Abstract
Children with Down syndrome (DS) up to the age of 4 years are at a 150-fold excess risk of developing myeloid leukemia (ML-DS). Approximately 4%-5% of newborns with DS develop transient myeloproliferative disorder (TMD). Blast cell structure and immunophenotype are similar in TMD and ML-DS. A mutation in the hematopoietic transcription factor GATA1 is present in almost all cases. Here, we show that simple techniques detect GATA1 mutations in the largest series of TMD (n = 134; 88%) and ML-DS (n = 103; 85%) cases tested. Furthermore, no significant difference in the mutational spectrum between the 2 disorders was seen. Thus, the type of GATA1 sequence mutation is not a reliable tool and is not prognostic of which patients with TMD are probable to develop ML-DS.
Introduction
Children with trisomy 21 (T21; Down syndrome [DS]) have ∼ 150-fold increased incidence of myeloid leukemia (ML-DS).1,2 Incidence of transient myeloproliferative disorder (TMD) is estimated at ∼ 4%-5% of neonates with DS.1,3 Approximately 20%-30% of these neonates develop ML-DS by 4 years of age.4,5 Both diseases are characterized by a clonal population of blasts, with similar immunophenotype and structure in blood and BM. However, TMD spontaneously regresses, whereas ML-DS is stably transformed.
In addition to T21, blast cells in TMD and ML-DS carry acquired mutations in the hematopoietic transcription factor GATA1.6-14 These mutations lead to expression of N-terminally truncated GATA1s protein. Mutations are detectable in disease but not in remission.3,6-14 Most reported mutations are found in GATA1 exon 2, including insertions, deletions, and point mutations. When TMD progresses to ML-DS, the same GATA1 mutation is usually present in blasts of both, showing their clonal relationship.5,14
Debate exists about whether the type of GATA1 mutation determines progression to ML-DS.9,15-17 To examine this, we analyzed GATA1 mutations in 134 TMD and 103 ML-DS cases, the largest patient cohort reported to date. Of these 8 paired TMD and follow-up ML-DS samples were available. GATA1 mutations were detected in 226 patients (95%). The lower limit blast percentage for successful detection of GATA1 mutations was 0.5%. No difference was observed in types of mutation between patients with TMD and with ML-DS. Contrary to previous data,15 we did not detect specific GATA1 mutation types more commonly in ML-DS. Therefore, the type of GATA1 mutation in our series is not prognostic of which patients with TMD will progress to ML-DS.
Methods
Mutation detection DNA was prepared from peripheral blood or BM with the use of the DNeasy Blood and Tissue kit (QIAGEN). PCR was performed with primers and conditions outlined in supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). PCR amplicons were analyzed by denaturing high-performance liquid chromatography (WAVE; Transgenomic) and direct sequencing. In sample subsets with blast percentage < 1%, blasts were sorted before DNA extraction (supplemental Methods), or PCR product was cloned with the pGEM-T-Easy vector system 1 kit (Promega) and sequenced.
Statistical analysis was performed with the Fisher exact test.
Results and discussion
GATA1 mutation screening was performed in the central reference of Acute Myeloid Leukemia Berlin-Frankfurt-Münster Study group in Hannover, Germany, and the Weatherall Institute of Molecular Medicine, Oxford, United Kingdom (134 TMD and 103 ML-DS samples). The mean age of patients with TMD was 0.78 months (range, 0-11 months). Mean blast count was 42% (range, 0.5%-95%), white blood cell count was 60.79 × 109/L (range, 1-1193.3 × 109/L), hemoglobin level was 13.77 g/dL (range, 2-21.3 g/dL), and platelet count was 201.78 × 109/L (range, 12-1800 × 109/L; Table 1). The mean age of the patients with DS-ML was 20.19 months (range, 1-60 months). Mean blast count was 27.8% (range, 1%-96%), white blood cell count was 13.95 × 109/L (range, 1-200.3 × 109/L), hemoglobin level was 9.06 g/dL (range, 3.1-15.1 g/dL) and platelet count was 52.2 × 109/L (range, 1.5-257 × 109/L; Table 2).
Summary of GATA1 mutations and patient characteristics for TMD samples
| Patient no. . | Blast count, % of total nucleated count . | WAVE* . | Mutation type . | No. of nucleotides changed, bp . | Position of mutation† . | Predicted effect of mutation on RNA . | Age at diagnosis, mo . | WBC count, ×109/L . | Hb level, g/dL . | Plt count, ×109/L . | Kanezaki et al15 class . | Karyotype . | Outcome . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 79 | Ex2 | Point | 1 | 4549 | Loss of start codon | 0 | 107 | 15 | 140 | 1st Met | 47, XY, +21c | CCR |
| 2‡ | 66 | Ex2 | Dup | 20 | 4722 | Frameshift and introduction of stop codon | 2 | 70 | 13.3 | 352 | PTC 1-3′ | 47, XX, +21c | ML-DS |
| 3 | 40 | Ex2 | Point | 1 | 4767 | Loss of splice acceptor site | 1.3 | 14 | 12.7 | 461 | Splice | 47, XY, +21c | CCR |
| 4 | 35 | Ex2 | Ins | 8 | 4701 | Frameshift and introduction of stop codon | 2 | 16 | 21.3 | 41 | PTC 1-5′ | 47, XY, +21c | CCR |
| 5 | 30 | Ex2 | Dup | 101 | 4653 | Frameshift and introduction of stop codon | 0.57 | 5 | 8.2 | 122 | PTC 1-5′ | 47, XY, +21c | CCR |
| 6 | 55 | Ex2 | Dup | 19 | 4733 | Frameshift and introduction of stop codon | 0.57 | 75.9 | 6.3 | 113 | PTC 1-3′ | 47, XX, +21c | CCR |
| 7 | 29 | Ex2 | Del | 2 | 4638 | Frameshift and introduction of stop codon | 0.28 | 44.6 | 17.4 | 45 | PTC 1-5′ | 47, XX, +21c | CCR |
| 8 | 92 | Ex2 | Dup | 2 | 4768 | Loss of splice acceptor site | 0.28 | 120 | 18.8 | 505 | Splice | N/A | CCR |
| 9‡ | 40 | Ex2 | Point | 1 | 4583 | Nonsynonymous change in amino acid sequence introducing a stop codon | 1 | 50 | N/A | N/A | PTC 1-5′ | N/A | ML-DS |
| 10 | 80 | Ex2 | Dup | 22 | 4735 | Frameshift and introduction of stop codon | 0 | 121 | 11.6 | 35 | PTC 1-3′ | 47, XY, +21 | Thrombo-cytopaenia |
| 11 | 11 | Ex2 | Dup | 17 | 4719 | Frameshift and introduction of stop codon | 0 | 38.2 | 18.7 | 132 | PTC 1-3′ | N/A | CCR |
| 12 | 21 | Ex2 | Point | 1 | 4768 | Loss of splice acceptor site | 2 | 19.4 | 10.8 | 52 | Splice | N/A | Unknown |
| 13 | 2 | Ex2 | Del | 1 | 4607 | Frameshift and introduction of stop codon | 0.71 | 9.5 | 12.7 | 54 | PTC 1-5′ | N/A | Unknown |
| 14 | 60 | Ex2 | Ins | 16 | 4721 | Frameshift and introduction of stop codon | 0.57 | 6.7 | 16.3 | 59 | PTC 1-3′ | N/A | Died |
| 15 | 5 | Ex2 | Dup | 22 | 4723 | Frameshift and introduction of stop codon | 0.71 | 12 | 14 | 88 | PTC 1-3′ | N/A | Unknown |
| 16 | 35 | Ex2 | Del | 26 | 4688 | Frameshift and introduction of stop codon | 2 | 39.3 | 13.8 | 335 | PTC 1-5′ | N/A | CCR |
| 17 | 82 | Ex2 | Del | 2 | 4458 | Frameshift and introduction of stop codon | 1.1 | 249 | 11.8 | 122 | PTC 1-5′ | 47, XY, +21c | CCR |
| 18 | 75 | Ex2 | Ins | 16 | 4706 | Frameshift and introduction of stop codon | N/A | 31.6 | 16.2 | 44 | PTC 1-5′ | N/A | CCR |
| 19 | 30 | Ex2 | Point | 1 | 4550 | Loss of start codon | 4 | 11.5 | 13.3 | 72 | 1st Met | 47, XY, +21c | CCR |
| 20 | N/A | Ex2 | Point | 1 | 4596 | Nonsynonymous change in amino acid sequence introducing a stop codon | N/A | N/A | N/A | N/A | PTC 1-5′ | N/A | Died |
| 21 | N/A | Ex2 | Point | 1 | 4766 | Loss of splice acceptor site | 1 | N/A | N/A | N/A | Splice | N/A | Unknown |
| 22 | 25 | Ex2 | Ins | 8 | 4709 | Frameshift and introduction of stop codon | 1 | 33.7 | 14.8 | 35 | PTC 1-5′ | N/A | Unknown |
| 23 | 45 | Ex2 | Substitution | 1 | 4744 | Nonsynonymous change in amino acid sequence introducing a stop codon | 2.7 | 20 | 2 | 51 | PTC 1-3′ | 47, XY, der(11)t(q23p15) t(1:11)(q23;q85), +21c | CCR |
| 24 | 50 | Ex2 | Del | 132 | 4671 | Loss of splice acceptor site | 0 | 58 | N/A | N/A | Splice | 47, XY, +21c | CCR |
| 25 | 76 | Ex2 | Ins | 1 | 4661 | Frameshift and introduction of stop codon | 2 | 124.9 | 9 | 28 | PTC 1-5′ | 47, XY, +21c | CCR |
| 26 | 25 | Ex2 | Del | 1 | 4662 | Frameshift and introduction of stop codon | 2 | 39.6 | 19.8 | 22 | PTC 1-5′ | 47, XY, +21c | CCR |
| 27‡ | 18 | Ex2 | Dup | 8 | 4717 | Frameshift and introduction of stop codon | 0 | 101 | 13.3 | 21 | PTC 1-5′ | 47, XY, t(5;11)(p15;q13), +21c | ML-DS |
| 28 | 70 | Ex2 | Del | 4 | 4684 | Frameshift and introduction of stop codon | 4 | 200 | 7.6 | 87 | PTC 1-5′ | N/A | CCR |
| 29 | 62 | Ex2 | Ins | 4 | 4643 | Frameshift and introduction of stop codon | 1 | N/A | N/A | N/A | PTC 1-5′ | N/A | Unknown |
| 30 | 11.4 | Ex2 | Dup | 8 | 4732 | Frameshift and introduction of stop codon | 0 | 33.5 | 9.1 | 695 | PTC 1-3′ | 47, XY, +21c | Unknown |
| 31 | 29 | Ex2 | Point | 1 | 4670 | Nonsynonymous change in amino acid sequence introducing a stop codon | 2 | 21.7 | 13 | 24 | PTC 1-5′ | N/A | Unknown |
| 32 | 60 | Ex2 | Del | 1 | 4697 | Frameshift and introduction of stop codon | 28 weeks gestation | 40 | 5.1 | 54 | PTC 1-5′ | N/A | Unknown |
| 33 | 79 | N/D | Del | 2 | 4638 | Frameshift and introduction of stop codon | 0 | 198.2 | 10.6 | 312 | PTC 1-5′ | N/A | Unknown |
| 34 | 66 | N/D | Dup | 40 | 4740 | Frameshift and introduction of stop codon | 0.25 | 9.8 | 12.7 | 60 | PTC 1-3′ | N/A | Unknown |
| 35 | 60 | N/D | Dup | 10 | 4710 | Frameshift and introduction of stop codon | 0.25 | 46.6 | 20.4 | 89 | PTC 1-5′ | N/A | Unknown |
| 36‡ | 35.5 | N/D | Ins | 14 | 4707 | Frameshift and introduction of stop codon | 0 | 55.6 | 17.3 | 570 | PTC 1-5′ | N/A | ML-DS |
| 37 | 54 | N/D | Point | 1 | 4768 | Loss of splice acceptor site | 0 | 55.6 | 17.3 | 570 | Splice | N/A | Unknown |
| 38 | 23 | N/D | Point | 1 | 4767 | Synonymous mutation | 1.25 | 19.2 | 6.7 | 125 | Unknown | N/A | Unknown |
| 39‡ | 84 | N/D | Del | 2 | 4604 | Frameshift and introduction of stop codon | 0 | 1193.3 | 14.7 | 255 | PTC 1-5′ | N/A | ML-DS |
| 40 | 10 | N/D | Del + Ins | 2 + 1 | 4770 + 4774 | Loss of splice donor site | 2 | 9.1 | 9.6 | 77 | Splice | N/A | Unknown |
| 41‡ | 88 | N/D | Del | 3 | 4680 | Frameshift and introduction of stop codon | 0 | 232 | 15.6 | 254 | PTC 1-5′ | N/A | ML-DS |
| 42 | 75 | N/D | Del + Ins | 1 + 2 | 4679 | Frameshift and introduction of stop codon | 0 | 172.8 | 16.2 | 101 | PTC 1-5′ | N/A | Unknown |
| 43 | 14 | N/D | Point | 1 | 4734 | Unknown | 0.25 | 16.9 | 14.9 | 353 | Unknown | N/A | Unknown |
| 44 | 13.5 | N/D | Dup | 36 | 4737 | Frameshift and introduction of stop codon | 0.5 | 3.2 | 11.1 | 16 | PTC 1-3′ | N/A | Unknown |
| 45 | 31 | N/D | Dup | 19 | 4722 | Frameshift and introduction of stop codon | 0 | 28.6 | 16.2 | 57 | PTC 1-3′ | N/A | Unknown |
| 46 | 43 | N/D | Ins + Dup | 2 + 13 | 4723 | Frameshift and introduction of stop codon | 0 | 61 | 17.6 | 149 | PTC 1-3′ | N/A | Unknown |
| 47 | 69 | N/D | Point | 1 | 4550 | Loss of start codon | 0 | 34 | 15 | 26 | 1st Met | N/A | Unknown |
| 48 | 10 | N/D | Dup | 21 | 4723 | Frameshift and introduction of stop codon | 0.25 | 7.7 | 13.2 | 78 | PTC 1-3′ | N/A | Unknown |
| 49 | 78 | N/D | Del + Ins | 19 + 17 | 4698 | Frameshift and introduction of stop codon | 0 | 68.6 | 10.9 | 212 | PTC 1-5′ | N/A | Unknown |
| 50 | 34 | N/D | Ins | 7 | 4713 | Frameshift and introduction of stop codon | 0.25 | 6.2 | 16.9 | 25 | PTC 1-5′ | N/A | Unknown |
| 51 | 60 | N/D | Del | 2 | 4638 | Frameshift and introduction of stop codon | 0 | 51.2 | 14.8 | 159 | PTC 1-5′ | N/A | Unknown |
| 52 | 75 | N/D | Point | 1 | 4747 | Nonsynonymous change in amino acid sequence introducing a stop codon | 0 | N/A | N/A | N/A | PTC 1-3′ | N/A | Unknown |
| 53 | 18.5 | N/D | Del | 162 | 4540 | Loss of start codon | 0 | 17.3 | 14 | 173 | 1st Met | N/A | Unknown |
| 54 | 1 | N/D | Del + Ins | 2 + 3 | 4698 | Frameshift and introduction of sop codon | 0.25 | 57.5 | 10.6 | 36 | PTC 1-5′ | N/A | Unknown |
| 55 | 8 | N/D | Ins + Dup | 3 + 36 | 4724 | Frameshift and introduction of stop codon | 0.25 | 16 | 18.5 | 48 | PTC 1-3′ | N/A | Unknown |
| 56 | 66.5 | N/D | Del | 2 | 4638 | Frameshift and introduction of stop codon | 0.75 | 62.9 | 12.3 | 93 | PTC 1-5′ | N/A | Unknown |
| 57‡ | 75 | N/D | Dup | 13 | 4744 | Frameshift and introduction of stop codon | 0 | 149.1 | 16.9 | 131 | PTC 1-3′ | 47, XY,+21.ish 21qter(D21S1446 × 3) | ML-DS |
| 58 | 85 | N/D | Point | 1 | 4762 | Nonsynonymous change in amino acid sequence introducing a stop codon | 0 | 46.4 | 20.6 | 197 | PTC 1-3′ | N/A | Unknown |
| 59 | 45 | N/D | Del + Ins | 7 + 10 | 4760 | Frameshift and introduction of stop codon | 0.5 | 78.3 | 15 | 1800 | PTC 1-3′ | N/A | Unknown |
| 60 | 38 | N/D | Ins + Dup | 3 + 9 | 4740 | Frameshift and introduction of stop codon | 0.25 | 32.9 | 11.9 | 220 | PTC 1-3′ | N/A | Unknown |
| 61‡ | 64.5 | N/D | Dup | 18 | 4733 | Frameshift and introduction of stop codon | 0 | 69.4 | 15.4 | 1178 | PTC 1-3′ | N/A | ML-DS |
| 62 | 58 | N/D | Del | 1 | 4698 | Frameshift and introduction of stop codon | 0.25 | 24.9 | 15.6 | 154 | PTC 1-5′ | N/A | Unknown |
| 63 | 34 | N/D | Del | 1 | 4690 | Frameshift and introduction of stop codon | 0 | 27.2 | 17 | 134 | PTC 1-5′ | N/A | Unknown |
| 64 | 85 | N/D | Del | 2 | 4604 | Frameshift and introduction of stop codon | 0 | 22 | 16.3 | 108 | PTC 1-5′ | N/A | Unknown |
| 65 | 11 | N/D | Point | 1 | 4549 | Loss of s | 1 | 7.4 | 12.3 | 241 | 1st Met | N/A | Unknown |
| 66 | 32 | N/D | Ins | 1 | 4654 | Frameshift and introduction of stop codon | 0 | 31.6 | 16.4 | 158 | PTC 1-5′ | N/A | Unknown |
| 67 | 3.5 | N/D | Point | 1 | 4597 | Nonsynonymous change in amino acid sequence | 2 | 8.4 | 9.3 | 105 | Unknown | N/A | Unknown |
| 68 | 22 | N/D | Del | 34 | 4693 | Frameshift and introduction of stop codon | 0.25 | 17.3 | 13.1 | 270 | PTC 1-5′ | N/A | Unknown |
| 69‡ | 87 | N/D | Point | 1 | 4552 | Nonsynonymous change in amino acid sequence | 0.25 | 133.9 | 13.8 | 32.9 | Unknown | N/A | ML-DS |
| 70 | 5.5 | N/D | Point | 1 | 4768 | Loss of splice acceptor site | 0 | 23.83 | 9.9 | 154 | Splice | N/A | Unknown |
| 71 | 32 | N/D | Ins + Dup | 1 + 12 | 4742 | Frameshift and introduction of stop codon | 0 | 27.6 | 14.6 | 46 | PTC 1-3′ | N/A | Unknown |
| 72 | 91 | N/D | Ins | 1 | 4647 | Frameshift and introduction of stop codon | 0 | 10.6 | 10.7 | 294 | PTC 1-5′ | N/A | Unknown |
| 73 | 16.5 | N/D | Del | 1 | 4653 | Frameshift and introduction of stop codon | 0.75 | 8.9 | 12.5 | 120 | PTC 1-5′ | N/A | Unknown |
| 74 | 14 | N/D | Dup | 19 | 4722 | Frameshift and introduction of stop codon | 11 | 3.39 | 10.5 | 91 | PTC 1-3′ | N/A | Unknown |
| 75 | 78 | N/D | Del | 1 | 4766 | Loss of splice acceptor site | 0 | 91.2 | 13.6 | 33 | Splice | 47, XY, t(21;21) (Q10;Q10) +21c | Unknown |
| 76 | 60.5 | N/D | Point | 1 | 4734 | Nonsynonymous change in amino acid sequence | 0 | 25.4 | 13.9 | 98 | Unknown | N/A | Unknown |
| 77 | 16 | N/D | Dup | 2 | 4734 | Frameshift and introduction of stop codon | 0.5 | 8.5 | 10.8 | 52 | PTC 1-3′ | N/A | Unknown |
| 78 | 12 | N/D | Del | 51 | 4706 | Frameshift and introduction of stop codon | 0 | 23.2 | 14.2 | 379 | PTC 1-5′ | N/A | Unknown |
| 79 | 95 | N/D | Ins + Dup | 6 + 7 | 4708 | Frameshift and introduction of stop codon | 0 | 410 | 11.1 | 261 | PTC 1-5′ | N/A | Unknown |
| 80 | 86 | N/D | Point | 1 | 4633 | Nonsynonymous change in amino acid sequence | 0 | 107.5 | 13.1 | 70 | Unknown | 47,XX,der(6)del(6) (p21)del(6)(q24, +der(6)del(6)(p12p22)t (6;10)(q16;q22),der(10) t(10;12)(q21;p12), der(11)t(11;12) (p15;q11),−12,+21c[17], 47,XX,+21c[2] | Unknown |
| 81‡ | 68 | N/D | Point | 1 | 4769 | Loss of splice acceptor site | 0 | 100 | 11.5 | 326 | Splice | N/A | ML-DS |
| 82 | 72 | N/D | Del | 12 | 4733 | Frameshift and introduction of stop codon | 0.5 | 49.8 | 14.2 | 43 | PTC 1-3′ | N/A | Unknown |
| 83 | 30 | N/D | Del | 4 | 4741 | Frameshift and introduction of top codon | 0 | 52 | 13.9 | 71 | PTC 1-3′ | N/A | Unknown |
| 84 | 35 | N/D | Ins | 1 | 4654 | Frameshift and introduction of stop codon | 0 | 28 | 12.8 | 274 | PTC 1-5′ | N/A | Unknown |
| 85 | 78 | N/D | Substitution | 40 | 4545 | Unknown | 0 | 72.7 | 19.4 | 606 | Unknown | N/A | Unknown |
| 86 | 79 | N/D | Ins | 1 | 4736 | Frameshift and introduction of stop codon | 0 | 100 | 10.9 | 70 | PTC 1-3′ | N/A | Unknown |
| 87 | 46.5 | N/D | Del | 2 | 4637 | Frameshift and introduction of stop codon | 0.25 | 23.3 | 17.5 | 80 | PTC 1-5′ | N/A | Unknown |
| 88‡ | 43 | N/D | Del | 2 | 4638 | Frameshift and introduction of stop codon | 0.25 | N/A | N/A | N/A | PTC 1-5′ | 47,XY,+21c[15] | ML-DS |
| 89 | 15 | N/D | Ins | 2 | 4725 | Frameshift and introduction of stop codon | 0.5 | 11 | 24.2 | 158 | PTC 1-3′ | N/A | Unknown |
| 90 | 16.5 | N/D | Del | 2 | 4604 | Frameshift and introduction of stop codon | 0.25 | 24.2 | 13.2 | 78 | PTC 1-5′ | N/A | Unknown |
| 91‡ | 66 | N/D | Dup | 5 | 4723 | Frameshift and introduction of stop codon | 0 | 26.1 | 16.5 | 48 | PTC 1-3′ | 47,XX,+21c | ML-DS |
| 92 | 38.5 | N/D | Dup | 43 | 4742 | Frameshift and introduction of stop codon | 0.5 | 20.2 | 12.3 | 12 | PTC 1-3′ | N/A | Unknown |
| 93 | 58 | N/D | Point | 1 | 4768 | Loss of splice acceptor site | 0 | 20.7 | 14.1 | 901 | Splice | N/A | Unknown |
| 94 | 1 | N/D | Point + Point | 1 + 1 | 92 + 126 | Unknown | 0.25 | 5.6 | 14.3 | 67 | Unknown | N/A | Unknown |
| 95 | 83 | N/D | Del + Del | 15 + 1 | 4888 + 4907 | Frameshift and introduction of stop codon | 0.25 | 114.7 | 16.6 | 141 | PTC 2 | N/A | Unknown |
| 96 | 20 | N/D | Dup | 6 | 4714 | Frameshift and introduction of stop codon | 0 | 14.12 | 18.6 | 15 | PTC 1-5′ | N/A | Unknown |
| 97 | 43 | N/D | Point | 1 | 4769 | Loss of splice acceptor site | 0 | 49.8 | 17 | 209 | Splice | N/A | Unknown |
| 98 | 49.9 | N/D | Del | 2 | 4638 | Frameshift and introduction of stop codon | 0.5 | 7.06 | 12 | 64 | PTC 1-5′ | N/A | Unknown |
| 99 | 61 | N/D | Dup | 12 | 4720 | Frameshift and introduction of stop codon | 0.75 | 91.6 | 16.6 | 150 | PTC 1-3′ | N/A | Unknown |
| 100 | 53.5 | N/D | Ins | 1 | 4598 | Frameshift and introduction of stop codon | 0 | 51 | 18.3 | 56 | PTC 1-5′ | N/A | Unknown |
| 101 | 34 | N/D | Del | 13 | 4706 | Frameshift and introduction of stop codon | 0 | 15.4 | 17.1 | 62 | PTC 1-5′ | N/A | Unknown |
| 102 | 25 | N/D | Ins | 1 | 4753 | Frameshift and introduction of stop codon | 0 | 19.9 | 18.3 | 115 | PTC 1-3′ | N/A | Unknown |
| 103 | 60 | N/D | Dup | 8 | 4707 | Frameshift and introduction of stop codon | 0 | 23.8 | 12.9 | 265 | PTC 1-5′ | N/A | Unknown |
| 104 | 51 | N/D | Point | 1 | 4748 | Synonymous mutation | 0 | 16.6 | 12.5 | 567 | Unknown | 47,XX,+21[5]/47,XX,der(7), +21[2] | |
| 105 | 15.5 | N/D | Del + Ins | 3 + 2 | 4742 | Frameshift and introduction of stop codon | 0.25 | 64.4 | 13.8 | 547 | PTC 1-3′ | N/A | Unknown |
| 106 | 50 | N/D | Dup | 19 | 4722 | Frameshift and introduction of stop codon | 0 | 43.5 | 15.4 | 128 | PTC 1-3′ | N/A | Known |
| 107 | 21 | N/D | Dup | 11 | 4735 | Frameshift and introduction of stop codon | 0 | 1 | 10.7 | 45 | PTC 1-3′ | N/A | Unknown |
| 108 | N/A | N/D | Ins | 1 | 4731 | Frameshift and introduction of stop codon | N/A | N/A | N/A | N/A | PTC -3′ | N/A | Unknown |
| 109 | 22 | N/D | Point | 1 | 4769 | Loss of splice acceptor site | 1.5 | 74.7 | 6 | 554 | Splice | N/A | Unknown |
| 110 | 57.5 | N/D | Dup | 105 | 4698 | Nonsynonymous change in amino acid sequence introducing a stop codon | 0 | 64.3 | 11.8 | 427 | PTC 1-5′ | N/A | Unknown |
| 111 | 62 | N/D | Del | 12 | 4764 | Loss of splice acceptor site | 1.5 | 21.9 | 6.7 | 102 | Splice | 47,XY,+21c | Unknown |
| 112 | 45 | N/D | Del | 4 | 4672 | Frameshift and introduction of stop codon | 1 | N/A | N/A | N/A | PTC 1-5′ | N/A | Unknown |
| 113 | 73 | N/D | Dup | 6 | 4706 | Frameshift and introduction of stop codon | 0 | 195 | 12 | 150 | PTC 1-5′ | N/A | Unknown |
| 114 | N/A | N/D | Del | 2 | 4586 | Frameshift and introduction of stop codon | N/A | N/A | N/A | N/A | PTC 1-5′ | N/A | Unknown |
| 115 | 77 | N/D | Dup | 10 | 4719 | Frameshift and introduction of stop codon | 0 | 170 | 10.6 | 655 | PTC 1-3′ | N/A | Unknown |
| 116‡ | 0.5 | N/D | Ins | 1 | 4733 | Frameshift and introduction of stop codon | 1.5 | 4 | 7 | 89 | PTC 1-3′ | 48, XY, +Y(q12),+11, +21c[8]/47, XY,+21c[7] | ML-DS |
| 117 | 30 | Ex2 | Not found | Unknown | Unknown | N/A | 1 | 4.2 | 12.9 | 25 | Unknown | 47, XX, del(16), +21c | Unknown |
| 118 | 10 | Ex2 | Not found | Unknown | Unknown | N/A | 1 | 20.4 | 13 | 119 | Unknown | N/A | Unknown |
| 119 | 2 | Ex2 | Not found | Unknown | Unknown | N/A | 0.38 | 21.4 | 15 | 218 | Unknown | 47, XY, +21c | Unknown |
| 120 | 4 | Ex2 | Not found | Unknown | Unknown | N/A | 3 | 20.7 | 16.1 | 63 | Unknown | 47, XY, +21c | Unknown |
| 121 | 3 | Ex2 | Not found | Unknown | Unknown | N/A | 4 | 6.86 | 6.4 | 83 | Unknown | 47, XY, +21c | Unknown |
| 122 | 30 | Ex2 | Not found | Unknown | Unknown | N/A | 2.5 | 8 | 13 | 1000 | Unknown | N/A | Unknown |
| 123 | 80 | Ex2 | Not found | Unknown | Unknown | N/A | 0 | 265.3 | 18 | 353 | Unknown | N/A | Unknown |
| 124 | 20 | Ex2 | Not found | Unknown | Unknown | N/A | 0.5 | 13.4 | 11.9 | 371 | Unknown | 47, XY, +21c | Unknown |
| 125 | 39 | Ex2 | Not found | Unknown | Unknown | N/A | 0 | 12.5 | 16.1 | 193 | Unknown | N/A | Unknown |
| 126 | 35 | Ex 3.1 | Del | 16 | 4871 | Frameshift and introduction of stop codon | 0 | 33.5 | 9.1 | 695 | PTC 2 | N/A | Unknown |
| 127 | 30 | Ex 3.1 | Dup | 2 | 4778 | Frameshift and introduction of stop codon | 2 | 21.7 | 13 | 24 | PTC 2 | N/A | Unknown |
| 128 | 13.5 | N/D | Not found | Unknown | Unknown | N/A | 0.25 | 8.91 | 18.4 | 46 | Unknown | 47,XX,+21c[17] | CCR |
| 129 | 3 | N/D | Not found | Unknown | Unknown | N/A | 0 | 10.6 | 18.9 | 65 | Unknown | N/A | CCR |
| 130 | 5 | N/D | Not found | Unknown | Unknown | N/A | 0.75 | 8.3 | 8.1 | 100 | Unknown | N/A | Unknown |
| 131 | 8 | N/D | Not found | Unknown | Unknown | N/A | 0 | 22.3 | 21 | 60 | Unknown | N/A | Unknown |
| 132 | 5 | N/D | Not found | Unknown | Unknown | N/A | 0.5 | 15.2 | 20.8 | 276 | Unknown | N/A | Unknown |
| 133 | 12 | N/D | Not found | Unknown | Unknown | N/A | 1 | N/A | N/A | N/A | Unknown | N/A | Unknown |
| 134 | 2 | N/D | Not found | Unknown | Unknown | N/A | 6 | 4.15 | 13 | 100 | Unknown | N/A | Unknown |
| Patient no. . | Blast count, % of total nucleated count . | WAVE* . | Mutation type . | No. of nucleotides changed, bp . | Position of mutation† . | Predicted effect of mutation on RNA . | Age at diagnosis, mo . | WBC count, ×109/L . | Hb level, g/dL . | Plt count, ×109/L . | Kanezaki et al15 class . | Karyotype . | Outcome . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 79 | Ex2 | Point | 1 | 4549 | Loss of start codon | 0 | 107 | 15 | 140 | 1st Met | 47, XY, +21c | CCR |
| 2‡ | 66 | Ex2 | Dup | 20 | 4722 | Frameshift and introduction of stop codon | 2 | 70 | 13.3 | 352 | PTC 1-3′ | 47, XX, +21c | ML-DS |
| 3 | 40 | Ex2 | Point | 1 | 4767 | Loss of splice acceptor site | 1.3 | 14 | 12.7 | 461 | Splice | 47, XY, +21c | CCR |
| 4 | 35 | Ex2 | Ins | 8 | 4701 | Frameshift and introduction of stop codon | 2 | 16 | 21.3 | 41 | PTC 1-5′ | 47, XY, +21c | CCR |
| 5 | 30 | Ex2 | Dup | 101 | 4653 | Frameshift and introduction of stop codon | 0.57 | 5 | 8.2 | 122 | PTC 1-5′ | 47, XY, +21c | CCR |
| 6 | 55 | Ex2 | Dup | 19 | 4733 | Frameshift and introduction of stop codon | 0.57 | 75.9 | 6.3 | 113 | PTC 1-3′ | 47, XX, +21c | CCR |
| 7 | 29 | Ex2 | Del | 2 | 4638 | Frameshift and introduction of stop codon | 0.28 | 44.6 | 17.4 | 45 | PTC 1-5′ | 47, XX, +21c | CCR |
| 8 | 92 | Ex2 | Dup | 2 | 4768 | Loss of splice acceptor site | 0.28 | 120 | 18.8 | 505 | Splice | N/A | CCR |
| 9‡ | 40 | Ex2 | Point | 1 | 4583 | Nonsynonymous change in amino acid sequence introducing a stop codon | 1 | 50 | N/A | N/A | PTC 1-5′ | N/A | ML-DS |
| 10 | 80 | Ex2 | Dup | 22 | 4735 | Frameshift and introduction of stop codon | 0 | 121 | 11.6 | 35 | PTC 1-3′ | 47, XY, +21 | Thrombo-cytopaenia |
| 11 | 11 | Ex2 | Dup | 17 | 4719 | Frameshift and introduction of stop codon | 0 | 38.2 | 18.7 | 132 | PTC 1-3′ | N/A | CCR |
| 12 | 21 | Ex2 | Point | 1 | 4768 | Loss of splice acceptor site | 2 | 19.4 | 10.8 | 52 | Splice | N/A | Unknown |
| 13 | 2 | Ex2 | Del | 1 | 4607 | Frameshift and introduction of stop codon | 0.71 | 9.5 | 12.7 | 54 | PTC 1-5′ | N/A | Unknown |
| 14 | 60 | Ex2 | Ins | 16 | 4721 | Frameshift and introduction of stop codon | 0.57 | 6.7 | 16.3 | 59 | PTC 1-3′ | N/A | Died |
| 15 | 5 | Ex2 | Dup | 22 | 4723 | Frameshift and introduction of stop codon | 0.71 | 12 | 14 | 88 | PTC 1-3′ | N/A | Unknown |
| 16 | 35 | Ex2 | Del | 26 | 4688 | Frameshift and introduction of stop codon | 2 | 39.3 | 13.8 | 335 | PTC 1-5′ | N/A | CCR |
| 17 | 82 | Ex2 | Del | 2 | 4458 | Frameshift and introduction of stop codon | 1.1 | 249 | 11.8 | 122 | PTC 1-5′ | 47, XY, +21c | CCR |
| 18 | 75 | Ex2 | Ins | 16 | 4706 | Frameshift and introduction of stop codon | N/A | 31.6 | 16.2 | 44 | PTC 1-5′ | N/A | CCR |
| 19 | 30 | Ex2 | Point | 1 | 4550 | Loss of start codon | 4 | 11.5 | 13.3 | 72 | 1st Met | 47, XY, +21c | CCR |
| 20 | N/A | Ex2 | Point | 1 | 4596 | Nonsynonymous change in amino acid sequence introducing a stop codon | N/A | N/A | N/A | N/A | PTC 1-5′ | N/A | Died |
| 21 | N/A | Ex2 | Point | 1 | 4766 | Loss of splice acceptor site | 1 | N/A | N/A | N/A | Splice | N/A | Unknown |
| 22 | 25 | Ex2 | Ins | 8 | 4709 | Frameshift and introduction of stop codon | 1 | 33.7 | 14.8 | 35 | PTC 1-5′ | N/A | Unknown |
| 23 | 45 | Ex2 | Substitution | 1 | 4744 | Nonsynonymous change in amino acid sequence introducing a stop codon | 2.7 | 20 | 2 | 51 | PTC 1-3′ | 47, XY, der(11)t(q23p15) t(1:11)(q23;q85), +21c | CCR |
| 24 | 50 | Ex2 | Del | 132 | 4671 | Loss of splice acceptor site | 0 | 58 | N/A | N/A | Splice | 47, XY, +21c | CCR |
| 25 | 76 | Ex2 | Ins | 1 | 4661 | Frameshift and introduction of stop codon | 2 | 124.9 | 9 | 28 | PTC 1-5′ | 47, XY, +21c | CCR |
| 26 | 25 | Ex2 | Del | 1 | 4662 | Frameshift and introduction of stop codon | 2 | 39.6 | 19.8 | 22 | PTC 1-5′ | 47, XY, +21c | CCR |
| 27‡ | 18 | Ex2 | Dup | 8 | 4717 | Frameshift and introduction of stop codon | 0 | 101 | 13.3 | 21 | PTC 1-5′ | 47, XY, t(5;11)(p15;q13), +21c | ML-DS |
| 28 | 70 | Ex2 | Del | 4 | 4684 | Frameshift and introduction of stop codon | 4 | 200 | 7.6 | 87 | PTC 1-5′ | N/A | CCR |
| 29 | 62 | Ex2 | Ins | 4 | 4643 | Frameshift and introduction of stop codon | 1 | N/A | N/A | N/A | PTC 1-5′ | N/A | Unknown |
| 30 | 11.4 | Ex2 | Dup | 8 | 4732 | Frameshift and introduction of stop codon | 0 | 33.5 | 9.1 | 695 | PTC 1-3′ | 47, XY, +21c | Unknown |
| 31 | 29 | Ex2 | Point | 1 | 4670 | Nonsynonymous change in amino acid sequence introducing a stop codon | 2 | 21.7 | 13 | 24 | PTC 1-5′ | N/A | Unknown |
| 32 | 60 | Ex2 | Del | 1 | 4697 | Frameshift and introduction of stop codon | 28 weeks gestation | 40 | 5.1 | 54 | PTC 1-5′ | N/A | Unknown |
| 33 | 79 | N/D | Del | 2 | 4638 | Frameshift and introduction of stop codon | 0 | 198.2 | 10.6 | 312 | PTC 1-5′ | N/A | Unknown |
| 34 | 66 | N/D | Dup | 40 | 4740 | Frameshift and introduction of stop codon | 0.25 | 9.8 | 12.7 | 60 | PTC 1-3′ | N/A | Unknown |
| 35 | 60 | N/D | Dup | 10 | 4710 | Frameshift and introduction of stop codon | 0.25 | 46.6 | 20.4 | 89 | PTC 1-5′ | N/A | Unknown |
| 36‡ | 35.5 | N/D | Ins | 14 | 4707 | Frameshift and introduction of stop codon | 0 | 55.6 | 17.3 | 570 | PTC 1-5′ | N/A | ML-DS |
| 37 | 54 | N/D | Point | 1 | 4768 | Loss of splice acceptor site | 0 | 55.6 | 17.3 | 570 | Splice | N/A | Unknown |
| 38 | 23 | N/D | Point | 1 | 4767 | Synonymous mutation | 1.25 | 19.2 | 6.7 | 125 | Unknown | N/A | Unknown |
| 39‡ | 84 | N/D | Del | 2 | 4604 | Frameshift and introduction of stop codon | 0 | 1193.3 | 14.7 | 255 | PTC 1-5′ | N/A | ML-DS |
| 40 | 10 | N/D | Del + Ins | 2 + 1 | 4770 + 4774 | Loss of splice donor site | 2 | 9.1 | 9.6 | 77 | Splice | N/A | Unknown |
| 41‡ | 88 | N/D | Del | 3 | 4680 | Frameshift and introduction of stop codon | 0 | 232 | 15.6 | 254 | PTC 1-5′ | N/A | ML-DS |
| 42 | 75 | N/D | Del + Ins | 1 + 2 | 4679 | Frameshift and introduction of stop codon | 0 | 172.8 | 16.2 | 101 | PTC 1-5′ | N/A | Unknown |
| 43 | 14 | N/D | Point | 1 | 4734 | Unknown | 0.25 | 16.9 | 14.9 | 353 | Unknown | N/A | Unknown |
| 44 | 13.5 | N/D | Dup | 36 | 4737 | Frameshift and introduction of stop codon | 0.5 | 3.2 | 11.1 | 16 | PTC 1-3′ | N/A | Unknown |
| 45 | 31 | N/D | Dup | 19 | 4722 | Frameshift and introduction of stop codon | 0 | 28.6 | 16.2 | 57 | PTC 1-3′ | N/A | Unknown |
| 46 | 43 | N/D | Ins + Dup | 2 + 13 | 4723 | Frameshift and introduction of stop codon | 0 | 61 | 17.6 | 149 | PTC 1-3′ | N/A | Unknown |
| 47 | 69 | N/D | Point | 1 | 4550 | Loss of start codon | 0 | 34 | 15 | 26 | 1st Met | N/A | Unknown |
| 48 | 10 | N/D | Dup | 21 | 4723 | Frameshift and introduction of stop codon | 0.25 | 7.7 | 13.2 | 78 | PTC 1-3′ | N/A | Unknown |
| 49 | 78 | N/D | Del + Ins | 19 + 17 | 4698 | Frameshift and introduction of stop codon | 0 | 68.6 | 10.9 | 212 | PTC 1-5′ | N/A | Unknown |
| 50 | 34 | N/D | Ins | 7 | 4713 | Frameshift and introduction of stop codon | 0.25 | 6.2 | 16.9 | 25 | PTC 1-5′ | N/A | Unknown |
| 51 | 60 | N/D | Del | 2 | 4638 | Frameshift and introduction of stop codon | 0 | 51.2 | 14.8 | 159 | PTC 1-5′ | N/A | Unknown |
| 52 | 75 | N/D | Point | 1 | 4747 | Nonsynonymous change in amino acid sequence introducing a stop codon | 0 | N/A | N/A | N/A | PTC 1-3′ | N/A | Unknown |
| 53 | 18.5 | N/D | Del | 162 | 4540 | Loss of start codon | 0 | 17.3 | 14 | 173 | 1st Met | N/A | Unknown |
| 54 | 1 | N/D | Del + Ins | 2 + 3 | 4698 | Frameshift and introduction of sop codon | 0.25 | 57.5 | 10.6 | 36 | PTC 1-5′ | N/A | Unknown |
| 55 | 8 | N/D | Ins + Dup | 3 + 36 | 4724 | Frameshift and introduction of stop codon | 0.25 | 16 | 18.5 | 48 | PTC 1-3′ | N/A | Unknown |
| 56 | 66.5 | N/D | Del | 2 | 4638 | Frameshift and introduction of stop codon | 0.75 | 62.9 | 12.3 | 93 | PTC 1-5′ | N/A | Unknown |
| 57‡ | 75 | N/D | Dup | 13 | 4744 | Frameshift and introduction of stop codon | 0 | 149.1 | 16.9 | 131 | PTC 1-3′ | 47, XY,+21.ish 21qter(D21S1446 × 3) | ML-DS |
| 58 | 85 | N/D | Point | 1 | 4762 | Nonsynonymous change in amino acid sequence introducing a stop codon | 0 | 46.4 | 20.6 | 197 | PTC 1-3′ | N/A | Unknown |
| 59 | 45 | N/D | Del + Ins | 7 + 10 | 4760 | Frameshift and introduction of stop codon | 0.5 | 78.3 | 15 | 1800 | PTC 1-3′ | N/A | Unknown |
| 60 | 38 | N/D | Ins + Dup | 3 + 9 | 4740 | Frameshift and introduction of stop codon | 0.25 | 32.9 | 11.9 | 220 | PTC 1-3′ | N/A | Unknown |
| 61‡ | 64.5 | N/D | Dup | 18 | 4733 | Frameshift and introduction of stop codon | 0 | 69.4 | 15.4 | 1178 | PTC 1-3′ | N/A | ML-DS |
| 62 | 58 | N/D | Del | 1 | 4698 | Frameshift and introduction of stop codon | 0.25 | 24.9 | 15.6 | 154 | PTC 1-5′ | N/A | Unknown |
| 63 | 34 | N/D | Del | 1 | 4690 | Frameshift and introduction of stop codon | 0 | 27.2 | 17 | 134 | PTC 1-5′ | N/A | Unknown |
| 64 | 85 | N/D | Del | 2 | 4604 | Frameshift and introduction of stop codon | 0 | 22 | 16.3 | 108 | PTC 1-5′ | N/A | Unknown |
| 65 | 11 | N/D | Point | 1 | 4549 | Loss of s | 1 | 7.4 | 12.3 | 241 | 1st Met | N/A | Unknown |
| 66 | 32 | N/D | Ins | 1 | 4654 | Frameshift and introduction of stop codon | 0 | 31.6 | 16.4 | 158 | PTC 1-5′ | N/A | Unknown |
| 67 | 3.5 | N/D | Point | 1 | 4597 | Nonsynonymous change in amino acid sequence | 2 | 8.4 | 9.3 | 105 | Unknown | N/A | Unknown |
| 68 | 22 | N/D | Del | 34 | 4693 | Frameshift and introduction of stop codon | 0.25 | 17.3 | 13.1 | 270 | PTC 1-5′ | N/A | Unknown |
| 69‡ | 87 | N/D | Point | 1 | 4552 | Nonsynonymous change in amino acid sequence | 0.25 | 133.9 | 13.8 | 32.9 | Unknown | N/A | ML-DS |
| 70 | 5.5 | N/D | Point | 1 | 4768 | Loss of splice acceptor site | 0 | 23.83 | 9.9 | 154 | Splice | N/A | Unknown |
| 71 | 32 | N/D | Ins + Dup | 1 + 12 | 4742 | Frameshift and introduction of stop codon | 0 | 27.6 | 14.6 | 46 | PTC 1-3′ | N/A | Unknown |
| 72 | 91 | N/D | Ins | 1 | 4647 | Frameshift and introduction of stop codon | 0 | 10.6 | 10.7 | 294 | PTC 1-5′ | N/A | Unknown |
| 73 | 16.5 | N/D | Del | 1 | 4653 | Frameshift and introduction of stop codon | 0.75 | 8.9 | 12.5 | 120 | PTC 1-5′ | N/A | Unknown |
| 74 | 14 | N/D | Dup | 19 | 4722 | Frameshift and introduction of stop codon | 11 | 3.39 | 10.5 | 91 | PTC 1-3′ | N/A | Unknown |
| 75 | 78 | N/D | Del | 1 | 4766 | Loss of splice acceptor site | 0 | 91.2 | 13.6 | 33 | Splice | 47, XY, t(21;21) (Q10;Q10) +21c | Unknown |
| 76 | 60.5 | N/D | Point | 1 | 4734 | Nonsynonymous change in amino acid sequence | 0 | 25.4 | 13.9 | 98 | Unknown | N/A | Unknown |
| 77 | 16 | N/D | Dup | 2 | 4734 | Frameshift and introduction of stop codon | 0.5 | 8.5 | 10.8 | 52 | PTC 1-3′ | N/A | Unknown |
| 78 | 12 | N/D | Del | 51 | 4706 | Frameshift and introduction of stop codon | 0 | 23.2 | 14.2 | 379 | PTC 1-5′ | N/A | Unknown |
| 79 | 95 | N/D | Ins + Dup | 6 + 7 | 4708 | Frameshift and introduction of stop codon | 0 | 410 | 11.1 | 261 | PTC 1-5′ | N/A | Unknown |
| 80 | 86 | N/D | Point | 1 | 4633 | Nonsynonymous change in amino acid sequence | 0 | 107.5 | 13.1 | 70 | Unknown | 47,XX,der(6)del(6) (p21)del(6)(q24, +der(6)del(6)(p12p22)t (6;10)(q16;q22),der(10) t(10;12)(q21;p12), der(11)t(11;12) (p15;q11),−12,+21c[17], 47,XX,+21c[2] | Unknown |
| 81‡ | 68 | N/D | Point | 1 | 4769 | Loss of splice acceptor site | 0 | 100 | 11.5 | 326 | Splice | N/A | ML-DS |
| 82 | 72 | N/D | Del | 12 | 4733 | Frameshift and introduction of stop codon | 0.5 | 49.8 | 14.2 | 43 | PTC 1-3′ | N/A | Unknown |
| 83 | 30 | N/D | Del | 4 | 4741 | Frameshift and introduction of top codon | 0 | 52 | 13.9 | 71 | PTC 1-3′ | N/A | Unknown |
| 84 | 35 | N/D | Ins | 1 | 4654 | Frameshift and introduction of stop codon | 0 | 28 | 12.8 | 274 | PTC 1-5′ | N/A | Unknown |
| 85 | 78 | N/D | Substitution | 40 | 4545 | Unknown | 0 | 72.7 | 19.4 | 606 | Unknown | N/A | Unknown |
| 86 | 79 | N/D | Ins | 1 | 4736 | Frameshift and introduction of stop codon | 0 | 100 | 10.9 | 70 | PTC 1-3′ | N/A | Unknown |
| 87 | 46.5 | N/D | Del | 2 | 4637 | Frameshift and introduction of stop codon | 0.25 | 23.3 | 17.5 | 80 | PTC 1-5′ | N/A | Unknown |
| 88‡ | 43 | N/D | Del | 2 | 4638 | Frameshift and introduction of stop codon | 0.25 | N/A | N/A | N/A | PTC 1-5′ | 47,XY,+21c[15] | ML-DS |
| 89 | 15 | N/D | Ins | 2 | 4725 | Frameshift and introduction of stop codon | 0.5 | 11 | 24.2 | 158 | PTC 1-3′ | N/A | Unknown |
| 90 | 16.5 | N/D | Del | 2 | 4604 | Frameshift and introduction of stop codon | 0.25 | 24.2 | 13.2 | 78 | PTC 1-5′ | N/A | Unknown |
| 91‡ | 66 | N/D | Dup | 5 | 4723 | Frameshift and introduction of stop codon | 0 | 26.1 | 16.5 | 48 | PTC 1-3′ | 47,XX,+21c | ML-DS |
| 92 | 38.5 | N/D | Dup | 43 | 4742 | Frameshift and introduction of stop codon | 0.5 | 20.2 | 12.3 | 12 | PTC 1-3′ | N/A | Unknown |
| 93 | 58 | N/D | Point | 1 | 4768 | Loss of splice acceptor site | 0 | 20.7 | 14.1 | 901 | Splice | N/A | Unknown |
| 94 | 1 | N/D | Point + Point | 1 + 1 | 92 + 126 | Unknown | 0.25 | 5.6 | 14.3 | 67 | Unknown | N/A | Unknown |
| 95 | 83 | N/D | Del + Del | 15 + 1 | 4888 + 4907 | Frameshift and introduction of stop codon | 0.25 | 114.7 | 16.6 | 141 | PTC 2 | N/A | Unknown |
| 96 | 20 | N/D | Dup | 6 | 4714 | Frameshift and introduction of stop codon | 0 | 14.12 | 18.6 | 15 | PTC 1-5′ | N/A | Unknown |
| 97 | 43 | N/D | Point | 1 | 4769 | Loss of splice acceptor site | 0 | 49.8 | 17 | 209 | Splice | N/A | Unknown |
| 98 | 49.9 | N/D | Del | 2 | 4638 | Frameshift and introduction of stop codon | 0.5 | 7.06 | 12 | 64 | PTC 1-5′ | N/A | Unknown |
| 99 | 61 | N/D | Dup | 12 | 4720 | Frameshift and introduction of stop codon | 0.75 | 91.6 | 16.6 | 150 | PTC 1-3′ | N/A | Unknown |
| 100 | 53.5 | N/D | Ins | 1 | 4598 | Frameshift and introduction of stop codon | 0 | 51 | 18.3 | 56 | PTC 1-5′ | N/A | Unknown |
| 101 | 34 | N/D | Del | 13 | 4706 | Frameshift and introduction of stop codon | 0 | 15.4 | 17.1 | 62 | PTC 1-5′ | N/A | Unknown |
| 102 | 25 | N/D | Ins | 1 | 4753 | Frameshift and introduction of stop codon | 0 | 19.9 | 18.3 | 115 | PTC 1-3′ | N/A | Unknown |
| 103 | 60 | N/D | Dup | 8 | 4707 | Frameshift and introduction of stop codon | 0 | 23.8 | 12.9 | 265 | PTC 1-5′ | N/A | Unknown |
| 104 | 51 | N/D | Point | 1 | 4748 | Synonymous mutation | 0 | 16.6 | 12.5 | 567 | Unknown | 47,XX,+21[5]/47,XX,der(7), +21[2] | |
| 105 | 15.5 | N/D | Del + Ins | 3 + 2 | 4742 | Frameshift and introduction of stop codon | 0.25 | 64.4 | 13.8 | 547 | PTC 1-3′ | N/A | Unknown |
| 106 | 50 | N/D | Dup | 19 | 4722 | Frameshift and introduction of stop codon | 0 | 43.5 | 15.4 | 128 | PTC 1-3′ | N/A | Known |
| 107 | 21 | N/D | Dup | 11 | 4735 | Frameshift and introduction of stop codon | 0 | 1 | 10.7 | 45 | PTC 1-3′ | N/A | Unknown |
| 108 | N/A | N/D | Ins | 1 | 4731 | Frameshift and introduction of stop codon | N/A | N/A | N/A | N/A | PTC -3′ | N/A | Unknown |
| 109 | 22 | N/D | Point | 1 | 4769 | Loss of splice acceptor site | 1.5 | 74.7 | 6 | 554 | Splice | N/A | Unknown |
| 110 | 57.5 | N/D | Dup | 105 | 4698 | Nonsynonymous change in amino acid sequence introducing a stop codon | 0 | 64.3 | 11.8 | 427 | PTC 1-5′ | N/A | Unknown |
| 111 | 62 | N/D | Del | 12 | 4764 | Loss of splice acceptor site | 1.5 | 21.9 | 6.7 | 102 | Splice | 47,XY,+21c | Unknown |
| 112 | 45 | N/D | Del | 4 | 4672 | Frameshift and introduction of stop codon | 1 | N/A | N/A | N/A | PTC 1-5′ | N/A | Unknown |
| 113 | 73 | N/D | Dup | 6 | 4706 | Frameshift and introduction of stop codon | 0 | 195 | 12 | 150 | PTC 1-5′ | N/A | Unknown |
| 114 | N/A | N/D | Del | 2 | 4586 | Frameshift and introduction of stop codon | N/A | N/A | N/A | N/A | PTC 1-5′ | N/A | Unknown |
| 115 | 77 | N/D | Dup | 10 | 4719 | Frameshift and introduction of stop codon | 0 | 170 | 10.6 | 655 | PTC 1-3′ | N/A | Unknown |
| 116‡ | 0.5 | N/D | Ins | 1 | 4733 | Frameshift and introduction of stop codon | 1.5 | 4 | 7 | 89 | PTC 1-3′ | 48, XY, +Y(q12),+11, +21c[8]/47, XY,+21c[7] | ML-DS |
| 117 | 30 | Ex2 | Not found | Unknown | Unknown | N/A | 1 | 4.2 | 12.9 | 25 | Unknown | 47, XX, del(16), +21c | Unknown |
| 118 | 10 | Ex2 | Not found | Unknown | Unknown | N/A | 1 | 20.4 | 13 | 119 | Unknown | N/A | Unknown |
| 119 | 2 | Ex2 | Not found | Unknown | Unknown | N/A | 0.38 | 21.4 | 15 | 218 | Unknown | 47, XY, +21c | Unknown |
| 120 | 4 | Ex2 | Not found | Unknown | Unknown | N/A | 3 | 20.7 | 16.1 | 63 | Unknown | 47, XY, +21c | Unknown |
| 121 | 3 | Ex2 | Not found | Unknown | Unknown | N/A | 4 | 6.86 | 6.4 | 83 | Unknown | 47, XY, +21c | Unknown |
| 122 | 30 | Ex2 | Not found | Unknown | Unknown | N/A | 2.5 | 8 | 13 | 1000 | Unknown | N/A | Unknown |
| 123 | 80 | Ex2 | Not found | Unknown | Unknown | N/A | 0 | 265.3 | 18 | 353 | Unknown | N/A | Unknown |
| 124 | 20 | Ex2 | Not found | Unknown | Unknown | N/A | 0.5 | 13.4 | 11.9 | 371 | Unknown | 47, XY, +21c | Unknown |
| 125 | 39 | Ex2 | Not found | Unknown | Unknown | N/A | 0 | 12.5 | 16.1 | 193 | Unknown | N/A | Unknown |
| 126 | 35 | Ex 3.1 | Del | 16 | 4871 | Frameshift and introduction of stop codon | 0 | 33.5 | 9.1 | 695 | PTC 2 | N/A | Unknown |
| 127 | 30 | Ex 3.1 | Dup | 2 | 4778 | Frameshift and introduction of stop codon | 2 | 21.7 | 13 | 24 | PTC 2 | N/A | Unknown |
| 128 | 13.5 | N/D | Not found | Unknown | Unknown | N/A | 0.25 | 8.91 | 18.4 | 46 | Unknown | 47,XX,+21c[17] | CCR |
| 129 | 3 | N/D | Not found | Unknown | Unknown | N/A | 0 | 10.6 | 18.9 | 65 | Unknown | N/A | CCR |
| 130 | 5 | N/D | Not found | Unknown | Unknown | N/A | 0.75 | 8.3 | 8.1 | 100 | Unknown | N/A | Unknown |
| 131 | 8 | N/D | Not found | Unknown | Unknown | N/A | 0 | 22.3 | 21 | 60 | Unknown | N/A | Unknown |
| 132 | 5 | N/D | Not found | Unknown | Unknown | N/A | 0.5 | 15.2 | 20.8 | 276 | Unknown | N/A | Unknown |
| 133 | 12 | N/D | Not found | Unknown | Unknown | N/A | 1 | N/A | N/A | N/A | Unknown | N/A | Unknown |
| 134 | 2 | N/D | Not found | Unknown | Unknown | N/A | 6 | 4.15 | 13 | 100 | Unknown | N/A | Unknown |
WBC indicates white blood cell; Hb, hemoglobin; Plt, platelet; Point, point substitution; CCR, complete clinical remission; Dup, duplication of nucleotides; Ins, insertion of nucleotides; Del, deletion of nucleotides; N/A, not available; and N/D, not done.
Exon number containing the mutation as defined by WAVE analysis.
Nucleotide 0 is the first nucleotide of GATA1 exon 1 including exons and introns. NCBI reference NT_079573.4 (Homo sapiens chromosome X genomic contig, starting position 11496706) was used.
Patients with TMD progressed to ML-DS.
Summary of GATA1 mutations and patient characteristics for ML-DS samples
| Patient no. . | Blast count, % of total nucleated count . | WAVE* . | Mutation . | Size, bp . | Position of mutation† . | Predicted effect of mutation on RNA . | Age at diagnosis, mo . | WBC count, ×109/L . | Hb level, g/dL . | Plt count, ×109/L . | Kanezaki et al15 class . | Karyotype . | Outcome . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 70 | Ex2 | Del | 56 | 4750 | Frameshift and introduction of stop codon | 24 | 12.6 | 5.7 | 22 | PTC 1-3′ | 47, XY, Del(7)(q32), +8, +21c | CCR |
| 2 | 15 | Ex2 | Dup | 13 | 4709 | Frameshift and introduction of stop codon | N/A | 5.3 | 7.6 | 22 | PTC 1-5′ | 47, XX, +21c | CCR |
| 3 | 84 | Ex2 | Ins | 11 | 4724 | Frameshift and introduction of stop codon | 23 | 27.7 | 8.5 | 19 | PTC 1-3′ | 47, XX, +21c | CCR |
| 4‡ | 90 | Ex2 | Dup | 20 | 4722 | Frameshift and introduction of stop codon | 36 | 8.5 | 11.3 | 20 | PTC 1-3′ | 56, XX, +2, +6, +8, +13, +13, +14, +14, −16, del(17)(q?22−24), +19, +del(19) (p13), +21, +21c[7], | CCR |
| 5 | 20 | Ex2 | Point | 1 | 4549 | Loss of start codon | 20 | 3.16 | 11 | 36 | 1st Met | 48, XY, +8, +21 | CCR |
| 6 | N/A | Ex2 | Del | 13 | 4753 | Frameshift and introduction of stop codon | 5 | 40 | 8.9 | 27 | PTC 1-3′ | 48, XY, der(1)add(1) (p32)dup(1)(q21q42), del(3)(q21q26),I(7) (q10),del(16)(q22q24), +21 | Died |
| 7 | 14 | Ex2 | Del | 10 | 4767 | Frameshift and introduction of stop codon | 21 | 27.8 | 5.4 | 85 | PTC 1-3′ | 47, XY, +21c | Died |
| 8 | N/A | Ex2 | Point | 1 | 4548 | Loss of splice acceptor site | 24 | 4.1 | 2.1 | 6 | Splice | 47XX,i(7)(q10),−16, +21c, +mar[7] | CCR |
| 9 | N/A | Ex2 | Dup | 20 | 4728 | Frameshift and introduction of stop codon | 1.2 | 11.9 | 7.5 | 18 | PTC 1-3′ | 47,XX, add(1)(q24), del(5)(p13), +21c | CCR |
| 10 | 50 | Ex2 | Point | 1 | 4768 | Loss of splice acceptor site | 31 | 2.8 | 9.7 | 21 | Splice | 48, XX, Add(8)(p23), +11, +21c[16]/ 47, XX, +21c [19] | CCR |
| 11 | N/A | Ex2 | Del | 29 | 4669 | Frameshift and introduction of stop codon | 2 | 14 | 13 | 112 | PTC 1-5′ | 47, XY, +21, +8 | CCR |
| 12 | N/A | Ex2 | Point | 1 | 4768 | Loss of splice acceptor site | 24 | 33.4 | 2.9 | 3.6 | Splice | 46,XX, der(7)add(7)(p?), −13,+21c[9]/47, XX,+21c | Died |
| 13 | 60 | Ex2 | Point | 1 | 4768 | Loss of splice acceptor site | 60 | 17.4 | 11 | 32 | Splice | 55∼57, XX, del(1)(q21;q25), +2, +3?adk1(5)(q33), i(7)(q10), +8, der(12)(t1;12)(q21;p13), +13, +14, +18, +19, +21, +22 | Died |
| 14 | 20 | Ex2 | Point | 1 | 4768 | Loss of splice acceptor site | 26 | 11.2 | 9.7 | 28 | Splice | 47. XX. +21c[36] | Unknown |
| 15 | 78 | Ex2 | Ins | 1 | 4736 | Frameshift and introduction of stop codon | 25 | 42.8 | 8.2 | 33 | PTC 1-3′ | 47, XY, dup(X) (p11.2,p21), +21c [2], 47, idem, der (4), t(2;4)(q31;q31), ?del(6)(q2?5; q2?7), +21c [5], 47, XY, +21c[3] | CCR |
| 16 | 37 | Ex2 | Ins | 1 | 4693 | Frameshift and introduction of stop codon | N/A | N/A | N/A | N/A | PTC 1-5′ | N/A | Relapsed |
| 17 | 16 | Ex2 | Point | 1 | 4597 | Nonsynonymous change in amino acid sequence introducing a stop codon | 31 | 1.7 | 8.7 | 14 | PTC 1-5′ | 48, XX, + 8, +21c | Unknown |
| 18 | N/A | Ex2 | Ins | 1 | 4632 | Frameshift and introduction of stop codon | 17 | 29 | 11.5 | 5 | PTC 1-5′ | 47 XX, +21c | Died |
| 19 | 36 | Ex2 | Del | 35 | 4684 | Frameshift and introduction of stop codon | 36 | 9.8 | 9 | 24 | PTC 1-5′ | 48, XX, +8, +21 | CCR |
| 20 | 93 | Ex2 | Ins | 1 | 4769 | Loss of splice acceptor site | N/A | 200.3 | 4.7 | 23 | Splice | 47, XX, t(3;17) (q25;q25), del(6)(q13;q22), +21c[19] | CCR |
| 21 | 20 | Ex2 | Del | 7 | 4705 | Frameshift and introduction of stop codon | 24 | 5.4 | 3.7 | 9 | PTC 1-5′ | 47, XY, +21, +8 | Died |
| 22‡ | 23 | Ex2 | Dup | 8 | 4717 | Frameshift and introduction of stop codon | 6 | 32 | 8.7 | 22 | PTC 1-5′ | 47, XY, t(5;11) (p15;q13), add(9)(q34), +21c[6]/ 47, XY, +21c[14] | CCR |
| 23 | 21 | Ex2 | Del | 2 | 4638 | Frameshift and introduction of stop codon | 11 | 6.4 | 15.1 | 5.1 | PTC 1-5′ | 47, XX, del (6), del (21), +21 | Died |
| 24 | 44 | Ex2 | Del | 23 | 4705 | Frameshift and introduction of stop codon | 13 | 8.3 | N/A | N/A | PTC 1-5′ | 48, XX, del (6) (q?13q?21) +21c, +21 [5] | CCR |
| 25 | 50 | Ex2 | Del | 2 | 4638 | Frameshift and introduction of stop codon | 12 | 7.3 | N/A | N/A | PTC 1-5′ | 50, XY, +10, +21c, +21, +22 [6] | CCR |
| 26‡ | 16 | N/D | Dup | 10 | 4710 | Frameshift and introduction of stop codon | 11 | 3.5 | 11.9 | 30 | PTC 1-5′ | 47∼49,XY, +?19,+21c, +21[cp7]/47, XY,+21c[5] | CCR |
| 27 | 20 | N/D | Dup | 15 | 4715 | Frameshift and introduction of stop codon | 15 | 6.4 | 7.6 | 60 | PTC 1-5′ | 47,XY,+21c 2/47, idem,t(4;15), (q?21;q?21), del(7)(q?31q?33) | Unknown |
| 28 | 8.5 | N/D | Del | 1 | 4698 | Frameshift and introduction of stop codon | 8 | 4 | 13.4 | 70 | PTC 1-5′ | N/A | Unknown |
| 29 | 31 | N/D | Del | 5 | 4704 | Frameshift and introduction of stop codon | 13 | 3.9 | 11.2 | 47 | PTC 1-5′ | 47,XX,der(5)t(1;5) (q12;p14)?del(1) (q23q25),+21c[4]/48, XX,+8,der (12)t(1;12)(q12;p12), +21c[2]/47, XX,+21c[9] | Unknown |
| 30 | 93 | N/D | Del | 2 | 4638 | Frameshift and introduction of stop codon | 24 | 26.4 | 9.1 | 147 | PTC 1-5′ | 47,XY,+21c[25] | Unknown |
| 31 | 39 | N/D | Del | 2 | 4638 | Frameshift and introduction of stop codon | 27 | 8.5 | 8.5 | 26 | PTC 1-5′ | N/A | Unknown |
| 32‡ | 4.5 | N/D | Del | 2 | 4604 | Frameshift and introduction of stop codon | 13 | 1 | 7.7 | 107 | PTC 1-5′ | N/A | Unknown |
| 33 | 19.5 | N/D | Ins + Dup | 2 + 14 | 4722 | Frameshift and introduction of stop codon | 21 | 26.4 | 7.2 | 151 | PTC 1-3′ | N/A | Unknown |
| 34 | 50.5 | N/D | Ins | 1 | 4768 | Frameshift and introduction of stop codon | 28 | 1.2 | 8 | 109 | Splice | 47∼48,XX,?add (5)(p1?5),−8, +21,+21c, +mar1, +mar2[cp8]/47, XX,+21c | Unknown |
| 35 | 10.5 | N/D | Point | 1 | 4768 | Loss of splice acceptor site | 35 | 3.4 | 10.4 | 17 | Splice | N/A | Unknown |
| 36 | 36 | N/D | Dup | 5 | 4734 | Frameshift and introduction of stop codon | 14 | 12.4 | 7.2 | 144 | PTC 1-3′ | 47,XX,+21c | Unknown |
| 37 | 34 | N/D | Del | 22 | 4704 | Frameshift and introduction of stop codon | 22 | 15.1 | 9.6 | 257 | PTC 1-5′ | 48,XY,+8,+21c | Unknown |
| 38 | 55 | N/D | Del + Ins | 31 + 6 | 4783 | Loss of splice donor site | 31 | 1.9 | 6.9 | 22 | Splice | N/A | Unknown |
| 39 | 35 | N/D | Dup | 3 | 4737 | Frameshift and introduction of stop codon | 31 | 4.2 | 9.2 | 20 | PTC 1-3′ | N/A | Unknown |
| 40 | 38 | N/D | Del + ins | 24 + 4 | 4683 | Frameshift and introduction of stop codon | 31 | 6.1 | 9.8 | 16 | PTC 1-5′ | N/A | Unknown |
| 41 | 43 | N/D | Dup | 21 | 4736 | Frameshift and introduction of stop codon | 13 | 4.9 | 6.7 | 38 | PTC 1-3′ | N/A | Unknown |
| 42 | 15 | N/D | Dup | 4 | 4668 | Frameshift and introduction of stop codon | 20 | 8.1 | 12.7 | 219 | PTC 1-5′ | 47,XX,+21c | Unknown |
| 43 | 16 | N/D | Dup | 7 | 4717 | Frameshift and introduction of stop codon | 36 | 160 | 5 | 1.5 | PTC 1-5′ | 47,XY,+21c,[3]/47, idem,t(1;8)(q32;q22) [2]/48,idem,t(1;8) (q32;q22),+der(8)t (1;8)(q32;q22) [10}] | Unknown |
| 44 | 24 | N/D | Ins | 5 | 4708 | Frameshift and introduction of stop codon | 10 | 6.88 | 9.7 | 35 | PTC 1-5′ | N/A | Unknown |
| 45 | 11 | N/D | Point | 1 | 4956 | Synonymous mutation | 16 | 2.5 | 8.1 | 101 | Unknown | N/A | Unknown |
| 46 | 28 | N/D | Ins | 1 | 4736 | Frameshift and introduction of stop codon | 31 | 12.9 | 3.1 | 76 | PTC 1-3′ | N/A | Unknown |
| 47 | 14 | N/D | Point | 1 | 4535 | Unknown | 31 | 22 | 8.5 | 37 | Unknown | 48, XY, +14, +21 | Unknown |
| 48 | 13 | N/D | Del | 9 | 4546 | Loss of Start codon | 19 | 6.2 | 10.6 | 131 | 1st Met | 48,XY,i(7)(q10), t(11;11)(p15;q13), +21,+21[13]/47,XY,+21[2] | Unknown |
| 49 | 15 | N/D | Point | 1 | 4768 | Loss of splice acceptor site | 12 | 5 | 12.1 | 45 | Splice | N/A | Unknown |
| 50 | 15 | N/D | Del | 7 | 4702 | Frameshift and introduction of stop codon | 20 | 2.74 | 10.7 | 110 | PTC 1-5′ | 50,XX,+8, +14,+21,+21c, inc[5]/47, XX,+21c[7] | Unknown |
| 51‡ | 16 | N/D | Point | 1 | 4552 | Nonsynonymous change in amino acid sequence | 12 | 3.4 | 11.1 | 27 | Unknown | 47,XY,?der(11) (q24∼25), +21c[2]/47, XY,+21c[8] | Unknown |
| 52 | 9 | N/D | Del + ins | 2 + 1 | 4619 | Frameshift and introduction of stop codon | 15 | 3.5 | 9.2 | 47 | PTC 1-5′ | 46,XY,i(7) (q10),der(21;21) (q10;q10)c, +21[6]/46,XY,der(21;21) (q10;q10)c, +21[5] | Unknown |
| 53 | 12 | N/D | Del + ins | 3 + 1 | 4654 | Frameshift and introduction of stop codon | 7 | 2.9 | 3.3 | 17 | PTC 1-5′ | 48,XX,der(1) (p?q?),+21c, +mar[14]/47,XX,+21c[4] | Unknown |
| 54 | 10.5 | N/D | Dup | 20 | 4727 | Frameshift and introduction of stop codon | 16 | 4.8 | 12 | 59 | PTC 1-3′ | N/A | Unknown |
| 55 | 17 | N/D | Not Found | Unknown | Unknown | Unknown | 21 | 5.9 | 11.4 | 42 | Unknown | 47,XX,+21c | Unknown |
| 56 | N/A | N/D | Dup | 16 | 4734 | Frameshift and introduction of stop codon | N/A | N/A | N/A | N/A | PTC 1-3′ | N/A | Unknown |
| 57 | 23 | N/D | Point | 1 | 4767 | Loss of splice acceptor site | 26 | 5.1 | 13.3 | 11 | Splice | 48∼50,XY, +8,+?14, +21,+21c[cp6]/47,XY,+21c[4] | Unknown |
| 58 | 21 | N/D | Point + Del | 1 + 1 | 4637 + 4639 | Frameshift and introduction of stop codon | 37 | 6.3 | 11.4 | 13.7 | Unknown | 47,XX,?del(2)(q35), del(7)(p12),+21c[7]/47,XX,+21c[5] | Unknown |
| 59 | 13 | N/D | Del | 1 | 4698 | Frameshift and introduction of stop codon | 11 | 6.3 | 10.3 | 84 | PTC 1-5′ | 47,XY,+21c[9]; 48,XY,del(5) (p11p14),r(7) (p22q11), +8,+21c[7] | Unknown |
| 60 | 35 | N/D | Del | 1 | 4548 | Unknown | 11 | 16 | 8 | 38 | Unknown | N/A | Unknown |
| 61 | 7 | N/D | Del | 1 | 4642 | Frameshift and introduction of stop codon | 15 | 1.8 | 9.3 | 43 | PTC 1-5′ | 47,XY,+21[1] | Unknown |
| 62 | 1 | N/D | Not Found | Unknown | Unknown | Unknown | 19 | 2.6 | 5.8 | 109 | Unknown | N/A | Unknown |
| 63 | 20.5 | N/D | Del + ins | 4 + 2 | 4656 | Frameshift and introduction of stop codon | 11 | 3.7 | 10.7 | 130 | PTC 1-5′ | 48,XX,+8,+21c[3]/47,XX,+21c[8] | Unknown |
| 64 | 2.5 | N/D | Del | 2 | 4656 | Frameshift and introduction of stop codon | 32 | 4.8 | 7.4 | 17 | PTC 1-5′ | 47,XX,+21c[6] | Unknown |
| 65 | 46 | N/D | Dup | 16 | 4719 | Frameshift and introduction of stop codon | 20 | 5.6 | 12 | 19 | PTC 1-3′ | N/A | Unknown |
| 66 | 19.5 | N/D | Point | 1 | 4767 | Loss of splice acceptor site | 32 | 5.46 | 13.6 | 52 | Splice | N/A | Unknown |
| 67 | 2 | N/D | Del | 1 | 4737 | Frameshift and introduction of stop codon | 26 | 3.3 | 7.1 | 69 | PTC 1-3′ | N/A | Unknown |
| 68 | 16 | N/D | Del | 2 | 4638 | Frameshift and introduction of stop codon | 35 | 2.8 | 9.5 | 20 | PTC 1-5′ | 48,XX,+8,+21c | Unknown |
| 69 | 23 | N/D | Del | 2 | 4638 | Frameshift and introduction of stop codon | 13 | 3.2 | 9 | 25 | PTC 1-5′ | 47,XY,+21c[10] | Unknown |
| 70 | 76 | N/D | Del | 2 | 4638 | Frameshift and introduction of stop codon | 17 | 23 | 6.5 | 28 | PTC 1-5′ | 46,XY,t(4;13) (q33∼34;q14), der(5)t(5;7) (p14∼15;q12), −7,+21c[cp11] | Unknown |
| 71 | 3.5 | N/D | Del | 2 | 4656 | Frameshift and introduction of stop codon | 13 | 3.3 | 11.4 | 46 | PTC 1-5′ | 47,XX,+21c[17] | Unknown |
| 72 | N/A | N/D | Point | 1 | 4637 | Unknown | N/A | N/A | N/A | N/A | Unknown | N/A | Unknown |
| 73 | 20 | N/D | Point | 1 | 4713 | Synonymous mutation | 19 | 4.1 | 9.5 | 13 | Unknown | 48∼49,XY,+21, +21c,inc[cp2]/47,XY,+21c[12] | Unknown |
| 74 | 42 | N/D | Not Found | Unknown | Unknown | Unknown | 24 | 5.4 | 10.3 | 21 | Unknown | 47,XY,+21c, ?der(22)(p12) [5]/47,XY, +21c[15] | Unknown |
| 75‡ | 25 | N/D | Dup | 13 | 4744 | Frameshift and introduction of stop codon | 26 | 2.7 | 11.8 | 20 | PTC 1-3′ | 47,XY,+21.ish 21qter(D21 S1446 × 3) | Unknown |
| 76 | 96 | N/D | Point | 1 | 4737 | Nonsynonymous change in amino acid sequence | 22 | 124 | 5.9 | 21 | Unknown | 47,X,−X, del(6)(q13q16), +21c,+21 | Unknown |
| 77 | 10.5 | N/D | Not Found | Unknown | Unknown | Unknown | 17 | 5.2 | 10.6 | 82 | Unknown | 47,XX,add(16) (p13),+21c[5]/47,XX,+21c[5] | Unknown |
| 78 | 26 | N/D | Point | 1 | 4956 | Synonymous mutation | 20 | 2 | 6.4 | 130 | Unknown | 48,XY,der(8)t (1;8)(q21; p22∼23), +11,+21c[7]/47,XY,+21c[8] | Unknown |
| 79 | 11 | N/D | Point | 1 | 4768 | Loss of splice acceptor site | 43 | 6.35 | 12 | 70 | Splice | N/A | Unknown |
| 80 | 13 | N/D | Point | 1 | 4633 | Nonsynonymous change in amino acid sequence | 16 | 3.3 | 10.5 | 84 | Unknown | N/A | Unknown |
| 81‡ | 26 | N/D | Del | 2 | 4638 | Frameshift and introduction of stop codon | 11 | 3.4 | 10.2 | 28 | PTC 1-5′ | 47,XY,+21c[15] | Unknown |
| 82 | 6 | N/D | Ins | 2 | 4725 | Frameshift and introduction of stop codon | 14 | 4 | 10.2 | 70 | PTC 1-3′ | N/A | Unknown |
| 83 | 4 | N/D | Dup | 6 | 4714 | Frameshift and introduction of stop codon | 8 | 8.75 | 11.4 | 69 | PTC 1-5′ | N/A | Unknown |
| 84 | N/A | N/D | Dup | 48 | 4771 | Frameshift and introduction of stop codon | N/A | N/A | N/A | N/A | PTC 1-3′ | N/A | Unknown |
| 85 | 17.5 | N/D | Del | 2 | 4638 | Frameshift and introduction of stop codon | 11 | 4.7 | 9.3 | 83 | PTC 1-5′ | N/A | Unknown |
| 86 | 45 | N/D | Dup | 10 | 4723 | Frameshift and introduction of stop codon | 14 | 11.5 | 8.6 | 44 | PTC 1-3′ | N/A | Unknown |
| 87 | 70.5 | N/D | Del | 2 | 4638 | Frameshift and introduction of stop codon | 16 | 32.5 | 7.9 | 103 | PTC 1-5′ | 46,XY[22]. nuc ish 8q22(ETOx2), 21q22(AML1 × 2), 13q14(RB1 × 2), 13q34 × 2), 11q23(MLLx2)[100] | Unknown |
| 88 | 31 | N/D | Del | 2 | 4638 | Frameshift and introduction of stop codon | 29 | 11.4 | 13.2 | 57.4 | PTC 1-5′ | 46,XX,del (9)(q?13q33), der(19)t(1;19) (q31;p13), der(21;21) (q10;q10)?c | Unknown |
| 89 | 7.5 | N/D | Del | 22 | 4706 | Frameshift and introduction of stop codon | 17 | 7.8 | 11.1 | 27 | PTC 1-5′ | 48,XY,add(7) (p2?2),+8, del(13)(q1?4), +21c[7]/46, XY,−21,+21c or 46,XY[3]/47,XY,+21c[2] | Unknown |
| 90 | 33 | N/D | Del + ins | 2 + 6 | 4707 | Frameshift and introduction of stop codon | 8 | 4.6 | 7.5 | 3 | PTC 1-5′ | N/A | Unknown |
| 91‡ | 24 | N/D | Ins | 1 | 4733 | Frameshift and introduction of stop codon | 11 | 3 | 7.1 | 57 | PTC 1-3′ | 48,X,add(Y) (q12),+11, +21c[8]/47,XY, +21c[7] | Unknown |
| 92 | 28 | N/D | Dup | 10 | 4719 | Frameshift and introduction of stop codon | 22 | 5.2 | 13.7 | 143 | PTC 1-3′ | N/A | Unknown |
| 93 | 16 | Ex2 | Not Found | N/A | N/A | N/A | 1 | 1.5 | 10.4 | 13 | Unknown | 47, XY, +21c | Died |
| 94 | 30 | Ex2 | Not Found | N/A | N/A | N/A | 15 | 14.47 | 4.1 | 38 | Unknown | 47, XX, t(3;3)(q21;q26), t(3;3)(p2?3;q13.3), +21c[13]/47, idem, der(3)t(3;3)add(3)(q29)[7] | CCR |
| 95 | 20 | Ex2 | Not Found | N/A | N/A | N/A | 17 | 11 | 9.9 | 52 | Unknown | 47, XY, der(11)t (q23p15)t (1:11)(q23;q85), +21c | CCR |
| 96 | 10 | Ex2 | Not Found | N/A | N/A | N/A | 11 | N/A | 8.6 | 86 | Unknown | 47, XY, del (7), +21 | Unknown |
| 97 | 21 | Ex2 | Not Found | N/A | N/A | N/A | 33 | 4.9 | 6 | 15 | Unknown | 48, XY, +21, +21c | CCR |
| 98 | 20 | Ex2 | Not Found | N/A | N/A | N/A | 15 | 15.2 | 9.9 | 18 | Unknown | 47 XX, ?dup(3) (p23p25), del(8) (p11.2p21), der(15)t(1:15) (q21;p11), del(16)(q22q24, del(20) | CCR |
| 99 | 6 | Ex2 | Not Found | N/A | N/A | N/A | 23 | 5.1 | 7.2 | 49 | Unknown | N/A | Unknown |
| 100 | 20 | Ex2 | Not Found | N/A | N/A | N/A | 38 | N/A | N/A | N/A | Unknown | 46, XX, +21, −7 | Unknown |
| 101 | 38 | Ex2 | Not Found | N/A | N/A | N/A | 21 | 11.9 | 7.3 | 34 | Unknown | 47, XX, +21c | CCR |
| 102 | 10 | Ex2 | Not Found | N/A | N/A | N/A | 10 | 3.9 | 10.9 | 65 | Unknown | N/A | Unknown |
| 103 | 20 | Ex2 | Not Found | N/A | N/A | N/A | 12 | 3.8 | 8.6 | 16 | Unknown | 47, XX, t(1;22)(q23;p13), +21c[9]/ 47, XX, +21c[4] | CCR |
| Patient no. . | Blast count, % of total nucleated count . | WAVE* . | Mutation . | Size, bp . | Position of mutation† . | Predicted effect of mutation on RNA . | Age at diagnosis, mo . | WBC count, ×109/L . | Hb level, g/dL . | Plt count, ×109/L . | Kanezaki et al15 class . | Karyotype . | Outcome . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 70 | Ex2 | Del | 56 | 4750 | Frameshift and introduction of stop codon | 24 | 12.6 | 5.7 | 22 | PTC 1-3′ | 47, XY, Del(7)(q32), +8, +21c | CCR |
| 2 | 15 | Ex2 | Dup | 13 | 4709 | Frameshift and introduction of stop codon | N/A | 5.3 | 7.6 | 22 | PTC 1-5′ | 47, XX, +21c | CCR |
| 3 | 84 | Ex2 | Ins | 11 | 4724 | Frameshift and introduction of stop codon | 23 | 27.7 | 8.5 | 19 | PTC 1-3′ | 47, XX, +21c | CCR |
| 4‡ | 90 | Ex2 | Dup | 20 | 4722 | Frameshift and introduction of stop codon | 36 | 8.5 | 11.3 | 20 | PTC 1-3′ | 56, XX, +2, +6, +8, +13, +13, +14, +14, −16, del(17)(q?22−24), +19, +del(19) (p13), +21, +21c[7], | CCR |
| 5 | 20 | Ex2 | Point | 1 | 4549 | Loss of start codon | 20 | 3.16 | 11 | 36 | 1st Met | 48, XY, +8, +21 | CCR |
| 6 | N/A | Ex2 | Del | 13 | 4753 | Frameshift and introduction of stop codon | 5 | 40 | 8.9 | 27 | PTC 1-3′ | 48, XY, der(1)add(1) (p32)dup(1)(q21q42), del(3)(q21q26),I(7) (q10),del(16)(q22q24), +21 | Died |
| 7 | 14 | Ex2 | Del | 10 | 4767 | Frameshift and introduction of stop codon | 21 | 27.8 | 5.4 | 85 | PTC 1-3′ | 47, XY, +21c | Died |
| 8 | N/A | Ex2 | Point | 1 | 4548 | Loss of splice acceptor site | 24 | 4.1 | 2.1 | 6 | Splice | 47XX,i(7)(q10),−16, +21c, +mar[7] | CCR |
| 9 | N/A | Ex2 | Dup | 20 | 4728 | Frameshift and introduction of stop codon | 1.2 | 11.9 | 7.5 | 18 | PTC 1-3′ | 47,XX, add(1)(q24), del(5)(p13), +21c | CCR |
| 10 | 50 | Ex2 | Point | 1 | 4768 | Loss of splice acceptor site | 31 | 2.8 | 9.7 | 21 | Splice | 48, XX, Add(8)(p23), +11, +21c[16]/ 47, XX, +21c [19] | CCR |
| 11 | N/A | Ex2 | Del | 29 | 4669 | Frameshift and introduction of stop codon | 2 | 14 | 13 | 112 | PTC 1-5′ | 47, XY, +21, +8 | CCR |
| 12 | N/A | Ex2 | Point | 1 | 4768 | Loss of splice acceptor site | 24 | 33.4 | 2.9 | 3.6 | Splice | 46,XX, der(7)add(7)(p?), −13,+21c[9]/47, XX,+21c | Died |
| 13 | 60 | Ex2 | Point | 1 | 4768 | Loss of splice acceptor site | 60 | 17.4 | 11 | 32 | Splice | 55∼57, XX, del(1)(q21;q25), +2, +3?adk1(5)(q33), i(7)(q10), +8, der(12)(t1;12)(q21;p13), +13, +14, +18, +19, +21, +22 | Died |
| 14 | 20 | Ex2 | Point | 1 | 4768 | Loss of splice acceptor site | 26 | 11.2 | 9.7 | 28 | Splice | 47. XX. +21c[36] | Unknown |
| 15 | 78 | Ex2 | Ins | 1 | 4736 | Frameshift and introduction of stop codon | 25 | 42.8 | 8.2 | 33 | PTC 1-3′ | 47, XY, dup(X) (p11.2,p21), +21c [2], 47, idem, der (4), t(2;4)(q31;q31), ?del(6)(q2?5; q2?7), +21c [5], 47, XY, +21c[3] | CCR |
| 16 | 37 | Ex2 | Ins | 1 | 4693 | Frameshift and introduction of stop codon | N/A | N/A | N/A | N/A | PTC 1-5′ | N/A | Relapsed |
| 17 | 16 | Ex2 | Point | 1 | 4597 | Nonsynonymous change in amino acid sequence introducing a stop codon | 31 | 1.7 | 8.7 | 14 | PTC 1-5′ | 48, XX, + 8, +21c | Unknown |
| 18 | N/A | Ex2 | Ins | 1 | 4632 | Frameshift and introduction of stop codon | 17 | 29 | 11.5 | 5 | PTC 1-5′ | 47 XX, +21c | Died |
| 19 | 36 | Ex2 | Del | 35 | 4684 | Frameshift and introduction of stop codon | 36 | 9.8 | 9 | 24 | PTC 1-5′ | 48, XX, +8, +21 | CCR |
| 20 | 93 | Ex2 | Ins | 1 | 4769 | Loss of splice acceptor site | N/A | 200.3 | 4.7 | 23 | Splice | 47, XX, t(3;17) (q25;q25), del(6)(q13;q22), +21c[19] | CCR |
| 21 | 20 | Ex2 | Del | 7 | 4705 | Frameshift and introduction of stop codon | 24 | 5.4 | 3.7 | 9 | PTC 1-5′ | 47, XY, +21, +8 | Died |
| 22‡ | 23 | Ex2 | Dup | 8 | 4717 | Frameshift and introduction of stop codon | 6 | 32 | 8.7 | 22 | PTC 1-5′ | 47, XY, t(5;11) (p15;q13), add(9)(q34), +21c[6]/ 47, XY, +21c[14] | CCR |
| 23 | 21 | Ex2 | Del | 2 | 4638 | Frameshift and introduction of stop codon | 11 | 6.4 | 15.1 | 5.1 | PTC 1-5′ | 47, XX, del (6), del (21), +21 | Died |
| 24 | 44 | Ex2 | Del | 23 | 4705 | Frameshift and introduction of stop codon | 13 | 8.3 | N/A | N/A | PTC 1-5′ | 48, XX, del (6) (q?13q?21) +21c, +21 [5] | CCR |
| 25 | 50 | Ex2 | Del | 2 | 4638 | Frameshift and introduction of stop codon | 12 | 7.3 | N/A | N/A | PTC 1-5′ | 50, XY, +10, +21c, +21, +22 [6] | CCR |
| 26‡ | 16 | N/D | Dup | 10 | 4710 | Frameshift and introduction of stop codon | 11 | 3.5 | 11.9 | 30 | PTC 1-5′ | 47∼49,XY, +?19,+21c, +21[cp7]/47, XY,+21c[5] | CCR |
| 27 | 20 | N/D | Dup | 15 | 4715 | Frameshift and introduction of stop codon | 15 | 6.4 | 7.6 | 60 | PTC 1-5′ | 47,XY,+21c 2/47, idem,t(4;15), (q?21;q?21), del(7)(q?31q?33) | Unknown |
| 28 | 8.5 | N/D | Del | 1 | 4698 | Frameshift and introduction of stop codon | 8 | 4 | 13.4 | 70 | PTC 1-5′ | N/A | Unknown |
| 29 | 31 | N/D | Del | 5 | 4704 | Frameshift and introduction of stop codon | 13 | 3.9 | 11.2 | 47 | PTC 1-5′ | 47,XX,der(5)t(1;5) (q12;p14)?del(1) (q23q25),+21c[4]/48, XX,+8,der (12)t(1;12)(q12;p12), +21c[2]/47, XX,+21c[9] | Unknown |
| 30 | 93 | N/D | Del | 2 | 4638 | Frameshift and introduction of stop codon | 24 | 26.4 | 9.1 | 147 | PTC 1-5′ | 47,XY,+21c[25] | Unknown |
| 31 | 39 | N/D | Del | 2 | 4638 | Frameshift and introduction of stop codon | 27 | 8.5 | 8.5 | 26 | PTC 1-5′ | N/A | Unknown |
| 32‡ | 4.5 | N/D | Del | 2 | 4604 | Frameshift and introduction of stop codon | 13 | 1 | 7.7 | 107 | PTC 1-5′ | N/A | Unknown |
| 33 | 19.5 | N/D | Ins + Dup | 2 + 14 | 4722 | Frameshift and introduction of stop codon | 21 | 26.4 | 7.2 | 151 | PTC 1-3′ | N/A | Unknown |
| 34 | 50.5 | N/D | Ins | 1 | 4768 | Frameshift and introduction of stop codon | 28 | 1.2 | 8 | 109 | Splice | 47∼48,XX,?add (5)(p1?5),−8, +21,+21c, +mar1, +mar2[cp8]/47, XX,+21c | Unknown |
| 35 | 10.5 | N/D | Point | 1 | 4768 | Loss of splice acceptor site | 35 | 3.4 | 10.4 | 17 | Splice | N/A | Unknown |
| 36 | 36 | N/D | Dup | 5 | 4734 | Frameshift and introduction of stop codon | 14 | 12.4 | 7.2 | 144 | PTC 1-3′ | 47,XX,+21c | Unknown |
| 37 | 34 | N/D | Del | 22 | 4704 | Frameshift and introduction of stop codon | 22 | 15.1 | 9.6 | 257 | PTC 1-5′ | 48,XY,+8,+21c | Unknown |
| 38 | 55 | N/D | Del + Ins | 31 + 6 | 4783 | Loss of splice donor site | 31 | 1.9 | 6.9 | 22 | Splice | N/A | Unknown |
| 39 | 35 | N/D | Dup | 3 | 4737 | Frameshift and introduction of stop codon | 31 | 4.2 | 9.2 | 20 | PTC 1-3′ | N/A | Unknown |
| 40 | 38 | N/D | Del + ins | 24 + 4 | 4683 | Frameshift and introduction of stop codon | 31 | 6.1 | 9.8 | 16 | PTC 1-5′ | N/A | Unknown |
| 41 | 43 | N/D | Dup | 21 | 4736 | Frameshift and introduction of stop codon | 13 | 4.9 | 6.7 | 38 | PTC 1-3′ | N/A | Unknown |
| 42 | 15 | N/D | Dup | 4 | 4668 | Frameshift and introduction of stop codon | 20 | 8.1 | 12.7 | 219 | PTC 1-5′ | 47,XX,+21c | Unknown |
| 43 | 16 | N/D | Dup | 7 | 4717 | Frameshift and introduction of stop codon | 36 | 160 | 5 | 1.5 | PTC 1-5′ | 47,XY,+21c,[3]/47, idem,t(1;8)(q32;q22) [2]/48,idem,t(1;8) (q32;q22),+der(8)t (1;8)(q32;q22) [10}] | Unknown |
| 44 | 24 | N/D | Ins | 5 | 4708 | Frameshift and introduction of stop codon | 10 | 6.88 | 9.7 | 35 | PTC 1-5′ | N/A | Unknown |
| 45 | 11 | N/D | Point | 1 | 4956 | Synonymous mutation | 16 | 2.5 | 8.1 | 101 | Unknown | N/A | Unknown |
| 46 | 28 | N/D | Ins | 1 | 4736 | Frameshift and introduction of stop codon | 31 | 12.9 | 3.1 | 76 | PTC 1-3′ | N/A | Unknown |
| 47 | 14 | N/D | Point | 1 | 4535 | Unknown | 31 | 22 | 8.5 | 37 | Unknown | 48, XY, +14, +21 | Unknown |
| 48 | 13 | N/D | Del | 9 | 4546 | Loss of Start codon | 19 | 6.2 | 10.6 | 131 | 1st Met | 48,XY,i(7)(q10), t(11;11)(p15;q13), +21,+21[13]/47,XY,+21[2] | Unknown |
| 49 | 15 | N/D | Point | 1 | 4768 | Loss of splice acceptor site | 12 | 5 | 12.1 | 45 | Splice | N/A | Unknown |
| 50 | 15 | N/D | Del | 7 | 4702 | Frameshift and introduction of stop codon | 20 | 2.74 | 10.7 | 110 | PTC 1-5′ | 50,XX,+8, +14,+21,+21c, inc[5]/47, XX,+21c[7] | Unknown |
| 51‡ | 16 | N/D | Point | 1 | 4552 | Nonsynonymous change in amino acid sequence | 12 | 3.4 | 11.1 | 27 | Unknown | 47,XY,?der(11) (q24∼25), +21c[2]/47, XY,+21c[8] | Unknown |
| 52 | 9 | N/D | Del + ins | 2 + 1 | 4619 | Frameshift and introduction of stop codon | 15 | 3.5 | 9.2 | 47 | PTC 1-5′ | 46,XY,i(7) (q10),der(21;21) (q10;q10)c, +21[6]/46,XY,der(21;21) (q10;q10)c, +21[5] | Unknown |
| 53 | 12 | N/D | Del + ins | 3 + 1 | 4654 | Frameshift and introduction of stop codon | 7 | 2.9 | 3.3 | 17 | PTC 1-5′ | 48,XX,der(1) (p?q?),+21c, +mar[14]/47,XX,+21c[4] | Unknown |
| 54 | 10.5 | N/D | Dup | 20 | 4727 | Frameshift and introduction of stop codon | 16 | 4.8 | 12 | 59 | PTC 1-3′ | N/A | Unknown |
| 55 | 17 | N/D | Not Found | Unknown | Unknown | Unknown | 21 | 5.9 | 11.4 | 42 | Unknown | 47,XX,+21c | Unknown |
| 56 | N/A | N/D | Dup | 16 | 4734 | Frameshift and introduction of stop codon | N/A | N/A | N/A | N/A | PTC 1-3′ | N/A | Unknown |
| 57 | 23 | N/D | Point | 1 | 4767 | Loss of splice acceptor site | 26 | 5.1 | 13.3 | 11 | Splice | 48∼50,XY, +8,+?14, +21,+21c[cp6]/47,XY,+21c[4] | Unknown |
| 58 | 21 | N/D | Point + Del | 1 + 1 | 4637 + 4639 | Frameshift and introduction of stop codon | 37 | 6.3 | 11.4 | 13.7 | Unknown | 47,XX,?del(2)(q35), del(7)(p12),+21c[7]/47,XX,+21c[5] | Unknown |
| 59 | 13 | N/D | Del | 1 | 4698 | Frameshift and introduction of stop codon | 11 | 6.3 | 10.3 | 84 | PTC 1-5′ | 47,XY,+21c[9]; 48,XY,del(5) (p11p14),r(7) (p22q11), +8,+21c[7] | Unknown |
| 60 | 35 | N/D | Del | 1 | 4548 | Unknown | 11 | 16 | 8 | 38 | Unknown | N/A | Unknown |
| 61 | 7 | N/D | Del | 1 | 4642 | Frameshift and introduction of stop codon | 15 | 1.8 | 9.3 | 43 | PTC 1-5′ | 47,XY,+21[1] | Unknown |
| 62 | 1 | N/D | Not Found | Unknown | Unknown | Unknown | 19 | 2.6 | 5.8 | 109 | Unknown | N/A | Unknown |
| 63 | 20.5 | N/D | Del + ins | 4 + 2 | 4656 | Frameshift and introduction of stop codon | 11 | 3.7 | 10.7 | 130 | PTC 1-5′ | 48,XX,+8,+21c[3]/47,XX,+21c[8] | Unknown |
| 64 | 2.5 | N/D | Del | 2 | 4656 | Frameshift and introduction of stop codon | 32 | 4.8 | 7.4 | 17 | PTC 1-5′ | 47,XX,+21c[6] | Unknown |
| 65 | 46 | N/D | Dup | 16 | 4719 | Frameshift and introduction of stop codon | 20 | 5.6 | 12 | 19 | PTC 1-3′ | N/A | Unknown |
| 66 | 19.5 | N/D | Point | 1 | 4767 | Loss of splice acceptor site | 32 | 5.46 | 13.6 | 52 | Splice | N/A | Unknown |
| 67 | 2 | N/D | Del | 1 | 4737 | Frameshift and introduction of stop codon | 26 | 3.3 | 7.1 | 69 | PTC 1-3′ | N/A | Unknown |
| 68 | 16 | N/D | Del | 2 | 4638 | Frameshift and introduction of stop codon | 35 | 2.8 | 9.5 | 20 | PTC 1-5′ | 48,XX,+8,+21c | Unknown |
| 69 | 23 | N/D | Del | 2 | 4638 | Frameshift and introduction of stop codon | 13 | 3.2 | 9 | 25 | PTC 1-5′ | 47,XY,+21c[10] | Unknown |
| 70 | 76 | N/D | Del | 2 | 4638 | Frameshift and introduction of stop codon | 17 | 23 | 6.5 | 28 | PTC 1-5′ | 46,XY,t(4;13) (q33∼34;q14), der(5)t(5;7) (p14∼15;q12), −7,+21c[cp11] | Unknown |
| 71 | 3.5 | N/D | Del | 2 | 4656 | Frameshift and introduction of stop codon | 13 | 3.3 | 11.4 | 46 | PTC 1-5′ | 47,XX,+21c[17] | Unknown |
| 72 | N/A | N/D | Point | 1 | 4637 | Unknown | N/A | N/A | N/A | N/A | Unknown | N/A | Unknown |
| 73 | 20 | N/D | Point | 1 | 4713 | Synonymous mutation | 19 | 4.1 | 9.5 | 13 | Unknown | 48∼49,XY,+21, +21c,inc[cp2]/47,XY,+21c[12] | Unknown |
| 74 | 42 | N/D | Not Found | Unknown | Unknown | Unknown | 24 | 5.4 | 10.3 | 21 | Unknown | 47,XY,+21c, ?der(22)(p12) [5]/47,XY, +21c[15] | Unknown |
| 75‡ | 25 | N/D | Dup | 13 | 4744 | Frameshift and introduction of stop codon | 26 | 2.7 | 11.8 | 20 | PTC 1-3′ | 47,XY,+21.ish 21qter(D21 S1446 × 3) | Unknown |
| 76 | 96 | N/D | Point | 1 | 4737 | Nonsynonymous change in amino acid sequence | 22 | 124 | 5.9 | 21 | Unknown | 47,X,−X, del(6)(q13q16), +21c,+21 | Unknown |
| 77 | 10.5 | N/D | Not Found | Unknown | Unknown | Unknown | 17 | 5.2 | 10.6 | 82 | Unknown | 47,XX,add(16) (p13),+21c[5]/47,XX,+21c[5] | Unknown |
| 78 | 26 | N/D | Point | 1 | 4956 | Synonymous mutation | 20 | 2 | 6.4 | 130 | Unknown | 48,XY,der(8)t (1;8)(q21; p22∼23), +11,+21c[7]/47,XY,+21c[8] | Unknown |
| 79 | 11 | N/D | Point | 1 | 4768 | Loss of splice acceptor site | 43 | 6.35 | 12 | 70 | Splice | N/A | Unknown |
| 80 | 13 | N/D | Point | 1 | 4633 | Nonsynonymous change in amino acid sequence | 16 | 3.3 | 10.5 | 84 | Unknown | N/A | Unknown |
| 81‡ | 26 | N/D | Del | 2 | 4638 | Frameshift and introduction of stop codon | 11 | 3.4 | 10.2 | 28 | PTC 1-5′ | 47,XY,+21c[15] | Unknown |
| 82 | 6 | N/D | Ins | 2 | 4725 | Frameshift and introduction of stop codon | 14 | 4 | 10.2 | 70 | PTC 1-3′ | N/A | Unknown |
| 83 | 4 | N/D | Dup | 6 | 4714 | Frameshift and introduction of stop codon | 8 | 8.75 | 11.4 | 69 | PTC 1-5′ | N/A | Unknown |
| 84 | N/A | N/D | Dup | 48 | 4771 | Frameshift and introduction of stop codon | N/A | N/A | N/A | N/A | PTC 1-3′ | N/A | Unknown |
| 85 | 17.5 | N/D | Del | 2 | 4638 | Frameshift and introduction of stop codon | 11 | 4.7 | 9.3 | 83 | PTC 1-5′ | N/A | Unknown |
| 86 | 45 | N/D | Dup | 10 | 4723 | Frameshift and introduction of stop codon | 14 | 11.5 | 8.6 | 44 | PTC 1-3′ | N/A | Unknown |
| 87 | 70.5 | N/D | Del | 2 | 4638 | Frameshift and introduction of stop codon | 16 | 32.5 | 7.9 | 103 | PTC 1-5′ | 46,XY[22]. nuc ish 8q22(ETOx2), 21q22(AML1 × 2), 13q14(RB1 × 2), 13q34 × 2), 11q23(MLLx2)[100] | Unknown |
| 88 | 31 | N/D | Del | 2 | 4638 | Frameshift and introduction of stop codon | 29 | 11.4 | 13.2 | 57.4 | PTC 1-5′ | 46,XX,del (9)(q?13q33), der(19)t(1;19) (q31;p13), der(21;21) (q10;q10)?c | Unknown |
| 89 | 7.5 | N/D | Del | 22 | 4706 | Frameshift and introduction of stop codon | 17 | 7.8 | 11.1 | 27 | PTC 1-5′ | 48,XY,add(7) (p2?2),+8, del(13)(q1?4), +21c[7]/46, XY,−21,+21c or 46,XY[3]/47,XY,+21c[2] | Unknown |
| 90 | 33 | N/D | Del + ins | 2 + 6 | 4707 | Frameshift and introduction of stop codon | 8 | 4.6 | 7.5 | 3 | PTC 1-5′ | N/A | Unknown |
| 91‡ | 24 | N/D | Ins | 1 | 4733 | Frameshift and introduction of stop codon | 11 | 3 | 7.1 | 57 | PTC 1-3′ | 48,X,add(Y) (q12),+11, +21c[8]/47,XY, +21c[7] | Unknown |
| 92 | 28 | N/D | Dup | 10 | 4719 | Frameshift and introduction of stop codon | 22 | 5.2 | 13.7 | 143 | PTC 1-3′ | N/A | Unknown |
| 93 | 16 | Ex2 | Not Found | N/A | N/A | N/A | 1 | 1.5 | 10.4 | 13 | Unknown | 47, XY, +21c | Died |
| 94 | 30 | Ex2 | Not Found | N/A | N/A | N/A | 15 | 14.47 | 4.1 | 38 | Unknown | 47, XX, t(3;3)(q21;q26), t(3;3)(p2?3;q13.3), +21c[13]/47, idem, der(3)t(3;3)add(3)(q29)[7] | CCR |
| 95 | 20 | Ex2 | Not Found | N/A | N/A | N/A | 17 | 11 | 9.9 | 52 | Unknown | 47, XY, der(11)t (q23p15)t (1:11)(q23;q85), +21c | CCR |
| 96 | 10 | Ex2 | Not Found | N/A | N/A | N/A | 11 | N/A | 8.6 | 86 | Unknown | 47, XY, del (7), +21 | Unknown |
| 97 | 21 | Ex2 | Not Found | N/A | N/A | N/A | 33 | 4.9 | 6 | 15 | Unknown | 48, XY, +21, +21c | CCR |
| 98 | 20 | Ex2 | Not Found | N/A | N/A | N/A | 15 | 15.2 | 9.9 | 18 | Unknown | 47 XX, ?dup(3) (p23p25), del(8) (p11.2p21), der(15)t(1:15) (q21;p11), del(16)(q22q24, del(20) | CCR |
| 99 | 6 | Ex2 | Not Found | N/A | N/A | N/A | 23 | 5.1 | 7.2 | 49 | Unknown | N/A | Unknown |
| 100 | 20 | Ex2 | Not Found | N/A | N/A | N/A | 38 | N/A | N/A | N/A | Unknown | 46, XX, +21, −7 | Unknown |
| 101 | 38 | Ex2 | Not Found | N/A | N/A | N/A | 21 | 11.9 | 7.3 | 34 | Unknown | 47, XX, +21c | CCR |
| 102 | 10 | Ex2 | Not Found | N/A | N/A | N/A | 10 | 3.9 | 10.9 | 65 | Unknown | N/A | Unknown |
| 103 | 20 | Ex2 | Not Found | N/A | N/A | N/A | 12 | 3.8 | 8.6 | 16 | Unknown | 47, XX, t(1;22)(q23;p13), +21c[9]/ 47, XX, +21c[4] | CCR |
WBC indicates white blood cell; Hb, hemoglobin; Plt, platelet; Del, deletion of nucleotides; CCR, complete clinical remission; Dup, duplication of nucleotides; N/A, not available; Ins, insertion of nucleotides; Point, point substitution; and N/D, not done.
Exon number containing the mutation as defined by WAVE analysis.
Nucleotide 0 is the first nucleotide of GATA1 exon 1 including exons and introns. NCBI reference NT_079573.4 (Homo sapiens chromosome X genomic contig, starting position 11496706) was used.
Patients with TMD progressed to ML-DS.
GATA1 mutations were analyzed by WAVE and direct sequencing of PCR products. GATA1 sequence mutations were determined in 118 of 134 patients with TMD (88.1%) and in 88 of 103 patients with ML-DS (85.4%). Mutations were detected by WAVE in a further 9 and 11 patients, respectively (Tables 1–2). The main reason for failure to detect mutations was low blast count. Alternative techniques such as high-resolution melt analysis or nested PCR may be able to detect mutations in some cases. The lower limit of blasts that allowed successful mutation detection was 0.5% (supplemental Methods). However, in one patient (blast count, 42%) failure to detect mutation suggests an uncommon mutation involving sequence outside the genomic area, spanning the PCR, or a deletion inside this area, affecting the primer annealing site.
Relative positions of sequence mutations are shown in Figure 1A. Insertion/deletion/duplications comprised 78% of mutations in both TMD and ML-DS (Figure 1B), consistent with previous reports.12 Point mutations were detected in 21% and 22% of TMD and ML-DS samples, respectively. Substitutions were rare, uniquely detected in 1% of patients with TMD. Therefore, there is little difference in mutational spectrum between TMD and ML-DS. Thirteen patients with TMD are known to have progressed to ML-DS. In these samples the spectrum of mutations was similar to the TMD group that did not progress to ML-DS (insertion/deletion/duplications, 77%; point mutations, 23%). Eight patients had paired TMD and follow-up ML-DS samples. The same mutation was present in both TMD and ML-DS for 7 of 8 samples, similar to previous observations.12,14
Position and types of GATA1 sequence mutations found in TMD and ML-DS samples. (A) A schematic diagram of GATA1 showing the positions and types of the sequence mutations found in TMD and ML-DS samples. Each arrow represents a different patient. (B) Diagram showing the mutational spectrum of patients with TMD and with ML-DS and (C) the effect that these mutations have on the sequence of GATA1.
Position and types of GATA1 sequence mutations found in TMD and ML-DS samples. (A) A schematic diagram of GATA1 showing the positions and types of the sequence mutations found in TMD and ML-DS samples. Each arrow represents a different patient. (B) Diagram showing the mutational spectrum of patients with TMD and with ML-DS and (C) the effect that these mutations have on the sequence of GATA1.
Predicted consequence of mutations was similar for both TMD and ML-DS (Figure 1C). Most mutations inserted a premature termination codon (PTC) either by introducing a stop codon or frameshift. Mutations affecting the splice site at GATA1 exon 2 exon/intron boundary were next most frequent. In some cases (3 TMD and 3 DS-ML) where a point mutation occurred in intronic sequence, predicted consequence was unclear. They may occur in splice site regulatory elements and may affect gene splicing.18 In 13 patients with TMD who progressed to ML-DS predicted consequences in the protein were similar.
Characterizing the sequence of GATA1 mutations provides an opportunity to develop patient mutation-specific quantitative PCR analysis to monitor resolution of TMD, persistence/re-emergence of GATA1 mutant clone leading to ML-DS, and therapy response in patients with ML-DS.13 Direct sequencing of DNA from blasts that underwent FACS was successful in identifying sequence mutation in 19 patients, whereas direct sequencing of DNA from unfractionated samples failed. In 20 cases it was necessary to subclone the GATA1 PCR product to pinpoint mutation sequence.
A previous study divided GATA1 mutations in TMD and ML-DS into 2 classes on the basis of levels of GATA1s protein expression.15 Mutations affecting gene splicing, the start codon (1st Met) or that introduced a PTC in the 3′ end of exon 2 (PTC 1-3′) resulted in high GATA1s protein levels; whereas a PTC in the 5′ end of exon 2 (PTC 1-5′) or in exon 3.1 (PTC-2) resulted in low GATA1s expression.15 Patients with TMD with mutations predicted to result in low GATA1s protein expression were more probable to develop ML-DS because this type of mutation had significantly higher incidence in ML-DS.15 We asked whether this was true for our sample cohort. No significant differences were found between numbers of samples in each of the mutation types when TMD and ML-DS were compared (supplementary Table 1; 1st Met, P = .7011; splice errors, P = .6741; PTC 1-3′, P = .3388; PTC 1-5′, P = .3021; PTC-2, P = .2667). Similarly, we found no significant increase in predicted GATA1s low-expressing mutations in ML-DS samples versus TMD (P = .5534; Tables 1–2). In 12 TMD samples that progressed to ML-DS whereby we obtained a sequence mutation predicted to affect protein expression, equal numbers were predicted to result in high and low GATA1s protein expression (ie, 6 of each). GATA1s cDNA qPCR for 36 TMD and 20 ML-DS patient samples showed no significant differences in GATA1s mRNA expression in patients with PTC 1-3′, Splice, PTC 1-5′, or unknown effect mutations (supplemental Figure 1).
Cytogenetic data were available for 31 patients with TMD and 68 patients with ML-DS (Tables 1–2). In neonates with TMD, no significant correlation was observed between not progressing to ML-DS and presenting with T21 as the only cytogenetic abnormality (P = .2533; supplemental Table 2). Therefore, karyotype does not predict patients who will progress to ML-DS.
In conclusion, simple techniques detect GATA1 mutations in most patients TMD and patients with ML-DS. It is not possible to predict progression of TMD to ML-DS on the basis of type of GATA1 mutation, at least in mainly white patients.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Esther Edlundh-Rose for technical help processing patient samples.
This work was supported in Oxford by the Leukemia & Lymphoma Research Specialist Program (award 08030; K.A.A., P.V., and I.R.), NIHR Biomedical Research Centres funding scheme (P.V.), the Medical Research Council Disease Team Award (P.V.), the MRC Molecular Hematology Unit (P.V.), and the Imperial College London Comprehensive Biomedical Research Center (I.R.). This work was supported in Hannover by German Research Foundation (grant DFG RE2580/1-1) and by a grant from the Madeleine-Schickedanz Leukemia Foundation.
Authorship
Contribution: K.A.A. performed and analyzed experiments and wrote the manuscript; A.N. and K.R. designed, performed, and analyzed experiments and data; C.G. performed experiments, analyzed data, and compiled figures; K.B., C.v.N., and A.K. performed and analyzed experiments; E.M. analyzed data; and P.V., D.R., J-H.K., H.H., and I.R. designed experiments, analyzed data, and wrote or reviewed the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
A complete list of the participants of the International ML-DS Study Group can be found in the supplemental Appendix.
Correspondence: Paresh Vyas, Molecular Haematology Unit, Weatherall Institute of Molecular Medicine, John Radcliffe Hospital, Oxford OX3 9DS, United Kingdom; e-mail: paresh.vyas@imm.ox.ac.uk; or Dirk Reinhardt, Department of Pediatric Hematology/Oncology, Medical School Hannover, Carl Neuberg Str 1, 30625 Hannover, Germany; e-mail: reinhardt.dirk@mh.hannover.de.
References
Author notes
K.A.A., K.R., C.G., and A.N. contributed equally to the work and are joint first authors.

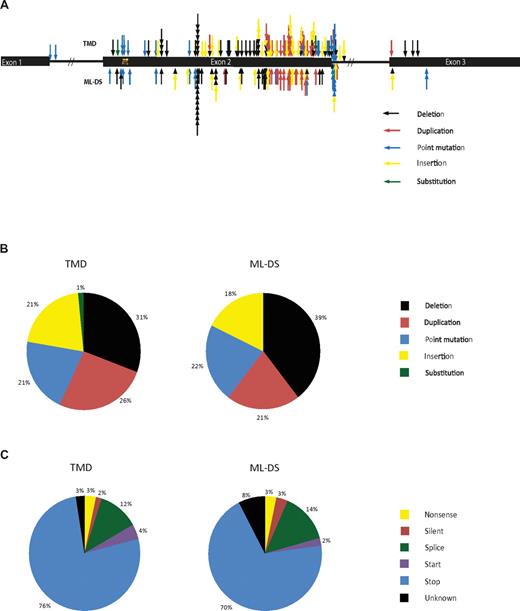
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal