1 | 2.04 | NM_021467 |
| Emid1 | 1.98 | NM_080595.1 |
| Tnni3 | 1.97 | NM_009406.2 |
| Slc6a13 | 1.85 | NM_144512.1 |
| Hbb-b1 | 1.79 | AK005442 |
| Tpcn1 | 1.73 | NM_145853.2 |
| Ddx25 | 1.72 | NM_013932.2 |
| Spns3 | 1.71 | XM_126365.3 |
| Igf2r | 1.56 | NM_010515.1 |
| Slc5a9 | 1.56 | NM_145551.2 |
| Slc14a1 | 0.68 | NM_028122.2 |
| Abcc5 | 0.67 | NM_176839.1 |
| Car2 | 0.53 | NM_009801.3 |
| Hebp1 | 0.51 | NM_013546.1 |
| Cytoskeleton organization | ||
| Myo18b† | 4.84 | XM_144513.3 |
| Tpm2 | 2.4 | NM_009416.2 |
| Cap1 | 2.07 | NM_007598.2 |
| Myo6† | 1.65 | NM_008662.1 |
| Ehd2 | 1.53 | AK029240 |
| Homer2 | 0.46 | XM_133550.4 |
| Ubiquitination | ||
| Usp20 | 1.56 | NM_028846.1 |
| Usp40 | 0.54 | XM_129956.2 |
| Metabolism | ||
| Chst1 | 4.71 | NM_023850.1 |
| Alox5 | 3.95 | XM_132832 |
| Abat | 2.31 | NM_172961.2 |
| Padi2 | 2.2 | NM_008812.1 |
| Hmgcs2 | 1.88 | NM_008256.2 |
| Acas2l | 1.65 | NM_080575.1 |
| Ltc4s | 1.65 | NM_008521.1 |
| Rsdr1-pending | 1.64 | AK087137 |
| Dhrs3 | 1.45 | NM_011303.2 |
| Gchfr | 0.71 | NM_177157.2 |
| Chsy3 | 0.65 | NM_178636 |
| Oxr1 | 0.63 | AK040881 |
| Hyi | 0.6 | XM_489058 |
| Mtap | 0.6 | NM_024433.1 |
| Serpinb1a | 0.56 | NM_025429.1 |
| Gucy1a3 | 0.47 | NM_021896.3 |
| Sdh1 | 0.45 | NM_146126.1 |
| Serpina3g | 0.4 | XM_354694.1 |
| Serpina3b | 0.38 | NM_173024.1 |
| Unidentified | ||
| 9830134C10Rik | 3.43 | AK036563 |
| 2310047A01Rik | 2.56 | XM_484355 |
| LOC381142 | 2.09 | XM_355058.1 |
| 6430559E15Rik | 1.99 | XM_131537.4 |
| C130080K17Rik | 1.71 | AK081831 |
| LOC382231 | 1.69 | XM_356343.1 |
| 1110012N22Rik | 1.68 | XM_126634.3 |
| BC023181 | 1.6 | XM_194180.3 |
| 4930427A07Rik | 1.51 | NM_134041.1 |
| BC057022 | 1.5 | NM_001004180.1 |
| 0610037L13Rik | 0.72 | NM_028754.1 |
| 5730446C15Rik | 0.69 | NM_146096 |
| B930074N03Rik | 0.68 | AK047478 |
| BC023823 | 0.68 | NM_153566.1 |
| C530050O22Rik | 0.68 | NM_172871.1 |
| AY078069 | 0.66 | NM_172142.1 |
| D8Ertd594e | 0.66 | NM_133791.3 |
| 2900008M13Rik | 0.63 | XM_110121 |
| 2010300G19Rik | 0.61 | NM_028097.2 |
| BC049975 | 0.59 | XM_138237.2 |
| 9030611O19Rik | 0.57 | NM_027828.2 |
| Gene symbol . | Fold change* . | Accession no. . |
|---|---|---|
| Adhesion/migration | ||
| Col18a1† | 7.77 | NM_009929.2 |
| Megf10 | 3.63 | NM_001001979 |
| Emilin2 | 3.29 | NM_145158 |
| Cd72 | 2.73 | NM_007654.1 |
| Serpine2† | 2.22 | AK045954 |
| Map17 | 2.12 | NM_026018.1 |
| Cd63† | 1.77 | NM_007653.1 |
| Emid1 | 1.75 | NM_080595.1 |
| Cxcr4† | 1.75 | NM_009911.2 |
| Ltbp1 | 1.61 | NM_019919.2 |
| Mfge8 | 1.45 | NM_008594 |
| Tens1 | 0.66 | XM_109868 |
| Elmo1 | 0.6 | NM_080288.1 |
| Esam1 | 0.55 | NM_027102.1 |
| Selp† | 0.4 | NM_011347.1 |
| Cell cycle/apoptosis | ||
| Cdkn1a† | 3.26 | NM_007669.2 |
| Tnfrsf18 | 1.97 | NM_009400.1 |
| Rassf2 | 1.64 | NM_175445.3 |
| Bin1 | 1.62 | AK041729.1 |
| Bcl2† | 1.59 | NM_177410.1 |
| Ndg2 | 1.5 | NM_175329.3 |
| Tmbim4 | 1.5 | NM_026617.1 |
| Cryab | 1.37 | NM_009964.1 |
| Nalp6 | 0.65 | NM_133946.1 |
| Catnal1 | 0.47 | NM_018761.2 |
| Transcription | ||
| Mta3 | 2.2 | NM_054082.1 |
| Carhsp1 | 2.04 | NM_025821.2 |
| Lyl1 | 1.77 | NM_008535.1 |
| Rcor1 | 1.66 | NM_054048.1 |
| Gata2 | 1.57 | NM_008090.3 |
| Hes6 | 1.52 | NM_019479.2 |
| Foxp1 | 1.48 | NM_053202.1 |
| Mta2 | 1.48 | NM_011842.2 |
| Chd7 | 0.71 | XM_149413.3 |
| Fhl1 | 0.69 | NM_010211.1 |
| Ccndbp1 | 0.68 | NM_010761.1 |
| Hoxb5 | 0.67 | NM_008268.1 |
| Mysm1 | 0.65 | NM_177239.1 |
| Fos | 0.61 | NM_010234.2 |
| Ctdspl2 | 0.59 | XM_283758.1 |
| Tle1 | 0.52 | NM_011599.2 |
| Ear10 | 0.35 | NM_053112.1 |
| Ear2 | 0.34 | NM_007895 |
| Tal1 | 0.2 | NM_011527.1 |
| Immune response | ||
| Igh-6 | 6.66 | XM_354710.1 |
| Cst7 | 2.57 | NM_009977.1 |
| Clecsf8 | 2.44 | NM_010819.1 |
| Cd59a | 0.54 | NM_007652.2 |
| Signal transduction | ||
| Fcer1g† | 6 | NM_010185.2 |
| P2ry14 | 2.84 | NM_133200.2 |
| Wbscr5 | 2.51 | NM_022964.2 |
| Ppp1r1c | 2.13 | NM_033264.1 |
| Cd69 | 1.98 | XM_132882.1 |
| Mrvi1† | 1.97 | NM_194464.1 |
| Itgb3bp | 1.88 | NM_026348.2 |
| Il15 | 1.87 | NM_008357.1 |
| Centd1† | 1.81 | XM_132099.5 |
| Rpel1 | 1.79 | NM_198419.2 |
| Dok4 | 1.75 | NM_053246.1 |
| Tas1r1 | 1.7 | NM_031867.1 |
| Fgf3 | 1.69 | NM_008007.1 |
| Irak3 | 1.69 | NM_028679.2 |
| Dusp4 | 1.69 | NM_176933 |
| Cysltr1 | 1.63 | NM_021476.2 |
| Diras2 | 1.68 | NM_001024474 |
| Lat† | 1.54 | NM_010689.2 |
| Plekhg5 | 1.5 | NM_001004156 |
| Agtrap | 1.48 | NM_009642.3 |
| Rab27a | 1.48 | NM_023635.2 |
| Ly6e | 1.46 | NM_008529 |
| Il18r1 | 1.45 | NM_008365.1 |
| Cd244 | 1.36 | NM_018729.1 |
| Dusp1 | 0.69 | NM_013642.1 |
| Grb10 | 0.67 | AK012646 |
| Gucy1a3 | 0.62 | NM_021896.3 |
| Rab11a | 0.62 | NM_017382 |
| Fcgr2b | 0.6 | NM_010187.1 |
| Pdgfb | 0.57 | NM_011057.2 |
| Drctnnb1a | 0.56 | NM_053090 |
| Il6ra | 0.48 | AK020663 |
| Gnaz† | 0.45 | NM_010311.2 |
| Pde10a | 0.37 | NM_011866.1 |
| Transport | ||
| Alox5 | 5.53 | XM_132832.3 |
| Tnni1 | 2.04 | NM_021467 |
| Emid1 | 1.98 | NM_080595.1 |
| Tnni3 | 1.97 | NM_009406.2 |
| Slc6a13 | 1.85 | NM_144512.1 |
| Hbb-b1 | 1.79 | AK005442 |
| Tpcn1 | 1.73 | NM_145853.2 |
| Ddx25 | 1.72 | NM_013932.2 |
| Spns3 | 1.71 | XM_126365.3 |
| Igf2r | 1.56 | NM_010515.1 |
| Slc5a9 | 1.56 | NM_145551.2 |
| Slc14a1 | 0.68 | NM_028122.2 |
| Abcc5 | 0.67 | NM_176839.1 |
| Car2 | 0.53 | NM_009801.3 |
| Hebp1 | 0.51 | NM_013546.1 |
| Cytoskeleton organization | ||
| Myo18b† | 4.84 | XM_144513.3 |
| Tpm2 | 2.4 | NM_009416.2 |
| Cap1 | 2.07 | NM_007598.2 |
| Myo6† | 1.65 | NM_008662.1 |
| Ehd2 | 1.53 | AK029240 |
| Homer2 | 0.46 | XM_133550.4 |
| Ubiquitination | ||
| Usp20 | 1.56 | NM_028846.1 |
| Usp40 | 0.54 | XM_129956.2 |
| Metabolism | ||
| Chst1 | 4.71 | NM_023850.1 |
| Alox5 | 3.95 | XM_132832 |
| Abat | 2.31 | NM_172961.2 |
| Padi2 | 2.2 | NM_008812.1 |
| Hmgcs2 | 1.88 | NM_008256.2 |
| Acas2l | 1.65 | NM_080575.1 |
| Ltc4s | 1.65 | NM_008521.1 |
| Rsdr1-pending | 1.64 | AK087137 |
| Dhrs3 | 1.45 | NM_011303.2 |
| Gchfr | 0.71 | NM_177157.2 |
| Chsy3 | 0.65 | NM_178636 |
| Oxr1 | 0.63 | AK040881 |
| Hyi | 0.6 | XM_489058 |
| Mtap | 0.6 | NM_024433.1 |
| Serpinb1a | 0.56 | NM_025429.1 |
| Gucy1a3 | 0.47 | NM_021896.3 |
| Sdh1 | 0.45 | NM_146126.1 |
| Serpina3g | 0.4 | XM_354694.1 |
| Serpina3b | 0.38 | NM_173024.1 |
| Unidentified | ||
| 9830134C10Rik | 3.43 | AK036563 |
| 2310047A01Rik | 2.56 | XM_484355 |
| LOC381142 | 2.09 | XM_355058.1 |
| 6430559E15Rik | 1.99 | XM_131537.4 |
| C130080K17Rik | 1.71 | AK081831 |
| LOC382231 | 1.69 | XM_356343.1 |
| 1110012N22Rik | 1.68 | XM_126634.3 |
| BC023181 | 1.6 | XM_194180.3 |
| 4930427A07Rik | 1.51 | NM_134041.1 |
| BC057022 | 1.5 | NM_001004180.1 |
| 0610037L13Rik | 0.72 | NM_028754.1 |
| 5730446C15Rik | 0.69 | NM_146096 |
| B930074N03Rik | 0.68 | AK047478 |
| BC023823 | 0.68 | NM_153566.1 |
| C530050O22Rik | 0.68 | NM_172871.1 |
| AY078069 | 0.66 | NM_172142.1 |
| D8Ertd594e | 0.66 | NM_133791.3 |
| 2900008M13Rik | 0.63 | XM_110121 |
| 2010300G19Rik | 0.61 | NM_028097.2 |
| BC049975 | 0.59 | XM_138237.2 |
| 9030611O19Rik | 0.57 | NM_027828.2 |
Fold change: ratio SclΔ/Δ MkPs/WT MkP.
Specific functions in megakaryopoiesis and/or platelet biology.
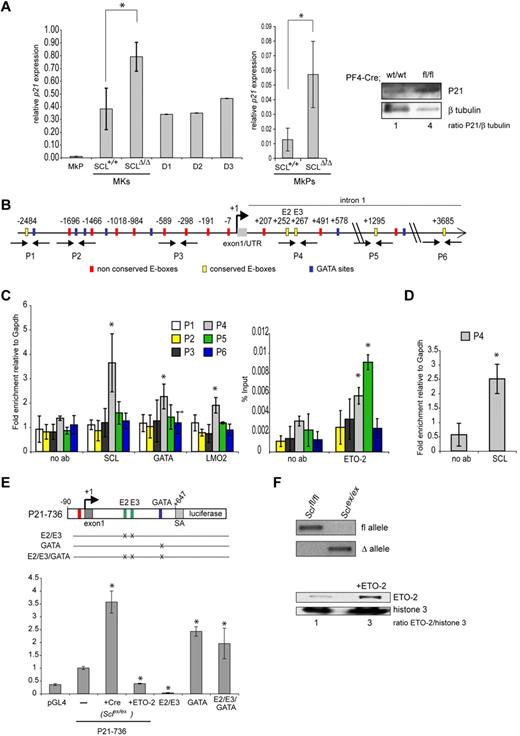
At least 14 genes (9.7% of total genes, footnoted in Table 1) show specific functions in megakaryopoiesis and/or platelet biology. One of them is the cyclin-dependent kinase inhibitor p21, which shows a 3.26-fold increase in expression in Cre;Sclfl/fl MKs. Given the role P21 plays in megakaryopoiesis (“Introduction”), we decided to investigate whether SCL-mediated regulation of p21 expression was functionally important in megakaryopoiesis.
The cyclin-dependent kinase inhibitor p21 is a direct target of SCL in MKs
As shown in Figure 6A, p21 is expressed at low levels in MkPs, and its expression is dramatically increased in more mature MKs (Scl+/+) and sustained at high levels in in vitro cultures (D1-D3). Confirming the microarray data, overexpression of p21 mRNA was observed in SCLΔ/Δ MkPs (Figure 6A middle panel, 4.5-fold increase), as well as in SCL-depleted MKs compared with control (Figure 6A left panel, 2.5-fold increase), suggesting that the transcriptional control exerted by SCL on p21 expression in MkPs extends to maturing MKs. Up-regulation of p21 mRNA was reflected at the protein level in mutant MKs (Figure 6A right panel).
p21 is a direct target of SCL. (A) Analysis of p21 levels in MKs. (Left and middle panels) Quantitative reverse transcription PCR was performed using p21-specific primers. The y-axis represents the enrichment in cDNA sequences normalized to Gapdh gene control sequences. Data are the mean ± SD of 2 independent experiments. *P = .04. (Left panel) RNA was isolated from wild-type MkPs and MKs subsequently cultured in vitro for the indicated number of days (D1-D3) as well as from Cre;Sclfl/fl and Cre;Sclwt/wt MKs (SclΔ/Δ and Scl+/+, respectively). (Middle panel) RNA was isolated from sorted Cre;Sclfl/fl and Cre;Sclwt/wt MkPs. (Right panel) Western blot analysis of P21 levels in Cre;Sclfl/fl and Cre;Sclwt/wt MKs showing a 4-fold increase in P21 protein in mutant MKs. β-tubulin served as loading control. (B) Schematic representation of the mouse p21 proximal promoter and part of the first intron. The location of E-box (CANNTG, red and yellow boxes) and GATA (WGATAR, blue boxes) motifs is indicated in base pairs relative to the transcription start site (+1). The E-boxes E2 (+252) and E3 (+267) and the GATA motif (+578) are the sites mutagenized in the transactivation assays (see panel E). P1 to P6 show the location of the primer pairs designed for real-time PCR (not to scale). (C) ChIP analysis over the p21 locus using material isolated from MKD1 cells and antibodies as indicated in the figure. Data are the mean ± SD of 5 to 7 independent experiments; enrichment over no antibody. *P < .044. P1-P6 represents the genomic regions analyzed, as shown in panel B. (D) Same as in panel C, but chromatin was isolated from primary MKs derived from 5-FU–treated mouse bone marrow. Data are the mean ± SD of 3 independent experiments (enrichment over no antibody). *P = .006. (E) Transactivation assays in MKD1 cells. (Top) The luciferase gene is under control of a 736-bp fragment of the p21 promoter, including part of the first intron (p21-736). SA is a 225-bp splice acceptor sequence from the p21 gene subcloned upstream of the luciferase gene to allow for splicing. The constructs E2/E3, GATA, and E2/E3/GATA bear mutations in the 2 conserved E-boxes and GATA motif located in intron 1 as shown. The graph represents relative luciferase activity measured in MKD1 cells nucleotransfected with the wild-type reporter (P21-736, —) or mutated versions as indicated. The P21-736 construct was also assayed in MKD1 cells on Cre-mediated excision of the Scl floxed alleles (+Cre, Sclex/ex) and overexpression of ETO2 (+ETO2). Data are mean ± SD of 3 or 4 independent experiments performed in duplicate. *P < .01 (vs control cells transfected with p21-736 construct). (F; Top) PCR showing amplification of the floxed (fl) and excised (Δ) alleles in MKD1 cells (Sclfl/fl) and after Cre-mediated excision (Sclex/ex). (Bottom) Western blot analysis of ETO2 expression in MKD1 cells and after overexpression of ETO2 (+ETO2, 3-fold increase in ratio ETO2/histone H3). Histone H3 served as a loading control.
p21 is a direct target of SCL. (A) Analysis of p21 levels in MKs. (Left and middle panels) Quantitative reverse transcription PCR was performed using p21-specific primers. The y-axis represents the enrichment in cDNA sequences normalized to Gapdh gene control sequences. Data are the mean ± SD of 2 independent experiments. *P = .04. (Left panel) RNA was isolated from wild-type MkPs and MKs subsequently cultured in vitro for the indicated number of days (D1-D3) as well as from Cre;Sclfl/fl and Cre;Sclwt/wt MKs (SclΔ/Δ and Scl+/+, respectively). (Middle panel) RNA was isolated from sorted Cre;Sclfl/fl and Cre;Sclwt/wt MkPs. (Right panel) Western blot analysis of P21 levels in Cre;Sclfl/fl and Cre;Sclwt/wt MKs showing a 4-fold increase in P21 protein in mutant MKs. β-tubulin served as loading control. (B) Schematic representation of the mouse p21 proximal promoter and part of the first intron. The location of E-box (CANNTG, red and yellow boxes) and GATA (WGATAR, blue boxes) motifs is indicated in base pairs relative to the transcription start site (+1). The E-boxes E2 (+252) and E3 (+267) and the GATA motif (+578) are the sites mutagenized in the transactivation assays (see panel E). P1 to P6 show the location of the primer pairs designed for real-time PCR (not to scale). (C) ChIP analysis over the p21 locus using material isolated from MKD1 cells and antibodies as indicated in the figure. Data are the mean ± SD of 5 to 7 independent experiments; enrichment over no antibody. *P < .044. P1-P6 represents the genomic regions analyzed, as shown in panel B. (D) Same as in panel C, but chromatin was isolated from primary MKs derived from 5-FU–treated mouse bone marrow. Data are the mean ± SD of 3 independent experiments (enrichment over no antibody). *P = .006. (E) Transactivation assays in MKD1 cells. (Top) The luciferase gene is under control of a 736-bp fragment of the p21 promoter, including part of the first intron (p21-736). SA is a 225-bp splice acceptor sequence from the p21 gene subcloned upstream of the luciferase gene to allow for splicing. The constructs E2/E3, GATA, and E2/E3/GATA bear mutations in the 2 conserved E-boxes and GATA motif located in intron 1 as shown. The graph represents relative luciferase activity measured in MKD1 cells nucleotransfected with the wild-type reporter (P21-736, —) or mutated versions as indicated. The P21-736 construct was also assayed in MKD1 cells on Cre-mediated excision of the Scl floxed alleles (+Cre, Sclex/ex) and overexpression of ETO2 (+ETO2). Data are mean ± SD of 3 or 4 independent experiments performed in duplicate. *P < .01 (vs control cells transfected with p21-736 construct). (F; Top) PCR showing amplification of the floxed (fl) and excised (Δ) alleles in MKD1 cells (Sclfl/fl) and after Cre-mediated excision (Sclex/ex). (Bottom) Western blot analysis of ETO2 expression in MKD1 cells and after overexpression of ETO2 (+ETO2, 3-fold increase in ratio ETO2/histone H3). Histone H3 served as a loading control.
We asked whether p21 could be a direct target of SCL using ChIP analyses. We first searched the mouse p21 proximal promoter and first intron (−2.5 kb/+5 kb) for SCL-binding motifs. We identified 28 E-boxes (CANNTG), 5 of them showing conservation between human and mouse sequences (Figure 6B). Anti-SCL ChIP assay was performed on material isolated from an immortalized MK cell line generated from Sclfl/fl mouse ES cells and presenting features of immature MkPs (thereafter referred to as MKD1 cells; H.C. and C.P., manuscript in preparation). We observed SCL binding exclusively in the 5′ region of the first intron of the gene, which contains 2 nonconserved and 2 conserved E-boxes (CAGGTG), as well as a GATA-binding site (TCTATCT) (Figure 6C left graph, region P4). Enrichment in GATA1 and, to a lesser extent, in LMO2 was also noticed in the P4 region, suggesting that the SCL multiprotein complex could be recruited to the p21 locus. Finally, we observed binding of SCL in the same region of the p21 gene in bone marrow-derived mature MKs, suggesting that SCL binds the p21 gene throughout MK differentiation (Figure 6D).
Because p21 is normally repressed by SCL in primary MkPs (Table 1), we determined whether corepressors could also bind the p21 promoter. A potential candidate was ETO2, a potent corepressor and known partner of SCL in MKs and erythroid cells.25,27,28 ETO2 was detected on P4 and P5 regions of p21 in MKD1 cells (Figure 6C right graph). These data suggested that SCL is recruited to the first intron of the p21 gene, possibly as part of a repressor complex composed of at least GATA1, LMO2, and ETO2, either through the E-boxes or the GATA site identified in this region.
We then assayed the transcriptional activity of the p21 promoter in MKD1 cell line. These cells present 2 important advantages: (1) the experiments are performed in a “megakaryocytic” environment and not in surrogate cells; and (2) the Scl locus is floxed in these cells, thereby allowing the assessment of its excision on p21 promoter activity. A 736-bp p21 promoter fragment (−90 to +647) encompassing the intronic region bound by SCL was used to drive luciferase expression (construct p21-736, Figure 6E top). This p21-736 reporter construct led to a 2.5-fold increase in transcriptional activity compared with the empty vector pGL4 (Figure 6E graph, condition p21-736). To check whether SCL was involved in this basal activity, we excised Scl floxed alleles on Cre-recombinase expression in MKD1 cells to obtain Sclex/ex cells (Figure 6F top panel) and repeated the assay. We observed a 3.5-fold increase in luciferase levels (Figure 6E graph, condition p21-736 +Cre, Sclex/ex), suggesting that SCL represses the p21 promoter. This is in agreement with our in vivo data (microarray data, Table 1; Figure 6A). To test whether ETO2 was involved in SCL-mediated repression, we overexpressed ETO2 protein in MKD1 cells (Figure 6F bottom panel). This significantly decreased p21 transcriptional activity by 2.5-fold (Figure 6E graph, condition p21-736 +ETO2).
To characterize the cis-elements required for SCL-mediated repression, we introduced point mutations in its potential binding sites (the conserved E2/E3 E-box motifs and GATA site; Figure 6E top). In contrast to what was observed in Sclex/ex cells, E2/E3 E-box mutation led to low transcriptional activity (Figure 6E graph, E2/E3), suggesting that they contribute to maintaining a basal expression level; this activity may be SCL independent and rely on other basic helix-loop-helix proteins given the functional difference observed when Scl alleles are excised. Remarkably, the GATA site mutation led to derepression of the luciferase activity to a level similar to that observed on Scl deletion (2.5- and 3.5-fold respectively, compared with p21-736 WT reporter gene, Figure 6E graph, compare p21-736 +Cre [Sclex/ex] with GATA condition), suggesting a functional link between the GATA motif and SCL activity. Finally, the E2/E3/GATA mutant construct exhibited luciferase levels similar to that of the GATA mutant alone (Figure 6E graph). This indicated that the E-boxes might not be critically required in the absence of transcriptional repression mediated through the GATA motif.
These data support a model by which p21 repression requires binding of SCL and possibly ETO2 through recruitment by GATA1. This activity seems dominant over the transcriptional activation mediated through the conserved E-boxes. However, in the light of the results obtained with the E2/E3 mutant construct, we propose that these E-boxes may be important to counterbalance SCL-mediated repression. The mechanism underlying this regulation remains to be determined.
P21 levels are critical in megakaryopoiesis
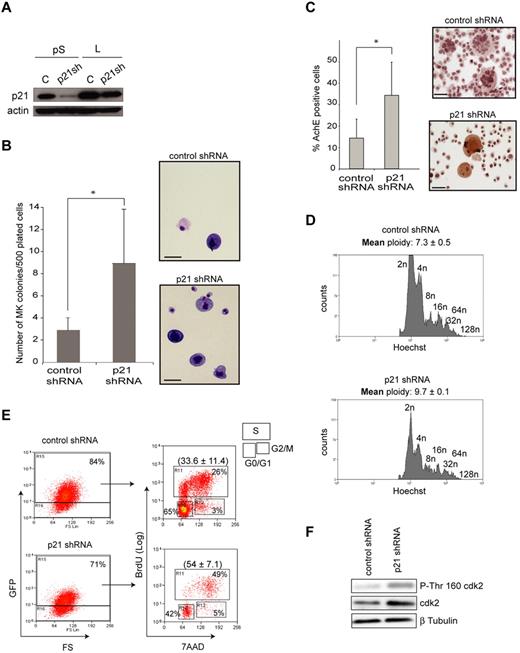
To further investigate whether SCL-mediated regulation of p21 was functionally required in megakaryopoiesis, we attempted a phenotypic rescue of the defects observed in Cre;Sclfl/fl MKs through shRNA-mediated knockdown of P21 levels. Cre;Sclfl/fl MkPs were transduced with a lentivirus expressing p21 shRNA sequences shown to significantly decrease P21 protein levels on expression in 3T3 cells (Figure 7A). Transduced cells were purified on the basis of GFP expression and the aspects of megakaryopoiesis perturbed in Cre;Sclfl/fl MKs reassessed.
p21 knock-down in Cre;Sclfl/fl restores aspects of megakaryopoiesis. (A) Western blot analysis showing reduced levels of P21 in NIH3T3 cells on transfection of p21 shRNA expression vectors: pS indicates pSuper vector; and L, lentiviral vector subsequently used to infect primary MKs. C indicates control shRNA (scrambled p21 shRNA sequence); p21sh, p21 shRNA; and actin, loading control. Efficacy of p21 shRNA sequences is observed with both vectors. (B) Colony assays. Bone marrow MkPs purified from Cre;Sclfl/fl homozygous mice were infected with p21 shRNA-expressing lentivirus or control vector. Day 2 GFP+ cells were plated in methylcellulose. MK colonies were scored at day 7. Data are the mean ± SD of 3 independent experiments performed in triplicate. *P = .01. (Right) Representative MK colonies. The photographs were taken using an Olympus BX60 microscope with a Qimaging camera. Openlab, Version 3 software (Improvision) was used for image acquisition, and images were exported into Adobe Photoshop, Version CS2 (Adobe Systems). Scale bars represent 20 μm. (C) Cre;Sclfl/fl MkPs were transduced with p21 shRNA-expressing lentivirus or control vector. Day 2 GFP+ cells were grown in TPO-containing medium and assayed for AChE activity 4 days later. Data are the mean ± SD of 3 independent experiments. *P = .08. (Right) Representative MKs. The photographs were taken using an Olympus BX60 microscope with a Qimaging camera. Openlab, Version 3 software (Improvision) was used for image acquisition, and images were exported into Adobe Photoshop, Version CS2 (Adobe Systems). Scale bars represent 40 μm. (D) Analysis of the ploidy of Cre;Sclfl/fl MKs transduced with p21 shRNA-expressing lentivirus or control vector on the GFP+CD41+ population. Peaks representing each ploidy class are labeled. One representative experiment of 3 is shown as well as the mean ploidy ± SD of the 3 independent experiments. P < .05. (E) Cell-cycle analysis. Day 2 transduced Cre;Sclfl/fl MkPs were sorted for GFP expression and expanded for 4 days. Cell-cycle phases were determined by analysis of BrdU incorporation and 7-amino-actinomycin D staining on CD41+ GFP+ gated cells. The percentage of cells in G0/G1, S, and G2M phases is shown. One representative experiment of 4 is shown. The mean (± SD) percentage of cells in S phase of the 4 experiments is shown in parentheses. P = .001. (F) Knockdown of P21 in mutant MkPs increases CDK2 phosphorylation on Thr 160 (P-Thr 160). Cre:Sclfl/fl MkPs expressing control shRNA or p21 shRNA sequences were lysed and subject to immunoblotting analyses using specific antibodies against CDK2 and P-Thr 160 CDK2. β-tubulin was used as a loading control. Increase in CDK2 phosphorylation in mutant sample (1.5- to 2-fold) was calculated after normalization for the amount of total CDK2.
p21 knock-down in Cre;Sclfl/fl restores aspects of megakaryopoiesis. (A) Western blot analysis showing reduced levels of P21 in NIH3T3 cells on transfection of p21 shRNA expression vectors: pS indicates pSuper vector; and L, lentiviral vector subsequently used to infect primary MKs. C indicates control shRNA (scrambled p21 shRNA sequence); p21sh, p21 shRNA; and actin, loading control. Efficacy of p21 shRNA sequences is observed with both vectors. (B) Colony assays. Bone marrow MkPs purified from Cre;Sclfl/fl homozygous mice were infected with p21 shRNA-expressing lentivirus or control vector. Day 2 GFP+ cells were plated in methylcellulose. MK colonies were scored at day 7. Data are the mean ± SD of 3 independent experiments performed in triplicate. *P = .01. (Right) Representative MK colonies. The photographs were taken using an Olympus BX60 microscope with a Qimaging camera. Openlab, Version 3 software (Improvision) was used for image acquisition, and images were exported into Adobe Photoshop, Version CS2 (Adobe Systems). Scale bars represent 20 μm. (C) Cre;Sclfl/fl MkPs were transduced with p21 shRNA-expressing lentivirus or control vector. Day 2 GFP+ cells were grown in TPO-containing medium and assayed for AChE activity 4 days later. Data are the mean ± SD of 3 independent experiments. *P = .08. (Right) Representative MKs. The photographs were taken using an Olympus BX60 microscope with a Qimaging camera. Openlab, Version 3 software (Improvision) was used for image acquisition, and images were exported into Adobe Photoshop, Version CS2 (Adobe Systems). Scale bars represent 40 μm. (D) Analysis of the ploidy of Cre;Sclfl/fl MKs transduced with p21 shRNA-expressing lentivirus or control vector on the GFP+CD41+ population. Peaks representing each ploidy class are labeled. One representative experiment of 3 is shown as well as the mean ploidy ± SD of the 3 independent experiments. P < .05. (E) Cell-cycle analysis. Day 2 transduced Cre;Sclfl/fl MkPs were sorted for GFP expression and expanded for 4 days. Cell-cycle phases were determined by analysis of BrdU incorporation and 7-amino-actinomycin D staining on CD41+ GFP+ gated cells. The percentage of cells in G0/G1, S, and G2M phases is shown. One representative experiment of 4 is shown. The mean (± SD) percentage of cells in S phase of the 4 experiments is shown in parentheses. P = .001. (F) Knockdown of P21 in mutant MkPs increases CDK2 phosphorylation on Thr 160 (P-Thr 160). Cre:Sclfl/fl MkPs expressing control shRNA or p21 shRNA sequences were lysed and subject to immunoblotting analyses using specific antibodies against CDK2 and P-Thr 160 CDK2. β-tubulin was used as a loading control. Increase in CDK2 phosphorylation in mutant sample (1.5- to 2-fold) was calculated after normalization for the amount of total CDK2.
In colony assays, GFP+Cre;Sclfl/fl MkPs transduced with p21 shRNA sequences showed a 3-fold increase in the number of MK colonies compared with cells infected with control lentivirus (p21 shRNA scrambled sequences). The colonies were smaller than those obtained from Cre;Sclwt/wt MkPs (2-10 cells as opposed to 10-15 respectively, not shown). Interestingly, we noticed the presence of many single cells whose megakaryocytic nature was confirmed on May-Grunwald-Giemsa staining, in the p21 knockdown dishes but not in the control (Figure 7B). Therefore, down-regulation of P21 in Cre;Sclfl/fl MkPs partially alleviated their block in forming MK colonies in vitro.
Next, we examined AChE activity. GFP+Cre;Sclfl/fl MkPs transduced with p21 shRNA sequences gave rise to a significantly higher number of AChE+ MKs compared with control cells (Figure 7C), indicating that P21 levels are critical for MK cytoplasmic maturation.
Polyploidization was analyzed next (Figure 7D). The mean ploidy in Cre;Sclfl/fl GFP+CD41+ MKs transduced with the p21 knockdown expression vector (N = 9.7) was increased compared with control (N = 7.4), confirming P21 role in polyploidization.
Next, we determined whether down-regulation of p21 in mutant MkPs could rescue the cell cycle defect (Figure 7E). Remarkably, 54% ± 7.1% of GFP+CD41+Cre;Sclfl/fl cells transduced with the p21 shRNA expression vector were in the S phase, as opposed to 33.6% ± 11.4% in the control (scrambled shRNA) population. This is similar to the percentage of Cre;Sclwt/wt CD41+ cells in S phase (52.7% ± 1.5%, Figure 4F), demonstrating a complete rescue of the cell cycle.
Finally, we checked whether the effects of P21 down-regulation on the cell cycle could correlate with an increase in the activity of cyclin-dependent kinase 2 (CDK2), a direct P21 downstream effector.39 Indeed, phosphorylation of threonine-160, a mark of CDK2 activation that is inhibited on association with P21, was increased in p21 knockdown/Cre;Sclfl/fl MkPs by 1.5- to 2-fold (Figure 7F).
Taken together, these results show that knockdown of P21 expression in Cre;Sclfl/fl MkPs partially alleviates the cytoplasmic and nuclear maturation blocks and fully restores cell cycle progression. Tight regulation of P21 levels by the SCL transcriptional complex is therefore required for normal megakaryocytic development.
Discussion
Despite SCL's established requirement in MK development, its exact function, mechanisms of action, and target genes in this lineage remained largely unknown. Through lineage-specific deletion, our study shows that SCL is instrumental for many aspects of MK biology in a steady-state situation, provides new insights into its role in proliferation, cellular maturation, and polyploidization, and identifies its genomic targets in MkPs.
Scl-deleted MkPs are impaired in their ability to form colonies confirming the critical role of SCL in the early stages of MK development.22,23,40 G1/S cell-cycle progression is blocked in Scl-null MKs, leading to reduced proliferation, polyploidization, and cytoplasmic maturation are impaired, suggesting a major regulatory role for SCL in MK biology. Finally, SCL is also involved in platelet formation, as platelet biogenesis (but not platelet function) is affected, as previously reported.24,40
Gene expression analysis established a molecular link between some of these phenotypic observations and deregulation of p21 expression. Increased p21 mRNA levels are detected in SclΔ/Δ MkPs as well as in maturing MKs. Phenotypic rescue experiments clearly demonstrate that this increase affects, above all, proliferation, and, to a certain extent, polyploidization and cytoplasmic maturation. In agreement with a functional role of P21 in MK development, endomitotic cell-cycle arrest is observed on overexpression of p21 in wild-type cultured MKs.41 Our study demonstrates that SCL is involved in the tight control of P21 levels in the early stages of MK differentiation to allow completion of polyploidization before the cells withdraw from the cell cycle and engage into terminal maturation.
Functional links between SCL and p21 have been reported in erythroid progenitors where p21 expression is repressed by the SCL core complex and corepressor ETO2 to allow cellular proliferation.27 Regulation of p21 expression in MKs by SCL is also probably direct as (1) binding of SCL is detected in the first intron of p21, and (2) we demonstrate that SCL exerts transcriptional repression on the p21 promoter. Importantly, SCL and ETO2 are present in multiprotein complexes in MKs.25,28 Here, we show that ETO2 overexpression decreases the transcriptional activity of p21 promoter sequences in transactivation assays, suggesting that SCL represses p21 through interaction with ETO2 in MKs. Interestingly, we observed a derepression of p21 promoter activity when the GATA motif was mutated, but not with mutations in the conserved E-boxes. Therefore, we propose that the SCL/ETO2 complex is recruited to the GATA site via interaction with GATA1 to exert its repressive activity on the p21 promoter. In agreement with this, GATA1 is a key determinant for SCL recruitment on GATA motifs in the absence of E-boxes.32 Interestingly, MkPs isolated from mice expressing a DNA-binding mutant form of SCL17 give rise to normal numbers of MK colonies in in vitro assays, suggesting DNA-binding independent mechanisms of action in this lineage (M.K. and C.P., unpublished observations, June 2009). Altogether, these data strongly suggest that SCL-mediated repression of p21 is independent of its direct DNA-binding activity and mediated through interaction with GATA1. Similarly, SCL's DNA binding activity is dispensable for binding and activating Nfe-2 promoter,24 a late target of SCL. Interestingly, it was suggested that SCL might be recruited through a GATA motif present in that region.
SCL binding on the p21 promoter is detected in immature MKs (MKD1 cells) as well as in mature primary MKs and, in both contexts, exerts a repressive activity. Therefore, SCL may control p21 expression throughout MK differentiation, and we anticipate that this regulation is complex. ETO2 expression is almost undetectable in mature MKs28 and therefore cannot account for SCL-repressive activities. One possible candidate is thenucleosome remodeling and deacetylase complex (NURD) nucleosome remodeling and deacetylase regulates megakaryopoiesis through activation and repression of select target genes of GATA1 and its partner FOG1.42 Strikingly, MkPs isolated from mice with defective interaction between NURD and FOG1 display defects similar to those described in this study: impaired in vitro MK colonies formation, no AChE activity and, similar to what we observed in mature MKs, down-regulation of GpVI expression.42 Whether SCL associates with the FOG1/NURD complex via GATA1 interaction to activate late megakaryocytic genes, such as GpVI, and repress p21 gene in mature MKs remains to be investigated.
A link between SCL and cell proliferation/quiescence has been recently documented in other cellular systems,43,44 stressing that one critical function of SCL in lineage maturation is to control cell-cycle progression. Moreover, recent studies have shown functional associations between other key transcription factors (RUNX1 and GATA1) and cell-cycle regulators (P19 and cyclin D1) in megakaryopoiesis.45,46 Therefore, an exquisite control of expression of cell-cycle regulatory genes by key transcription factors, to either promote or arrest cell-cycle progression, is necessary for MK development.
Complete restoration of polyploidization and differentiation from SclΔ/Δ MKs is not achieved in our rescue experiments. This could be the result of incomplete down-regulation of p21 levels and/or to the involvement of other putative targets of SCL in MK development. Interestingly, some of the genes identified in our expression analysis are involved in MK-specific processes, linking SCL to maturation mechanisms. As examples, Myo18b and Myo6 are involved in cytoskeleton organization and platelet function; Col18a1, Rab11a, and Gnaz have previously been implicated in MK differentiation47 ; Cxcr4, Ltbp1, Selp, and Cd63, involved in proplatelet formation and platelet functions, have also recently been linked to nuclear maturation.3 Supporting a role for SCL in the latest stages of MK maturation, an independent study described Nfe-2 as a target of SCL in the terminal phase of platelet production.24 Therefore, SCL seems to control numerous pathways in megakaryopoiesis, from proliferation of MkPs to terminal maturation processes.
In conclusion, the in vivo lineage-specific gene deletion approach used in this study has allowed to bypass the early defects observed when Scl is deleted in the whole adult hematopoietic system. This has revealed the key functions of SCL in megakaryopoiesis and provided a mechanistic link between SCL, the cell-cycle regulator P21, and megakaryocytic development.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank S. Soneji for microarray data analysis, G. Juban for technical advice, A. Schmitt for analyzing the electronic microscopy images, W. Vainchenker for helpful discussion, A. Elefanty and C.G. Begley for providing the Scl floxed targeting vector (AE655), the Genomic Services Group (Wellcome Trust Center of Human Genetics, Oxford) for Illumina expression profiling, and the Biomedical Services (University of Oxford) for mouse blastocyst injection and chimera production.
This work was supported by the Medical Research Council. P.V. was supported by the Medical Research Council Disease Team Award, the Medical Research Council Molecular Hematology Unit, and the Oxford Partnership Comprehensive Biomedical Research Center with funding from the Department of Health's NIHR Biomedical Research Centres.
Wellcome Trust
Authorship
Contribution: H.C. designed and performed the experiments, analyzed the data, and wrote the paper; M.K. and S.B. performed experiments; N.G., K.C., and A.A. provided assistance with FACS sorting; R.C.S. provided the Pf4-Cre mouse strain; A.C.P. and S.P.W. performed the work on platelet function; D.J.P.F. performed the electronic microscopy; P.V. analyzed the data; and C.P. designed the experiments, analyzed the data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for A.C.P. is Novartis Horsham Research Centre, Horsham, United Kingdom. The current affiliation for A.A. is Institute of Molecular Medicine, Trinity College Dublin, Dublin, Ireland.
Correspondence: Catherine Porcher, Medical Research Council Molecular Haematology Unit, Weatherall Institute of Molecular Medicine, John Radcliffe Hospital, Oxford, OX3 9DS, United Kingdom; e-mail: catherine.porcher@imm.ox.ac.uk.
References
Author notes
M.K. and S.B. contributed equally to this study.

![Figure 3. Impaired megakaryocytic colony formation from Cre;Sclfl/fl MkPs. Bone marrow progenitors (CMP [A], MEP [B], MkP [C]) purified from Cre;Sclwt/wt wild-type (black bars), Cre;Sclwt/fl heterozygous (open bars), and Cre;Sclfl/fl homozygous (gray bars) mice were plated in methylcellulose. Colony-forming units-erythroid (CFUE), burst-forming units-erythroid (BFUE), MK, mixed, and myeloid colonies were scored at day 7. Data are the mean ± SD of 2 independent experiments performed in triplicate. *P < .01. (Bottom) Representative MK colonies grown from Cre;Sclwt/wt and Cre;Sclfl/fl MkPs. The photographs were taken using an inverted IX51 Olympus microscope with a Jenoptik C14 camera. Openlab, Version 3 software (Improvision) was used for image acquisition, and images were exported into Adobe Photoshop, Version CS2 (Adobe Systems). Scale bars represent 20 μm. Note the difference in the morphology and size of the colonies. A control MK shading platelet (arrow) is shown. Scale bar represents 50 μm.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/118/3/10.1182_blood-2011-01-328765/4/m_zh89991174200003.jpeg?Expires=1765030616&Signature=WhWYcrMuWSDz8cAZhYib6uobN6awAc0UgB2mKDuU8dGSek42g2EWkVPj915LksFiuRK0pMKVnkCw5dMQj8cMaRAwQ4ys-cX-H1VyMXc5yzGKREG-WApqMe8yajkr1eMhmMYOB1jTeeqvn-isdEfievRhZHjQ1PxTRbN~f1Mp9eLRCsdrxn7GQc-sq7d2SJssN1v0-Dgn1nrSqjkgQsZQ1VYLuNsgtrYog4Vu2pocrC7-o9rpe7g1wNSKp8vX8clF5upixX5fDN~pdPDkKhklE4w8kQDSz4cOSsV5eB~X1xrR8trT07Yl7RElQyt4ZK~-Bf6ZnYjeOOfYGF0L-efM5A__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal