Abstract
In this study, we have addressed how Lyn kinase signaling mediates nilotinib-resistance by quantitative phospho-proteomics using Stable Isotope Labeling with Amino acid in Cell culture. We have found an increased tyrosine phosphorylation of 2 additional tyrosine kinases in nilotinib-resistant cells: the spleen tyrosine kinase Syk and the UFO family receptor tyrosine kinase Axl. This increased tyrosine phosphorylation involved an interaction of these tyrosine kinases with Lyn. Inhibition of Syk by the inhibitors R406 or BAY 61-3606 or by RNA interference restored the capacity of nilotinib to inhibit cell proliferation. Conversely, coexpression of Lyn and Syk were required to fully induce resistance to nilotinib in drug-sensitive cells. Surprisingly, the knockdown of Syk also strongly decreased tyrosine phosphorylation of Lyn and Axl, thus uncovering interplay between Syk and Lyn. We have shown the involvement of the adaptor protein CDCP-1 in resistance to nilotinib. Interestingly, the expression of Axl and CDCP1 were found increased both in a nilotinib-resistant cell line and in nilotinib-resistant CML patients. We conclude that an oncogenic signaling mediated by Lyn and Syk can bypass the need of Bcr-Abl in CML cells. Thus, targeting these kinases may be of therapeutic value to override imatinib or nilotinib resistance in CML.
Introduction
Chronic myeloid leukemia (CML) is characterized by the presence of the Philadelphia (Ph) chromosome that results from a t(9;22)(q34;q11) reciprocal translocation.1,2 Twenty-five percent of acute lymphoblastic leukemias (ALL) in adults are also characterized by this translocation. The Ph chromosome contains a BCR-ABL hybrid gene, the molecular hallmark of CML.3 BCR-ABL encodes an oncogenic fusion protein of 190, 210 or 230 kDa, depending on the breakpoint on the BCR gene. The unifying feature of all these Bcr-Abl fusion proteins is their deregulated protein tyrosine kinase activity that is responsible for leukemogenesis in vitro and in vivo.4,5 Targeting the tyrosine kinase activity of Bcr-Abl is an attractive therapeutic strategy in CML or in Bcr-Abl positive ALL that has recently found success with the development of new drugs such as tyrosine kinase inhibitors (TKI). Imatinib, a Bcr-Abl TKI that competes with ATP for binding to the Abl kinase domain, is now the front-line therapy of CML in chronic phase. However, imatinib resistance is now a well-recognized problem, particularly in the advanced phase of the disease. One of the well known mechanisms of resistance in CML patients is mutations in the Bcr-Abl tyrosine kinase domain. Nilotinib and dasatinib, the second-generation tyrosine kinase inhibitors, have been developed to override this phenomenon,6,7 however in vitro and in vivo resistance to these 2 TKI is also observed.
We have previously generated and characterized nilotinib-resistant cells from different Ph-positive cell lines (K562, AR230, and LAMA84) showing that, in addition to Bcr-Abl and/or multidrug resistance P-glycoprotein (Pgp) overexpression, the up-regulated expression of the Src family kinase p53/56 Lyn defines an additional mechanism of cell resistance to nilotinib.8 However the mechanism by which Lyn induces this cellular process is still not known. In an attempt to identify downstream effectors of Lyn involved in the resistance to nilotinib, a quantitative phosphoproteomic analysis was performed using nilotinib-sensitive and resistant cells by Stable Isotope Labeling with Amino acid in Cell culture (SILAC). This approach allowed the identification of 2 additional protein tyrosine kinases involved in this biologic process: Syk and Axl. Moreover, we have identified interplay between Syk and Lyn in the signaling process leading to tyrosine phophorylation of Axl and the adaptor CDCP-1 and eventually to resistance to nilotinib. The results uncovered a new role for Syk and Axl in TKI resistance that could be of therapeutic value in the treatment of CML resistant to imatinib or nilotinib.
Methods
Reagents
RPMI 1640 medium, FCS, PBS, trypan blue and antibiotics were purchased from Invitrogen. Imatinib and nilotinib were kindly provided by Novartis Pharma. MTS was purchased from Promega. The Syk inhibitors piceatannol and BAY 61-3606 were purchased from Sigma-Aldrich. The Syk inhibitor R406 and the Axl inhibitor R428 were provided by Rigel Inc and are under MTA approval. The inhibitors PP1 and PP2 were purchased from Calbiochem. All siRNAs were purchased from Dharmacon.
Cell lines
The Bcr-Abl positive human erythroleukemic K562 cell line has been rendered nilotinib-resistant as previously described for imatinib, and designated K562-rn.9 Cells were maintained in RPMI 1640 medium supplemented with 10% FCS, 2mM l-glutamine, 100 U/mL penicillin, and 0.1 mg/mL streptomycin (referred to in this article as ‘RF-10′) at 37°C in a humidified atmosphere containing 5% CO2. Aliquots were taken at 24 hour intervals for assessment of cell viability by Trypan blue exclusion (Sigma-Aldrich). For SILAC, K562 cells were grown in “light” medium whereas K562-rn were grown in “heavy” medium for 7 days leading to 95% of isotope incorporation as previously described.10
CD34 cells isolation
Mononuclear cells were isolated from bone marrow or blood by Ficoll gradient. CD34 positive cells were purified according to the manufacturer's instructions (Miltenyi Biotech) and purity was analyzed by flow cytometry using phycoerythrin-conjugated anti-CD34 antibody (Becton Dickinson).
Cell proliferation assays
Cell proliferation was measured using MTS tetrazolium (Cell Titer96 Aqueous; Promega). Cells were washed and plated (104 cells) in quadruplicate into microtiter-plate wells in RF-10 medium plus various doses of inhibitors as indicated. Cell viability was measured every day during 4 days. After addition of 20 μL of MTS to the wells followed by a 2 hour incubation at 37°C, the 490 nm absorbance was read using a microplate autoreader (Dynex Technologies). The mean results of the 4-well set were standardized in comparison to the initial optical density at day 0. All the experiments were repeated at least 3 times.
Phosphotyrosine immunoaffinity purification and mass spectrometry analysis
SILAC (13C615N4-Arg- and 13C615N2-Lys as heavy labeling), phosphotyrosine immunoaffinity purification (using a mixture of 4G10 and pY100 antibodies), and tryptic digests were essentially performed as described Leroy et al.10 Briefly, cells were lysed in immunoprecipitation buffer. Heavy and light cleared lysates, 5 mg each, were mixed at a 1:1 ratio and incubated overnight at 4°C with 1 mL of anti-phosphotyrosine immunoaffinity beads. Elution was performed with 100mM phenyl phosphate (Sigma-Aldrich) and proteins were precipitated with cold acetone. The proteins were separated by 4%-15% gradient SDS-PAGE and the gel was stained with the Colloidal Brilliant Blue G (Sigma-Aldrich). Each lane was cut in 45 pieces and protein bands were in-gel digested using trypsin (sequencing grade; Promega). Mass spectrometry (MS) analysis of the samples was performed on a QSTAR pulsar-i quadripole-time-of-flight mass spectrometer (Applied Biosystems) coupled to an Ultimate 3000 (Dionex) nanoflow system driven by Chromeleon software. MS/MS spectra were searched against the human entries of UniProt Knowledgebase Release 10.2 database (http://www.expasy.ch) using the Mascot v2.1 algorithm (http://www.matrixscience.com). Search parameters were mass accuracy 0.1 Da for MS and MS/MS data; 1 miscleavage; variable modifications: oxidized methionine, Phospho (ST), Phospho (Y), SILAC-labels: Lys-8 and Arg-10. Identification was done on the basis of at least one specific peptide. All significant hits (P < .05) were manually inspected. Quantification was done on at least 2 MS spectra per protein using MSQuant v1.4.1 software (http://msquant.sourceforge.net). All MS spectra used for quantification were manually verified.
Tyrosine kinase silencing by siRNA
To inhibit Syk protein expression, 2 × 106 cells were washed 3 times in cold PBS. siRNA used was from Dharmacon smart pool (5′-GAGCAAAUUGUCCUGAUAG-3′). Transfection was performed using the Amaxa system according to the manufacturer's instructions using the protocol T03 (Amaxa AG). Cells were seeded at 2 × 105/mL in RF-10 for 24 hours after which cells were separated in 2 batches and incubated in the absence or in the presence of 20nM nilotinib. Aliquots of the culture were harvested daily for protein expression analysis and measurement of cell viability by triplicate trypan blue exclusion counts. Experiments were performed no longer than 4 days and were repeated 3 times. Cell viability is presented as the mean of the triplicate for one experiment representative of 3.
Lentiviral Production, Titration, and Cell Transduction
Production and titration of infectious lentiviral particles were as detailed previously.11 Primers for the short hairpin anti-Syk were designed as followed: forward 5′-AGCTTCCGAGCAAATTGTCCTGATAGTTCAAGAGACTATCAGGACAATTTGCTCTTTTTGGAAG-3′, reverse 5′-TCGACTTCCAAAAAGAGCAAATTGTCCTGATAGTCTCTTGAACTATCAGGACAATTTGCTCGGA-3′. Ligation was performed in pTRIPΔU3EF1α-DsRed-WPRE-MSCΔU3 vector and used in with PAX2 and VSVG vectors to transfect HEK293T cells for lentiviral production. Lentiviral particles were added to the target cells and incubated for 24 hours. Then, the cells were washed twice in PBS and grown in the presence of medium for 6 days before experimental use. Cells in which Syk had been silenced by Syk ShRNA expression were selected by cell sorting using DsRed expression as a threshold, analyzed by flow cytometry as a homogenous cell population with purity > 98%. Syk silencing was verified by Western blotting.
Ectopic tyrosine kinase expression
To coexpress Lyn and Syk proteins in a similar ratio than in K562-rn cells, we transfected K562-s cells (ie sensitive to TKI) with cDNA coding for Lyn and Syk at a 4:1 ratio. Lyn cDNA coded for the wild type form or the constitutively activated mutant (Y508) as described.10 Syk cDNA was kindly provided by Dr P. Coopman as the wild-type, the kinase dead form (K402R), and the constitutively activated form (Y130E).12 K562 cells (2 × 106) were washed 3 times in cold PBS. Transfection was performed using the Amaxa system according to the manufacturer's instructions using the protocol T03 (Amaxa AG). Cells were seeded at 2 × 105/mL in RF-10 for 24 hours after which cells were separated in 2 batches and incubated in the absence or in the presence of 20nM nilotinib. Aliquots of the culture were harvested daily for protein expression analysis and measurement of cell viability by triplicate trypan blue exclusion counts. Experiments were performed no longer than 4 days and were repeated 3 times. Results are presented as 1 experiment representative of 3.
Flow cytometry
For detection of apoptosis, 105 cells were incubated for 15 minutes in 500 μL of PBS with 2mM Ca2+, 2 μL of annexin V–FITC or APC before flow cytometry analysis on Facscalibur. Five thousand events were acquired for statistical analysis. Results are expressed as the percent of annexin V positive cells in comparison to control (untreated cells). Annexin V–FITC or APC-conjugated were purchased from Beckman Coulter.
For the detection of Axl and Syk phosphorylation, 105 cells were incubated for 10 minutes in 500 μl of PBS-paraformaldehyde (4% wt/vol) and 10 minutes in PBS-Triton (1% wt/vol). After saturation for 30 minutes in PBS-BSA (2% wt/vol), cells were incubated for 1 hour with control rabbit IgG or specific antibody (anti-Axl or anti–p-Syk525/526, 3 μg/mL). After 1 wash, cells were incubated with Alexa 488-conjugated anti–rabbit antibody for 1 hour. After the last wash, cells were analyzed by flow cytometry.
Western blot analysis
Protein lysates were prepared according to Pocaly et al.13 Protein concentration was measured by the BCA Protein Assay (Pierce) and the lysates were stored at −80°C. Approximately 25 μg of protein were resolved on 10% SDS-PAGE gels, transferred onto PVDF membranes (Bio-Rad) by semi-dry electrophoretic transfer, probed with individual antibodies, and visualized by the ECL system (Perkin Elmer). For immunoprecipitation, 400 μg of protein lysates were precleared with 30 μL of 50% slurry protein A-sepharose by incubation for 1 hour at 4°C. After centrifugation, the supernatant was incubated with 2 μg of antibody overnight and then incubated for 1 hour with 30 μL of protein A-sepharose. After 3 washes with lysis buffer, the pellet of protein A-sepharose was solubilized with SDS loading buffer and samples were resolved by SDS-PAGE.
Quantification by Q RT-PCR in sensitive and resistant cells
Seven CML patients in chronic phase (n = 2) or in accelerated phase (n = 5; 2 males, 5 females) with Ph chromosome and BCR-ABL–positive CML were investigated after failure of imatinib or nilotinib. The duration of therapy was 3 to 36 months and all patients were analyzed before nilotinib and at the time of failure. Resistance was defined by progression or absence of hematologic and cytogenetic response. Total RNA was extracted from cell lines or primary CML cells, and Q RT-PCR performed using MX3005P from Stratagene according to the manufacturer's instructions. Primers used for CDCP1 were forward: 5′-TGGTT CCACCCCAGAAATGT-3′ and reverse: 5′-GATGATGCACAGACGTTTTATAGATGA-3′, as previously described.14 For Axl these were forward: 5′-GTTTGGAGCTGTGATGGAAGGC-3′ and reverse: 5′-CGCTTCACTCAGGAAATCCTCC-3′. With a 10-fold serial dilution series for cDNA, the assay was found to be linear over at least 5 orders of magnitude (slope, −3.414; intercept, 35.81). Quantification of relative expression was performed using GUS as endogenous control, as previously described.15 Analysis was done by comparative Ct method giving the amount of target normalized to the endogenous reference and relative to the same pool of mononuclear cells as the calibrator.
Statistical analysis
A Mann Whitney test was used to calculate differences between means; differences were considered significant when P < .05.
Results
Tyrosine kinase signaling in nilotinib resistant cells by quantitative phosphoproteomics
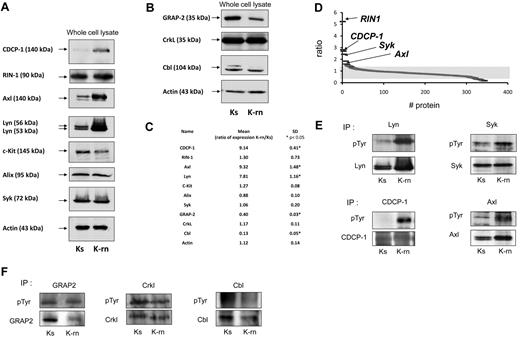
Comparing the tyrosine phosphorylations of K562 sensitive to TKI (Ks) with K562 resistant to nilotinib (K-rn) by quantitative phosphoproteomics showed that K-rn cells overexpressed the tyrosine kinase Lyn (Figure 1A) with a 7.8-fold increase (n = 6, P < .05) compared with Ks. We then investigated Lyn substrates in nilotinib resistant cells using a SILAC approach in which K-rn cells were labeled with “heavy” 13C615N4-Arg– and 13C615N2-Lys–containing medium, whereas K-s cells were cultured in “light” normal conditions. Cell lysates were next combined, and phosphotyrosine-containing proteins were purified using antiphosphotyrosine antibodies. Proteins were separated by SDS-PAGE, digested with trypsin, and subjected to liquid chromatography-tandem Mass Spectrometry. Quantification was made from the relative intensities of Arg/Lys labeled tryptic peptides. With this approach, we identified 344 proteins, 9 of which exhibiting a significant increased and 91 reduced tyrosine phosphorylation, in Lyn overexpressing cells (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). As expected, Bcr-Abl was identified in this screen with a quantification ratio of 1. This data confirmed that resistance to nilotinib was not because of a change in expression of Bcr-Abl in addition to the absence of mutation in the kinase domain as already published.8 Surprisingly, Lyn was not obtained with our analysis, indicating that our MS methodology is not extensive in this case. Nevertheless, Western blot analyses confirmed that Lyn was still up-regulated in K-rn cells (Figure 1A and supplemental Figure 1).
Validation of SILAC results by Western-blotting. Proteins identified by SILAC were detected in nilotinib-sensitive Ks or nilotinib-resistant K-rn cell lysates. After transfer, specific proteins were individually detected for proteins over detected in K-rn (A) or down detected (B). After stripping, membranes were probed for actin as loading control. Proteins detected are indicated by an arrow on the left. Quantification of protein expression was performed by densitometry (C). Protein expression level was normalized using actin as loading control. Results are expressed as the mean fold increase expression for each protein by calculating the ratio of the K-rn cells to their sensitive counterpart Ks from at least 3 independent experiments. Significance was calculated by Mann Whitney test and indicated by an asterisk. Proteins identified by mass spectrometry and scored by SILAC were graphically represented as the ratio between the heavy and light isotope (D). Indicated proteins were immunoprecipitated from Ks or K-rn cell lysates. Tyrosine phosphorylated proteins were detected followed by the specific detection of immunoprecipitated proteins over detected in K-rn (E) or down detected (F). Results shown are from one independent experiment representative of 3.
Validation of SILAC results by Western-blotting. Proteins identified by SILAC were detected in nilotinib-sensitive Ks or nilotinib-resistant K-rn cell lysates. After transfer, specific proteins were individually detected for proteins over detected in K-rn (A) or down detected (B). After stripping, membranes were probed for actin as loading control. Proteins detected are indicated by an arrow on the left. Quantification of protein expression was performed by densitometry (C). Protein expression level was normalized using actin as loading control. Results are expressed as the mean fold increase expression for each protein by calculating the ratio of the K-rn cells to their sensitive counterpart Ks from at least 3 independent experiments. Significance was calculated by Mann Whitney test and indicated by an asterisk. Proteins identified by mass spectrometry and scored by SILAC were graphically represented as the ratio between the heavy and light isotope (D). Indicated proteins were immunoprecipitated from Ks or K-rn cell lysates. Tyrosine phosphorylated proteins were detected followed by the specific detection of immunoprecipitated proteins over detected in K-rn (E) or down detected (F). Results shown are from one independent experiment representative of 3.
We next focused on tyrosine phosphorylated proteins involved in signaling showing a clear over (ratio > 1.5) or under (ratio < 0.66) phosphorylation in K-rn cells (supplemental Tables 2-3). With this setting, 6 proteins had increased tyrosine phosphorylation > 1.5 fold (Figure 1D): the Ras and Rab interactor 1 (RIN1); the CUB domain-containing protein 1 precursor, an adaptor protein also called CDCP-1; the programmed cell death 6-interacting protein, PDCD6IP or Alix and 3 additional tyrosine kinases: the spleen tyrosine kinase Syk; the UFO receptor Axl and the stem cell factor receptor c-kit. These MS data have systematically been confirmed by Western blot showing increased tyrosine phosphorylation of Lyn, Syk, Axl and CDCP-1 in K-rn cells. Interestingly increased phosphorylation of Axl and CDCP-1 was accompanied by an increase in the expression of these proteins in K-rn cells (mean ± SD ratio of expression: 9.3 ± 1.4 and 9.1 ± 0.4, respectively; Figure 1A-C). We also focused on 7 proteins among the 91 whose ratio was < 0.66 (supplemental Table 1). This included proteins involved in cytoskeleton regulation, such as the tyrosine kinase Fak, Wave2, Nap1 and desmoplakin and signaling proteins such as Gab1, Cbl, CrkL and GRAP2. This data were confirmed by Western blot for some of them (Figure 1B). The decreased tyrosine phosphorylation of GRAP2 and Cbl in K-rn cells was correlated with/accompanied by a reduced expression of these proteins (Figure 1F; mean ± SD ratio of expression: 0.4 and 0.13, respectively; Figure 1C). In contrast, CrkL has a similar level of expression and phosphorylation (Figure 1F).
Syk and Axl interact with Lyn
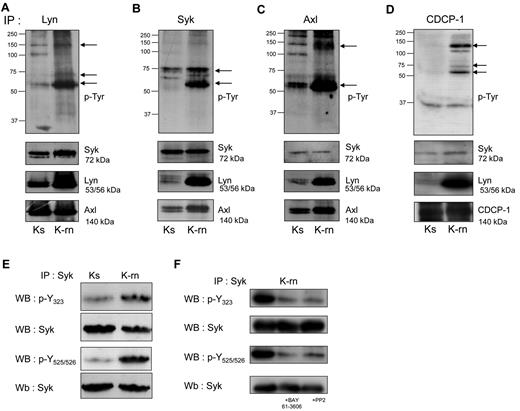
We next investigated whether Lyn was involved in tyrosine phosphorylation of Syk and Axl. As shown in Figure 2, Lyn interacted with Syk and Axl in both cell-lines, as demonstrated by coimmunoprecipitation of these proteins. In contrast, Syk coprecipitated significantly higher amounts of Lyn and Axl in K-rn compared with Ks cells. Finally Axl also coprecipitated Lyn in K-rn cells. To further investigate the interactions between these kinases, Syk was immunoprecipitated from Lyn immunoprecipitate followed by the detection of Axl. The detection of Axl in the last immunoprecipitate confirmed the existence of a Lyn/Syk/Axl ternary complex whose tyrosine phosphorylation is increased in nilotinib resistant cells (supplemental Figure 2A).
Detection of tyrosine phosphorylated proteins associated with the kinases Lyn, Syk and Axl. Proteins (A: Lyn; B: Syk and C: Axl, D: CDCP-1) were immunoprecipitated from Ks and K-rn cell lysates and subjected to phosphotyrosine Western blotting using a mix of 4G10 and pY-100 antibodies (top panels). Differentially tyrosine phosphorylated proteins are indicated by arrows on the right. After stripping, membranes were cut and blotted against Syk (top), Lyn (middle), Axl (bottom panels). Results are from one experiment representative of 5. (E) Syk was immunoprecipitated from Ks and K-rn cell lysates. Immunprecipitates were subjected to Western-blotting using phosphospecific antibodies against residues Y323 and Y525/526, and reprobed for whole Syk. (F) Syk was immunoprecipitated from K-rn cell lysates from untreated, BAY 61-3606–treated (2μM 2 hours) and PP2-treated (20μM 2 hours) K-rn cells. Immunoprecipitates were subjected to Western blotting using phosphospecific antibodies as described in panel E.
Detection of tyrosine phosphorylated proteins associated with the kinases Lyn, Syk and Axl. Proteins (A: Lyn; B: Syk and C: Axl, D: CDCP-1) were immunoprecipitated from Ks and K-rn cell lysates and subjected to phosphotyrosine Western blotting using a mix of 4G10 and pY-100 antibodies (top panels). Differentially tyrosine phosphorylated proteins are indicated by arrows on the right. After stripping, membranes were cut and blotted against Syk (top), Lyn (middle), Axl (bottom panels). Results are from one experiment representative of 5. (E) Syk was immunoprecipitated from Ks and K-rn cell lysates. Immunprecipitates were subjected to Western-blotting using phosphospecific antibodies against residues Y323 and Y525/526, and reprobed for whole Syk. (F) Syk was immunoprecipitated from K-rn cell lysates from untreated, BAY 61-3606–treated (2μM 2 hours) and PP2-treated (20μM 2 hours) K-rn cells. Immunoprecipitates were subjected to Western blotting using phosphospecific antibodies as described in panel E.
We next asked whether Syk was constitutively activated in K-rn cells. Syk levels were not modified on nilotinib resistance induction (mean ± SD ratio of expression: 1.06 ± 0.2; Figure 1C). The use of phosphospecific antibodies showed an increased Syk phosphorylation on Tyr323, known to be involved in the interaction with Cbl and Tyr525/526 involved in the activation Syk (Figure 2E). This data suggested that Syk is activated during nilotinib resistance induction. We then wondered whether this activation was regulated by Lyn. As shown in Figure 2F, both the SFK inhibitor PP2 (20μM) and the Syk inhibitor BAY 61-3606 (2μM) induced a large decrease of Syk phosphorylation. Similar results were obtained with the dual Abl/Src inhibitor dasatinib which, like PP2, did not affect Syk kinase activity (supplemental Figure 2B-C). Overall, this data suggests that Lyn deregulation mediates Syk activation in K-rn cells.
Inhibition of Syk tyrosine kinase restores nilotinib sensitivity
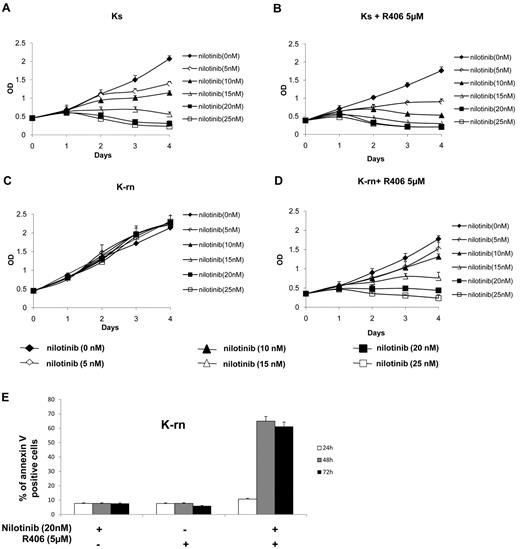
Having shown that Syk activity was higher in nilotinib resistant cells, we wanted to confirm that the function of Syk was instrumental in determining the resistance to nilotinib. We tested the Syk inhibitor R406 used in several diseases including chronic lymphocytic leukemia.16,17 R406 (5μM) has no significant effect on Ks and K-rn cell proliferation as assessed by 4-day MTS experiments (supplemental Figure 3A,B). Ks cells responded to increasing dose of nilotinib by a decrease of proliferation accompanied by a loss of viability (Figure 3A) whereas K-rn cells were fully resistant (Figure 3C). The incubation with 5μM of R406 did not significantly modify the response of Ks cells to nilotinib (Figure 3B) although a slight decrease was observed. In contrast, nilotinib sensitivity was fully restored in K-rn cells in the presence of R406 with decreased MTS viability (Figure 3D) confirmed by increased detection of apoptotic cells (Figure 3E). Similar results were obtained with 1 or 10μM of the Syk inhibitor; BAY 61-3606 (supplemental Figure 3C-D), thus confirming that Syk activation is crucial for nilotinib resistance in these cells.
Inhibition of Syk tyrosine kinase activity restores nilotinib sensitivity. Proliferation of the nilotinib-sensitive (Ks) and resistant cell lines (K-rn) were tested in MTS assays in the presence of increasing concentrations of nilotinib in the absence (A,C) or the presence (B,D) of R406 (5μM). Results are expressed as the mean of the optical density (OD) of the 4-well set standardized in comparison with the starting optical density at day 0, which is directly proportional to the number of viable cells. K-rn cells were grown in the presence of nilotinib (20nM), R406 (5μM) or a combination of nilotinib plus R406. Apoptosis was detected by flow cytometry using annexin V–FITC at 24, 48, and 72 hours. Results, expressed as the percentage of annexin V positive cells, are from one experiment representative of 3 (E).
Inhibition of Syk tyrosine kinase activity restores nilotinib sensitivity. Proliferation of the nilotinib-sensitive (Ks) and resistant cell lines (K-rn) were tested in MTS assays in the presence of increasing concentrations of nilotinib in the absence (A,C) or the presence (B,D) of R406 (5μM). Results are expressed as the mean of the optical density (OD) of the 4-well set standardized in comparison with the starting optical density at day 0, which is directly proportional to the number of viable cells. K-rn cells were grown in the presence of nilotinib (20nM), R406 (5μM) or a combination of nilotinib plus R406. Apoptosis was detected by flow cytometry using annexin V–FITC at 24, 48, and 72 hours. Results, expressed as the percentage of annexin V positive cells, are from one experiment representative of 3 (E).
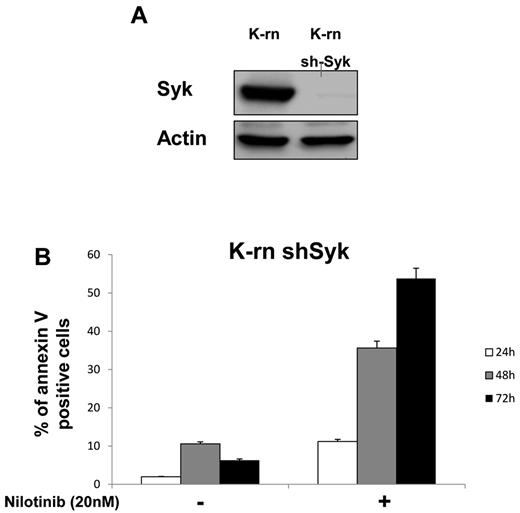
To further address the role of Syk in Lyn-mediated resistance, its expression was inhibited by a specific siRNA in K-rn cells. In contrast to a scrambled version of the siRNA (negative control siRNA Neg), the Syk-targeted siRNA (siRNA Syk) induced a large decrease of Syk expression at after 3 days of transfection (supplemental Figure 4A). This inhibition of Syk expression was associated with an increased sensitivity to nilotinib as measured by an increased cell mortality (+ 34%; supplemental Figure 4B). The involvement of Syk was also confirmed by a Syk shRNA that was stably introduced into K-rn cells by lentiviral infection. This approach led to complete inhibition of Syk expression without disturbing cell proliferation (Figure 4A). After purification of the Syk-null cell population by sorting Ds-Red-positive cells, the K-rn–Syk shRNA cell line was tested for nilotinib sensitivity. The inhibition of Syk expression fully restored the sensitivity to nilotinib accompanied by the apoptosis of K-rn cells (Figure 4B).
Silencing Syk restores nilotinib sensitivity. K-rn cells were infected by lentivirus coding for a shRNA anti-Syk, grown for 10 days, selected for DsRed expression and purified by cell sorting. Syk silencing was confirmed by Western blotting (A) and apoptosis in response to nilotinib was measured on both K-rn and K-rn ShRNA Syk cells by cytometry using annexin V-FITC (B). Results are expressed as the percentage of annexin V positive cells.
Silencing Syk restores nilotinib sensitivity. K-rn cells were infected by lentivirus coding for a shRNA anti-Syk, grown for 10 days, selected for DsRed expression and purified by cell sorting. Syk silencing was confirmed by Western blotting (A) and apoptosis in response to nilotinib was measured on both K-rn and K-rn ShRNA Syk cells by cytometry using annexin V-FITC (B). Results are expressed as the percentage of annexin V positive cells.
Coexpression of Lyn and Syk triggers resistance to nilotinib
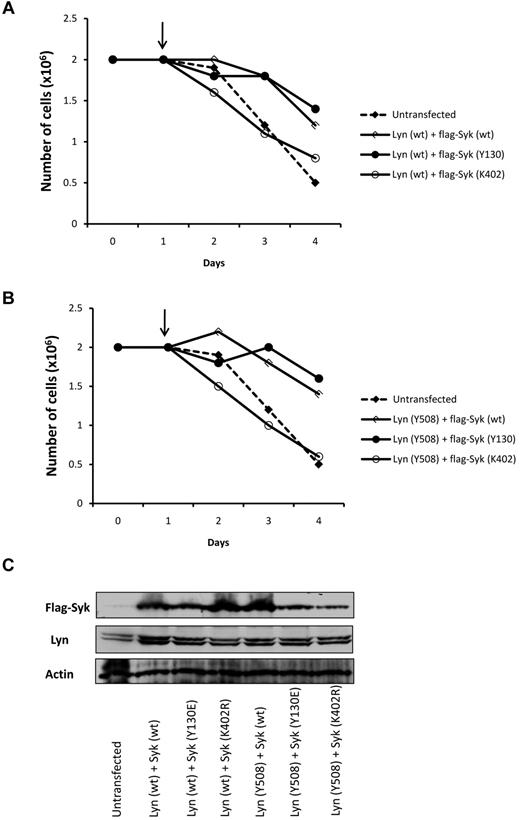
To further confirm the role of Syk and Lyn in nilotinib resistance, Lyn and Syk were coexpressed in Ks cells. As described, a ratio of 4:1 Lyn to Syk was used to mimic the over expression of Lyn observed in K-rn cells. Both the wild-type (wt; Figure 5A) and the constitutively activated form of LynY508F (Figure 5B) were used in transient transfection experiments. Nilotinib induced death of Ks cells (supplemental Figure 5A-B) when cotransfected with Lyn (both wt and Y508E) and a catalytic-inactive form of Syk (SykK402R) confirming that overexpression of Lyn alone is not enough to induce nilotinib resistance. In contrast, the coexpression of Lyn (wt) with Syk (Syk wt) or constitutively activated Syk (Syk-Y130E) induced resistance to nilotinib as shown by cell proliferation (Figure 5) and cell death measurements (supplemental Figure 5A). A similar resistance to nilotinib was observed on coexpression of the constitutively activated form of Lyn (Y508) with Syk wt or Y130E (Figure 5B and supplemental Figure 5B). Interestingly, the expression of Syk alone did not change the sensitivity to nilotinib in Ks cells whereas it increases resistance in K-rn cells (supplemental Figure 5C-D). These results indicate that Syk or Lyn per se are not sufficient to ensure cell resistance to nilotinib and emphasize the interplay between Syk and Lyn in this biologic process.
Coexpression of Lyn and Syk induced nilotinib resistance in Ks cells. Ks cells were transfected with Lyn and Syk constructs with a ratio of 4:1 to mimic the over expression of Lyn in K-rn cells. Both Lyn wild type (Lyn wt panel A) and the constitutively activated Lyn (Lyn Y508 panel B) were used in combination with Syk wild type (wt), constitutively activated (Y130) and kinase dead (K402). On day 1 at the time of transfection nilotinib was added (indicated by the arrow, 20nM) and cell proliferation was quantified over a 4 days time course experiment by triplicate cell counting. Results are from 1 experiment representative of 2. (C) Samples taken at day 4 and analyzed for Lyn and Syk expression.
Coexpression of Lyn and Syk induced nilotinib resistance in Ks cells. Ks cells were transfected with Lyn and Syk constructs with a ratio of 4:1 to mimic the over expression of Lyn in K-rn cells. Both Lyn wild type (Lyn wt panel A) and the constitutively activated Lyn (Lyn Y508 panel B) were used in combination with Syk wild type (wt), constitutively activated (Y130) and kinase dead (K402). On day 1 at the time of transfection nilotinib was added (indicated by the arrow, 20nM) and cell proliferation was quantified over a 4 days time course experiment by triplicate cell counting. Results are from 1 experiment representative of 2. (C) Samples taken at day 4 and analyzed for Lyn and Syk expression.
Syk regulates Lyn, Axl and CDCP-1 phosphorylation
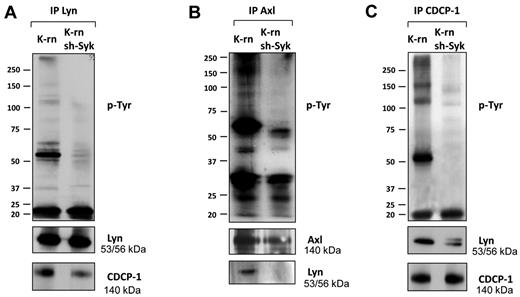
We next searched for Syk dependent phosphorylations involved in the acquisition of cell resistance to nilotinib. We first observed a significant decrease of tyrosine phosphorylation content in K-rn sh-Syk cells (supplemental Figure 6A). Then, we addressed the impact of the absence of Syk on the tyrosine phosphorylation of Lyn, Axl and CDCP-1. The inhibition of Syk expression led to a decreased Lyn phosphorylation without altering its level of expression (Figure 6A and supplemental Figure 6A). CDCP-1 tyrosine phosphorylation was also decreased on Syk depletion (Figure 6C) and was accompanied by a reduction in the association with Lyn (Figure 6A-C). Similarly, Syk knock-down led to a decreased Axl tyrosine phosphorylation and association with Lyn without modification of its expression (Figure 6B and supplemental Figure 6A). These data suggest that Syk regulates tyrosine phosphorylation of Lyn, Axl and CDCP-1 and their interaction.
Inhibition of Syk expression altered phosphorylation and interaction of Lyn, Axl and CDCP-1. Lyn (A), Axl (B) and CDCP-1 (C) were immunoprecipitated from K-rn and K-rn sh-Syk cells. After transfer, membranes were probed for tyrosine phosphorylated proteins using a mix of 4G10 and pY-100 antibodies (top panels). After stripping, membranes were cut and reprobed as indicated. Results are from 1 experiment representative of at least 3.
Inhibition of Syk expression altered phosphorylation and interaction of Lyn, Axl and CDCP-1. Lyn (A), Axl (B) and CDCP-1 (C) were immunoprecipitated from K-rn and K-rn sh-Syk cells. After transfer, membranes were probed for tyrosine phosphorylated proteins using a mix of 4G10 and pY-100 antibodies (top panels). After stripping, membranes were cut and reprobed as indicated. Results are from 1 experiment representative of at least 3.
Axl and CDCP-1 are up-regulated and involved in nilotinib resistance in vitro and in vivo
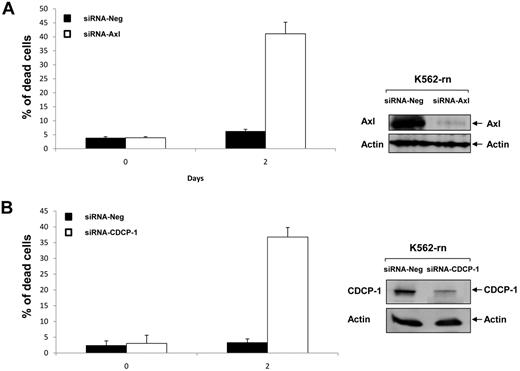
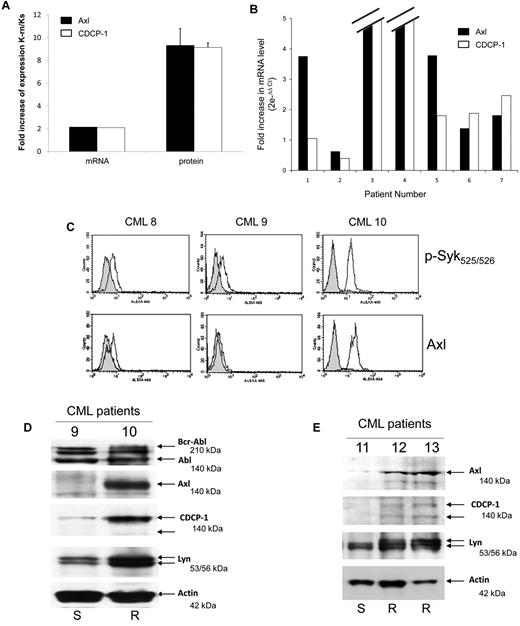
Finally, we investigated the role of Axl and CDCP-1 in cell resistance to nilotinib. Axl or CDCP-1 expression was inhibited by specific siRNA in K-rn cells. The efficiency of these siRNA was shown by a large decreased protein level at day 2 (Figure 7A-B). Both the inhibition of Axl or CDCP-1 expression induced a partial recovery of cell sensitivity to nilotinib (∼ 40% dead cells after 2 days of nilotinib treatment). This data suggests that Axl and CDCP1 may be important mediators of the Syk/Lyn signaling process leading to cell resistance to nilotinib. Finally, we wished to confirm the involvement of this signaling process in vivo. Firstly, we observed that both CDCP-1 and Axl transcripts were increased by 2-fold and respective protein levels by 8- to 9-fold in K-rn cells (Figure 8A). We searched for the existence of similar molecular events in nilotinib-resistant cells from CML patients overexpressing Lyn.8 Messenger RNAs of Axl and CDCP-1 were increased from 2 to 180-fold in 5 nilotinib-resistant CML cells (Figure 8B).
siRNA targeting Axl or CDCP-1 decrease nilotinib resistance. K-rn cells were transfected with 200nM of siRNA corresponding to a negative control siRNA (siRNA Neg) or targeted siRNA; siRNA Axl (A) or siRNA-CDCP-1 (B). Cells were seeded at 2 × 105/mL in RF-10 for 24 hours after incubated in the presence of 20nM nilotinib. Aliquots of the culture were harvested at day 2 for measurement of cell death by triplicate trypan blue exclusion counts and protein expression analysis. Results are from one experiment representative of 2.
siRNA targeting Axl or CDCP-1 decrease nilotinib resistance. K-rn cells were transfected with 200nM of siRNA corresponding to a negative control siRNA (siRNA Neg) or targeted siRNA; siRNA Axl (A) or siRNA-CDCP-1 (B). Cells were seeded at 2 × 105/mL in RF-10 for 24 hours after incubated in the presence of 20nM nilotinib. Aliquots of the culture were harvested at day 2 for measurement of cell death by triplicate trypan blue exclusion counts and protein expression analysis. Results are from one experiment representative of 2.
Axl and CDCP1 are over expressed in nilotinib resistant CML cells. (A) Total mRNA (1 μg) from Ks and K-rn cells were reverse transcribed into cDNA and used in Q-PCR to quantify the mRNA expression of Axl (black box) and CDCP-1 (white box) between Ks and K-rn cells. In parallel, the level of protein expression was quantified by densitometry analysis of 5 separate Western-blots. Results are expressed as a fold increase expression for the mRNA by the ΔΔ Ct method and the protein by the ratio of K-rn/Ks. (B) Seven CML patients (2 males, 5 females) were investigated as previously described.8 All patients were analyzed at the beginning or before nilotinib treatment and under treatment at the moment of nilotinib failure in chronic phase (number 6 and 7) or in accelerated phase (number 1 to 5). Quantitative Real-time PCR to amplify Axl (black box) and CDCP-1 (white box) transcripts was carried out on the cDNA. Analysis was done by comparison of the 2 time points (before nilotinib and under treatment at the moment of failure) by the comparative ΔΔCt method giving the fold increase of the amount of target normalized to the endogenous reference as 2- ΔΔCt. (C) Purified CD34 positive cells from blood samples of CML patients (8 and 9 responding to nilotinib, 10 resistant to nilotinib) were analyzed by flow cytometry as described in “Methods” for Syk phosphorylation (phosphospecific antibody p-Syk 525/526) and Axl expression. Control using non immune IgG (gray histogram) and specific antibody (white histogram) are shown. (D) Bcr-Abl, Axl, CDCP-1 and Lyn were detected by Western-blotting using protein samples from CML 9 and 10 patients. (E) CD34 positive cells from nilotinib-responding (11: S) or resistant (12 and 13: R) CML patients were isolated and solubilized in Laemli buffer. Proteins were separated by SDS-PAGE and specific proteins were detected by Western-blotting as indicated. Actin was used as a loading control.
Axl and CDCP1 are over expressed in nilotinib resistant CML cells. (A) Total mRNA (1 μg) from Ks and K-rn cells were reverse transcribed into cDNA and used in Q-PCR to quantify the mRNA expression of Axl (black box) and CDCP-1 (white box) between Ks and K-rn cells. In parallel, the level of protein expression was quantified by densitometry analysis of 5 separate Western-blots. Results are expressed as a fold increase expression for the mRNA by the ΔΔ Ct method and the protein by the ratio of K-rn/Ks. (B) Seven CML patients (2 males, 5 females) were investigated as previously described.8 All patients were analyzed at the beginning or before nilotinib treatment and under treatment at the moment of nilotinib failure in chronic phase (number 6 and 7) or in accelerated phase (number 1 to 5). Quantitative Real-time PCR to amplify Axl (black box) and CDCP-1 (white box) transcripts was carried out on the cDNA. Analysis was done by comparison of the 2 time points (before nilotinib and under treatment at the moment of failure) by the comparative ΔΔCt method giving the fold increase of the amount of target normalized to the endogenous reference as 2- ΔΔCt. (C) Purified CD34 positive cells from blood samples of CML patients (8 and 9 responding to nilotinib, 10 resistant to nilotinib) were analyzed by flow cytometry as described in “Methods” for Syk phosphorylation (phosphospecific antibody p-Syk 525/526) and Axl expression. Control using non immune IgG (gray histogram) and specific antibody (white histogram) are shown. (D) Bcr-Abl, Axl, CDCP-1 and Lyn were detected by Western-blotting using protein samples from CML 9 and 10 patients. (E) CD34 positive cells from nilotinib-responding (11: S) or resistant (12 and 13: R) CML patients were isolated and solubilized in Laemli buffer. Proteins were separated by SDS-PAGE and specific proteins were detected by Western-blotting as indicated. Actin was used as a loading control.
To confirm these results, both tyrosine Y525/526 phosphorylation of Syk and expression of Axl were investigated in CD34 positive cells isolated from CML patient's blood samples. Detection of Syk phosphorylation and Axl expression by flow cytometry has been previously validated using Ks and K-rn cells but also pervanadate-treated CD34+ cells (supplemental Figure 7A-B). As shown in Figure 8C and D, 2 patients responding to nilotinib (CML 8 and 9) share basal phosphophorylation of Syk and expression of Axl. In contrast, patient CML 10 who is resistant to nilotinib has a high tyrosine Y525/526 phosphorylation of Syk and Axl expression.
Discussion
The present work was performed to investigate the tyrosine kinase signaling involved in nilotinib resistance using a resistant Bcr-Abl positive cell line, K-rn, overexpressing the tyrosine kinase Lyn.8 Because the overexpression of Lyn has already been demonstrated both in imatinib and nilotinib resistant CML cells, we identified and quantified by SILAC the tyrosine phosphorylated proteins in the nilotinib-resistant cell line in comparison to the sensitive counterpart. Among the 350 proteins identified and quantified, we focused on the role of 2 tyrosine kinases overexpressed and/or hyperphosphorylated, Syk and Axl. The spleen tyrosine kinase Syk is known to participate in immune receptor signaling, lymphocyte proliferation or to have tumor suppressor roles in melanoma and breast cancer.18-21 It has also been reported overexpressed in acute myeloid leukemias suggesting a potential oncogenic role.22 We detected an increase in Syk phosphorylation in K-rn cells (with no significant difference of expression) which is in agreement to the report of an unidentified 75-kDa phosphoprotein in Lyn overexpressing imatinib resistant cells.23 Indeed, phosphorylation of Syk on tyrosine residues Y323 and Y525/526 in K-rn cells was decreased by PP2, a Src kinase inhibitor and was fully abolished by dasatinib, confirming that a PP2 and dasatinib sensitive tyrosine kinase is involved in Syk phosphorylation.
The role of Syk in nilotinib resistance was confirmed both by the inhibition of Syk kinase activity using R406 (or BAY61-3606) and by inhibition of its own expression by a shRNA. Inhibitors of Syk may be not very specific and could inhibit other kinases involved in cell proliferation. Nevertheless, our results suggest that Syk is also involved. Our results indicate a crosstalk between these 2 kinases. Their coexpression in Ks cells generated resistance to nilotinib confirming that both kinases are required. Lyn and Syk induced resistance is only observed when Syk can be activated as the kinase dead form (Syk K402) did not induce resistance in combination with Lyn. Overall, results show that the mechanisms of resistance to nilotinib use at least a signaling involving a Syk-Lyn pathway.
The second tyrosine kinase we focused on was the UFO receptor Axl, a member of the receptor tyrosine kinase family implicated in GAS6 signaling and originally discovered in myeloid leukemia.24,25 Although Axl expression is increased in K-rn cells, it is similarly detected in Lyn immunoprecipitates from Ks and K-rn cells suggesting that the overexpression of Lyn did not change its interaction with Axl. This interaction could be mediated by the SH2 domains of the p85 subunit of PI3-kinase as several tyrosine residues are phosphorylated in Axl.26 An in vitro interaction has also been reported between Axl and the SH2 domain of Src.27 This is in agreement with the interplay existing between Src kinases and receptor tyrosine kinases suggesting a possible association and a phosphorylation of Axl by Lyn and/or Syk.28 Indeed, Axl detection from Syk immunoprecipitate performed on Lyn immunoprecipitate confirmed that Lyn, Syk and Axl are present in a ternary complex in K-rn cells that is also emphasized by the absence of Lyn in Axl immunoprecipitate from ShRNA Syk cells.
Looking at the role of the identified kinases, a multistep mechanism may be hypothesized with Syk-Lyn as a starter and downstream signaling partners such as Axl and the adaptor CDCP-1. This was confirmed by silencing Axl and CDCP-1 expression. In addition, overexpression of Axl and CDCP-1 has also been shown in vivo. Indeed, a large increase of expression of Lyn, CDCP-1 and Axl was detected in CD34 positive cells of nilotinib-resistant CML patients. However, the initiating event leading to nilotinib resistance through these overexpressions has still to be elucidated. A dysregulated methylation may be proposed as the expression of these proteins is regulated by methylation. Indeed, incubation of K-rn cells with temozolomide, a methylating agent, decrease Axl expression and partially restore sensitivity to nilotinib (R.G., unpublished results). On the other side, the overexpression of Lyn has been associated with a decreased expression of c-Cbl suggesting it may be taken into account for the overexpression of tyrosine kinase such as Lyn or Axl.23 This down-regulation of c-Cbl could also contribute to the hyperphosphorylation of Syk.29,30 These overexpressions could be linked to c-Cbl mutation itself although no c-Cbl mutation has been detected in K-rn cells (R.G., unpublished results).31 All these deregulated mechanisms may contribute to the difference observed between the mRNA and protein level of Axl and CDP-1 measured in CML patient samples. This difference may also be related to a Lyn-dependent regulation of miRNA such as miRNA 181 associated with a Mcl-1 up-regulation leading to imatinib resistance.32 The characterization of miRNA regulated by Lyn overexpression in K-rn cells will provided information about their potential roles in the overexpression of Axl and CDCP-1.
Even if the Syk-Lyn dual kinase complex controls resistance, downstream elements such as Axl or CDCP-1 are playing critical roles. Inhibition of Axl or CDCP-1 expression confirmed their participation in nilotinib resistance. How Axl is involved in resistance is not yet known. It could be through an inhibitory effect on the NFkB signaling.33 On other hand, the role of CDCP-1 in the Syk-Lyn pathway is suspected to be required for providing a multi-substrate docking-site and thus preventing nilotinib-induced cell death through a PKCδ pathway.34 This role could also be played by Axl itself.27 Axl being already a drug target in other cancers, it could be a pertinent kinase to target in resistant CML and this should be confirmed.35,36 Moreover, this will give a better understanding of how resistance in cancer may be overcome.
The role of the Syk-Lyn kinase complex with the participation of CDCP-1 and Axl is emphasized by the detection of such mechanism in CD34 positive cells from nilotinib resistant CML patients. Moreover, nilotinib resistance mediated by Lyn overexpression and hyperactivation may be overcome by Lyn expression silencing or tyrosine kinase activity inhibition confirming previous reports.37,38 It should be worthwhile to investigate the expression and activation of the newly identified candidates in Lyn-mediated imatinib resistance as nilotinib resistance has always been associated with an increased expression and/or activation of Syk, Axl and CDCP-1. Although this resistance mechanism is not so frequent in CML, this study demonstrates it has occurred in vitro and in vivo. To the authors' knowledge, this is the first time that Syk and Axl kinases have been reported to play a role in CML and in nilotinib resistance. Although dual tyrosine kinase inhibitors like dasatinib or ponatinib are able to overcome Lyn-mediated resistance, one would take advantage that Syk and Axl inhibitors have been recently developed. It could be worthwhile to validate their targeting in this kind of resistance by preclinical studies to increase the panel of therapeutic tools. Because CML cells can evade the inhibitory effect of tyrosine kinase inhibitors by recruiting downstream kinases, they could be the next targets for a combined therapy of CML disease.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Elisabeth Buchdunger and Paul Manley (Novartis Pharma) for providing us nilotinib. This work was supported by la Ligue Nationale Contre le Cancer (LNCC) Comité de la Dordogne et des Landes, l'INCA, l'Association pour la Recherche sur le Cancer (ARC), la Fondation de France, l'Association Laurette Fugain, la Cancéropôle du Grand Sud-Ouest, l'Université Victor Segalen Bordeaux 2 and Inserm.
R.G. has a fellowship from INCA and ARC, and C.D. has a fellowship from LNCC. The Syk inhibitor R406 is provided by RIGEL Inc, and is controlled by material transfer agreement. Syk constructs were kindly provided by Dr P. Coopman (CNRS 5237, Montpellier, France).
Authorship
Contribution: R.G., C.L., C.D., and V.L. performed research and analyzed data; G.E., S.D., and E.L. analyzed data from CML patients; S.R. and F.-X.M. designed the research, analyzed data and wrote the paper; and J.-M.P. designed and performed the research, analyzed data and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jean-Max Pasquet, Hématopoïèse Leucémique et Cibles Thérapeutiques, Inserm U1035, Université Victor Segalen, 146 rue Léo Saignat Bat TP 4e étage, 33076 Bordeaux cedex, France; e-mail: jean-max.pasquet@u-bordeaux2.fr.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal