Abstract
Fanconi anemia (FA) nuclear core complex is a multiprotein complex required for the functional integrity of the FA-BRCA pathway regulating DNA repair. This pathway is inactivated in FA, a devastating genetic disease, which leads to hematologic defects and cancer in patients. Here we report the isolation and characterization of a novel 20-kDa FANCA-associated protein (FAAP20). We show that FAAP20 is an integral component of the FA nuclear core complex. We identify a region on FANCA that physically interacts with FAAP20, and show that FANCA regulates stability of this protein. FAAP20 contains a conserved ubiquitin-binding zinc-finger domain (UBZ), and binds K-63–linked ubiquitin chains in vitro. The FAAP20-UBZ domain is not required for interaction with FANCA, but is required for DNA-damage–induced chromatin loading of FANCA and the functional integrity of the FA pathway. These findings reveal critical roles for FAAP20 in the FA-BRCA pathway of DNA damage repair and genome maintenance.
Introduction
Fanconi anemia (FA) is characterized by developmental defects, bone marrow (BM) failure, and higher predisposition to both hematologic and nonhematologic cancers.1 The primary reason for morbidity and mortality in FA patients is progressive BM failure because of the depletion of hematopoietic stem cells.1,2 Although BM transplant significantly reduces hematologic deficiencies and improves outcomes, FA patients still have a greater risk of developing myelodysplastic syndrome, acute myeloid leukemia, and other solid tumors, such as squamous cell carcinomas.1-3 Diagnostic features of the disease are increased chromosomal breaks and hypersensitivity of FA cells to DNA interstrand cross-linking (ICL) agents.1
FA is a genetically heterogeneous disease, comprising 15 complementation groups; the genes mutated in these groups have been identified.3 Eight of the FA proteins (FANCA, -B,-C, -E, -F, -G, -L, and -M) and 5 associated factors (FAAP100, FAAP24, HES1, MHF1, and MHF2) form the FA nuclear core complex. The core complex is required for mono-ubiquitination of FANCD2-FANCI dimer on DNA damage, which results in activation of downstream DNA repair and tolerance reactions.3,4 The downstream FA proteins include FANCD1/BRCA2, FANCJ/BACH1, FANCN/PALB2, FANCO/RAD51C, and FANCP/SLX4, along with FA-associated proteins FAN1, RAD18, and RAD51.3 Together, these proteins function in the “FA-BRCA” pathway, which facilitates DNA cross-link repair and coordinates other DNA damage-responsive events, thereby stabilizing stalled replication forks, conveying signals to DNA checkpoint pathways, and facilitating recovery of replication forks.3,4
Discovery of several new members of the core complex in the past decade contributed much to the understanding of this pathway. Despite the isolation and characterization of several core complex members, a clear understanding of the core complex is far from clear. To better understand the functions of the core complex, it is necessary to isolate and characterize all core-complex proteins and associated subcomplexes. Our previous attempt to better define the composition of the core complex led to the discovery of FANCB, FANCL, FANCM, FAAP100, MHF1, and MHF2.5-9 In this study, we report the isolation and characterization of a novel core complex protein, FAAP20.
Methods
Cloning and constructs
The pMIEG3 retroviral vector was used for protein expression in mammalian cells.9 The pMYFP retroviral vector was made by replacing the EGFP of pMIEG3 with yellow fluorescent protein gene (YFP). Mammalian expression constructs and their construction strategy are discussed in detail in supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Mammalian cells and cell culture procedures
Adherent epithelial cells were cultured in Dulbecco modified Eagle medium (DMEM) and lymphoblast cells were cultured in Iscove modified Dulbecco medium (IMDM) with 10% fetal bovine serum (FBS) and 5% CO2. Details of the cell lines are described in supplemental Methods.
Antibodies
Rabbit FAAP20 polyclonal antibody was raised against fusion protein containing full-length FAAP20 with maltose binding protein (MBP). The fusion protein was expressed in and purified from Escherichia coli, in accordance with the manufacturer's protocols. Anti-FANCM, -FANCA, -FANCL, -FANCG, -FAAP100, -FANCB, -FANCD2, -actin, -ATR, and -H2A antibodies have been previously described.9-11
Purification and mass spectrometry analyses of protein complexes
FA core complex was isolated from nuclear extracts of HeLaS3:His6FANCA/FLAGFAAP100 or HeLaS3:His6-FLAGFAAP20 cells, using 2-step affinity chromatography; a schematic of the purification procedure is presented in supplemental Figure 1B. Core-complex proteins were identified by mass spectrometry (MS) analysis, using a procedure previously described.12
siRNA or shRNA knockdown
For transient knockdown of FAAP20, 2 independent siRNA oligos were used, siFAAP20#1 (5′-CGGAGCCCACUGAAGUCUUUU-3′) or siFAAP20#2 (5′-CCAUGUGCCAGAAGGAGUUUU-3′), and also siControl (D-001810-01-20), all purchased from Dharmacon. Oligos were transfected using Lipofectamine 2000, according to the manufacturer's protocol. More than 70% knockdown of FAAP20 was observed within 24 hours posttransfection. See supplemental Methods for shRNA constructs.
In vitro ubiquitin-binding assay
Ubiquitin-binding assay was carried out as previously described.13 Briefly, M2-agarose-bound His6-FLAGFAAP20 wild-type (WT) or mutants, or FLAGRAD18 were incubated for 4 hours at 4°C with 7 μg mono-Ub, Ub-K48 or Ub-K63 (Boston-Biochem) in 200 μL of binding buffer (50mM Tris, pH 7.5, 150mM NaCl, 10% glycerol, 0.1% triton, 5mM β-mercaptoethanol, and 2mM N-ethylmaleimide. Resin was washed 5 times with binding buffer, and the proteins eluted using 3X-FLAG peptide (Sigma-Aldrich). To study the effect of FANCA on FAAP20 ubiquitin binding, purified MBPFANCA1065-1455 was incubated with M2-agarose-bound His6-FLAGFAAP20 overnight at 4°C. After the binding reaction, unbound FANCA was removed by washing 5 times with binding buffer. The resulting FAAP20-FANCA complex was incubated with ubiquitin and processed as previously described. Samples were boiled with 2X-SDS loading buffer, resolved in a 10%-20% gradient Tricine Gel (Invitrogen), and immunoblotted with ubiquitin antibody (Boston-Biochem).
Results
Identification of FAAP20 as a novel integral component of the FA nuclear core complex
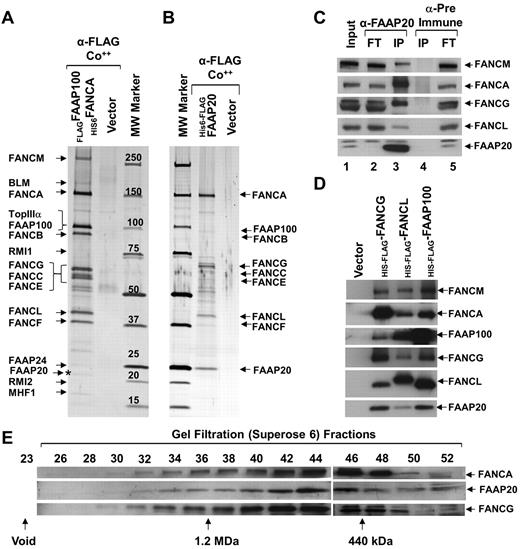
To isolate the FA nuclear core complex to homogeneity, we generated a HeLaS3 cell line stably expressing N-terminally histidine-tagged FANCA (His6FANCA) and N-terminally FLAG-tagged FAAP100 (FLAGFAAP100; supplemental Figure 1A). To identify FA nuclear core-complex proteins, we used 2-step affinity purification of His6FANCA and FLAGFAAP100, from nuclear extracts, coupled with mass spectrometry (MS data available on request; supplemental Figure 1B). In the resulting purified complex, we found known members of the BLM complex and FA nuclear core complex (Figure 1A). In addition, we identified a 20 kDa polypeptide, which we named FAAP20 (FA-associated protein, 20 kDa; Figure 1A). Database searches revealed that FAAP20 is similar to an uncharacterized protein named C1orf86 (chromosome 1 open reading frame 86; NM_182533). FAAP20 was consistently found by MS analysis in several independent purifications performed as previously described, under normal conditions and also in the presence of replication blockers, such as hydroxyurea (HU), or ICL agents, such as mitomycin C (MMC). To confirm that FAAP20 is part of the core complex, we generated a cell line stably expressing FAAP20 containing an N-terminally–tagged histidine (6X) followed by a FLAG tag in tandem (His6-FLAGFAAP20). Using the previously described method, we purified core complex associated with ectopically expressed His6-FLAGFAAP20 (Figure 1B). MS and immunoblot analysis of the purified FAAP20 containing complex identified known core-complex proteins, suggesting that ectopically expressed FAAP20 was associated with the core complex (Figure 1B, supplemental Figure 1C).
FAAP20 is a novel component of the FA nuclear core complex. (A) Silver-stained gel showing polypeptide bands isolated by 2-step purification, first purification using anti-FLAG (α-FLAG) and second purification using talon beads (Co++), from nuclear extracts of HeLaS3:vector or HeLaS3:His6FANCA/FLAG-FAAP100 cells. Polypeptides identified by MS analysis are indicated, including the novel 20 kDa polypeptide, FAAP20 (asterisk). (B) Silver-stained gel showing the polypeptide bands purified from nuclear extracts of HeLaS3:vector or HeLaS3:His6-FLAGFAAP20 using 2-step purification as described in panel A. Polypeptides identified by MS analysis are indicated. (C) Immunoblot of immunoprecipitated HeLa cell extract showing core-complex proteins present in IPs with anti-FAAP20 (lane 3), but absent in IPs with pre-immune serum (lane 4). FT is the flow-thru fraction. (D) Immunoblot showing FAAP20 coprecipitated in FLAG-IPs from HeLaS3:His6-FLAGFANCG, HeLaS3: His6-FLAGFANCL, and HeLaS3:His6-FLAGFAAP100 and not in HeLaS3:vector. (E) Immunoblot showing identical gel filtration profiles on cofractionation of FANCA, FANCG, and FAAP20 using a superose 6 gel filtration column.
FAAP20 is a novel component of the FA nuclear core complex. (A) Silver-stained gel showing polypeptide bands isolated by 2-step purification, first purification using anti-FLAG (α-FLAG) and second purification using talon beads (Co++), from nuclear extracts of HeLaS3:vector or HeLaS3:His6FANCA/FLAG-FAAP100 cells. Polypeptides identified by MS analysis are indicated, including the novel 20 kDa polypeptide, FAAP20 (asterisk). (B) Silver-stained gel showing the polypeptide bands purified from nuclear extracts of HeLaS3:vector or HeLaS3:His6-FLAGFAAP20 using 2-step purification as described in panel A. Polypeptides identified by MS analysis are indicated. (C) Immunoblot of immunoprecipitated HeLa cell extract showing core-complex proteins present in IPs with anti-FAAP20 (lane 3), but absent in IPs with pre-immune serum (lane 4). FT is the flow-thru fraction. (D) Immunoblot showing FAAP20 coprecipitated in FLAG-IPs from HeLaS3:His6-FLAGFANCG, HeLaS3: His6-FLAGFANCL, and HeLaS3:His6-FLAGFAAP100 and not in HeLaS3:vector. (E) Immunoblot showing identical gel filtration profiles on cofractionation of FANCA, FANCG, and FAAP20 using a superose 6 gel filtration column.
We next wished to immunoprecipitate endogenous FAAP20 from HeLaS3 cells. To do this, we first generated an antibody against FAAP20 which proved suitable for immunoblot analysis of both His6-FLAGFAAP20 and endogenous FAAP20 (Figure 1C, supplemental Figure 1C). Using this antibody, we immunoprecipitated FAAP20 from HeLaS3- and HSC93-cell lysates (Figure 1C, supplemental Figure 1D). Immunoblot analysis revealed the presence of other known core-complex proteins – such as FANCM, FANCA, FANCG, and FANCL (Figure 1C, supplemental Figure 1D). Interestingly, the amount of immunoprecipitated FANCA and FANCG was greater than the amount of other core-complex proteins (eg, FANCM and FANCL; Figure 1C, supplemental Figure C-D). In addition, FANCA and FANCG were reduced in the flow-through fraction (Figure 1C), suggesting that FAAP20 may also exist in subcomplex with FANCA, and FANCG. In a reciprocal experiment, we immunoprecipitated FANCG, FANCL, and FAAP100 from nuclear extracts of HeLaS3: His6-FLAGFANCG, His6-FLAGFANCL, and His6-FLAGFAAP100, respectively. In each instance, FAAP20 was present in the immunoprecipitate (Figure 1D). The association between FAAP20 and core complex members was not because of DNA contamination of the lysate, because an immunoprecipitation (IP) done in the presence or absence of ethidium bromide, which precipitates DNA, were not visibly different (supplemental Figure 1E).14 These IP data uniformly suggest that FAAP20 is an integral component of the core complex.
To determine whether FAAP20 cofractionates with known core-complex proteins, we performed gel filtration experiments using superose-6. The gel filtration profile for FAAP20 was coincident with that for FANCA and FANCG (Figure 1F), suggesting that these 3 proteins exist in the same complex. The data from these 4 sets of experiments provide very strong evidence that FAAP20 is an integral component of FA nuclear core complex.
FAAP20 is unstable in the absence of FANCA
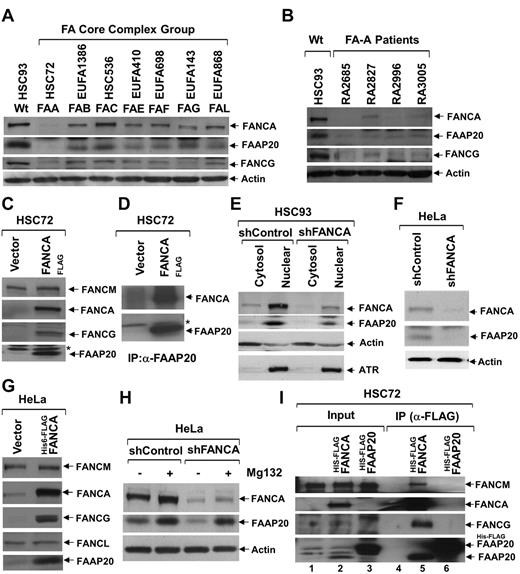
To check the fate of FAAP20 in the absence of other core-complex proteins, we examined levels of FAAP20 in total lysates of patient-derived FA cells from complementation groups defective in several core-complex proteins: HSC72 (FA-A), EUFA1386 (FA-B), HSC536 (FA-C), EUFA410 (FA-E), EUFA698 (FA-F), EUFA143 (FA-G), and EUFA868 (FA-L; Figure 2A). The immunoblots revealed varied amounts of FAAP20 in different complementation groups, with a negligible amount in FANCA deficient HSC72 cells (Figure 2A). To determine whether this is an artifact of the HSC72 cells, or if the lack of FANCA in these cells results in reduced levels of FAAP20, we obtained several patient-derived FA-A cell lines from the International Fanconi Anemia Registry, in which FANCA protein is reduced or absent (Figure 2B). Immunoblot analysis of total lysate from these FA-A cells, revealed FAAP20 levels that were low or negligible (Figure 2B). We hypothesized that the reduced levels of FAAP20 are because of the absence of FANCA. To test this hypothesis, we complemented the HSC72 cells with FANCAFLAG; the resulting overexpression of FANCA, lead to increased levels of FAAP20 protein (Figure 2C). In addition, FANCG levels, which are also reduced in HSC72 cells, were restored by FANCA overexpression (Figure 2C).15 These findings received further confirmation in FA-A fibroblast cells (PD220), in which like FANCA, FAAP20 levels are also reduced; in these cells, FANCA overexpression also led to increased expression of FAAP20 (supplemental Figure 2A). To confirm the reduced levels of FAAP20 in HSC72 cells, and to see whether overexpressed FANCA binds FAAP20, we performed IP/immunoblot analysis of total lysates from HSC72 and HSC72-complemented cells using the FAAP20 antibody. The immunoblots revealed negligible amounts of FAAP20 and overexpressed FANCA was able to copurify with FAAP20 (Figure 2D). To further confirm that FAAP20 is unstable in the absence of FANCA, and to determine the subcellular localization of FAAP20, we made cytosol and nuclear extracts from HSC93 WT lymphoblast cells stably expressing control shRNA (shControl) or shRNA targeted against FANCA (shFANCA; Figure 2E). Although FAAP20 was found in both compartments, its localization was predominantly nuclear (Figure 2E). Knockdown of FANCA resulted in reduced levels of both cytosol and nuclear FAAP20 (Figure 2E). These findings confirm the observation made in HSC72 cells that FAAP20 is unstable in the absence of FANCA.
FANCA is required for stability of FAAP20. (A) Immunoblot showing levels of FANCA, FAAP20, and FANCG in various FA-patient-derived cells defective in one of the complementation groups (indicated above each lane); actin serves as loading control. In the absence of FANCA, FAAP20 amounts were negligible. (B) Immunoblot showing levels of FANCA, FAAP20, and FANCG from HSC93 WT cells and various FA-A patient-derived cells obtained from IFAR. FAAP20 amounts were negligible in the absence of FANCA. (C) Immunoblot showing that FAAP20 and FANCG but not FANCM are stabilized in FANCA-overexpressing HSC72 cells. A nonspecific band is indicated by an asterisk. (D) Immunoblot showing levels of FANCA and FAAP20 in IPs of HSC72 and HSC72:FANCAFLAG cell extracts. Negligible amount of FAAP20 was immunoprecipitated from HSC72 cells in the absence of FANCA. A nonspecific band is indicated by an asterisk. (E) Immunoblot showing levels of FANCA and FAAP20 in cytosol and nuclear extracts of HSC93:shControl and HSC93:shFANCA cells; actin and ATR serve as controls. FANCA and FAAP20 are predominantly in the nuclear fraction, and FANCA knockdown results in reduced levels of FAAP20. (F) Immunoblot showing levels of FANCA and FAAP20 in total lysates of HeLa:vector and HeLa:shFANCA cells. Knockdown of FANCA results in decreased FAAP20. (G) Immunoblot of HeLa cell lysates showing that FAAP20 and FANCG, but not FANCL or FANCM, are induced by overexpression of FANCA. (H) Immunoblot showing levels of FANCA and FAAP20 in total lysates of HeLa:shControl and HeLa:shFANCA cells cultured in the presence (+) or absence (−) of Mg132 protease inhibitor. Inhibition of the proteasome pathway results in increased FAAP20 levels, despite low levels of FANCA. (I) Immunoblot showing core-complex proteins in input and IP samples from HSC72 cells stably expressing His6-FLAGFANCA or His6-FLAGFAAP20. FAAP20 induced on FANCA expression was able to coprecipitate with FANCA and FAAP20 failed to interact with other core-complex proteins in the absence of FANCA.
FANCA is required for stability of FAAP20. (A) Immunoblot showing levels of FANCA, FAAP20, and FANCG in various FA-patient-derived cells defective in one of the complementation groups (indicated above each lane); actin serves as loading control. In the absence of FANCA, FAAP20 amounts were negligible. (B) Immunoblot showing levels of FANCA, FAAP20, and FANCG from HSC93 WT cells and various FA-A patient-derived cells obtained from IFAR. FAAP20 amounts were negligible in the absence of FANCA. (C) Immunoblot showing that FAAP20 and FANCG but not FANCM are stabilized in FANCA-overexpressing HSC72 cells. A nonspecific band is indicated by an asterisk. (D) Immunoblot showing levels of FANCA and FAAP20 in IPs of HSC72 and HSC72:FANCAFLAG cell extracts. Negligible amount of FAAP20 was immunoprecipitated from HSC72 cells in the absence of FANCA. A nonspecific band is indicated by an asterisk. (E) Immunoblot showing levels of FANCA and FAAP20 in cytosol and nuclear extracts of HSC93:shControl and HSC93:shFANCA cells; actin and ATR serve as controls. FANCA and FAAP20 are predominantly in the nuclear fraction, and FANCA knockdown results in reduced levels of FAAP20. (F) Immunoblot showing levels of FANCA and FAAP20 in total lysates of HeLa:vector and HeLa:shFANCA cells. Knockdown of FANCA results in decreased FAAP20. (G) Immunoblot of HeLa cell lysates showing that FAAP20 and FANCG, but not FANCL or FANCM, are induced by overexpression of FANCA. (H) Immunoblot showing levels of FANCA and FAAP20 in total lysates of HeLa:shControl and HeLa:shFANCA cells cultured in the presence (+) or absence (−) of Mg132 protease inhibitor. Inhibition of the proteasome pathway results in increased FAAP20 levels, despite low levels of FANCA. (I) Immunoblot showing core-complex proteins in input and IP samples from HSC72 cells stably expressing His6-FLAGFANCA or His6-FLAGFAAP20. FAAP20 induced on FANCA expression was able to coprecipitate with FANCA and FAAP20 failed to interact with other core-complex proteins in the absence of FANCA.
In HeLa cells, knockdown of FANCA resulted in reduced levels of FAAP20 (Figure 2F). Importantly, overexpression of FLAGFANCA in HeLa cells resulted in increased stabilization of FAAP20 and FANCG, but not other core-complex proteins (Figure 2G). This phenotype, increased levels of FAAP20, was unique to FANCA, because overexpression of other core-complex proteins in HeLa cells did not result in such a dramatic increase in FAAP20 (supplemental Figure 2B). Based on our observations that FAAP20 is unstable in the absence of FANCA, and overexpression of FANCA results in increased FAAP20 levels, we hypothesized that FAAP20 is degraded in the absence of FANCA, possibly via the proteasome-mediated degradation pathway. To test this hypothesis we cultured FANCA-knockdown HeLa cells in the presence or absence of proteasome inhibitor MG132. Inhibiting the proteasome pathway resulted in restoration of FAAP20 levels, even though FANCA levels were depleted by RNAi (Figure 2H). These data suggest that FANCA stabilizes FAAP20, and in the absence of FANCA, FAAP20 is degraded via the proteasome-mediated protein degradation pathway.
FANCA is required for interaction of FAAP20 with other core-complex proteins
To see whether FANCA is required for interaction of FAAP20 with other core-complex proteins, we overexpressed His6-FLAGFAAP20 in HSC72 cells (Figure 2I lane 3), and then used anti-FLAG to IP FAAP20 from total cell lysates. Interestingly, neither FANCM nor FANCG were apparent in immunoblot (Figure 2I lane 6), suggesting that FANCA is required for FAAP20 interaction with other core complex members. Moreover, HSC72 cells expressing His6-FLAGFAAP20 failed to correct the FANCD2 defect in these cells (supplemental Figure 2C).
C-terminal region of FANCA binds FAAP20
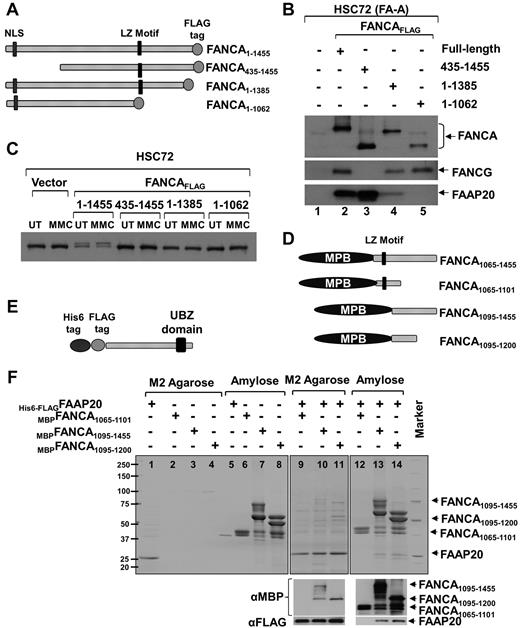
The experiments above demonstrate that higher amounts of FANCA and FANCG are complexed with FAAP20 (Figure 1C), FANCA, FANCG, and FAAP20 have similar gel filtration profiles (Figure 1E), FANCA, but not FANCG, is required for stability of FAAP20 (Figure 2), and absence of FANCA results in loss of FAAP20 interaction with other core-complex proteins (Figure 2I). Based on these observations, we hypothesized that FAAP20 interact directly with FANCA. To test this hypothesis, we made 3 deletion constructs of FANCA with a C-terminal FLAG tag and expressed them in HSC72 cells along with full-length FANCA (Figure 3A). Immunoblot analysis of FLAG-purified complex revealed that full-length FANCA interacts with both FANCG and FAAP20 (Figure 3B lane 2). Deletion of 435 amino acids (aa) from the N-terminus resulted in loss of FANCA interaction with FANCG,16 but not with FAAP20, suggesting that the N-terminus of FANCA is not required for binding FAAP20 (Figure 3B lane 3). Deletion of 70 aa from the C-terminus resulted in weak interaction that was completely abolished by extending the deletion to 393 aa, suggesting that the C-terminus region of FANCA is required for interaction of FAAP20 and FANCA (Figure 3B lanes 4-5). All 3 deletion constructs failed to activate the FA pathway, as indicated by their failure to mono-ubiquitinate FANCD2 (Figure 3C).
FAAP20 interact with FANCA in vivo and in vitro. (A) Schematic of FANCA deletion constructs expressed in HSC72 cells. Nuclear localization signal (NLS), leucine-zipper (LZ) motif and C-terminal FLAG tag are shown. (B) Immunoblot of FLAG IP showing that full-length FANCA and the N-terminal deletion interact with FAAP20, but the C-terminal deletion shows weak or no interaction. (C) Immunoblot showing FANCD2 mono-ubiquitination in MMC treated (MMC) or untreated (UT) HSC72 cells expressing various FANCA deletion fragments. Although full-length FANCA corrected the FANCD2 monoubiquitination defect in HSC72 cells, all 3 deletion constructs failed to do so. (D) Schematic of FANCA deletion constructs expressed in E coli cells. MBP is fused to N-terminus. (E) Schematic of FAAP20 full-length construct expressed in E coli. (F) Top panel: Coomassie-stained gel showing FAAP20 and FANCA purified from E coli cell lysate expressing His6-FLAGFAAP20 or MBPFANCA fragment or both using M2 agarose (FAAP20 target) or amylose (FANCA target) beads. MBPFANCA1065-1101 construct with the leucine-zipper motif failed to interact with FAAP20. MBPFANCA1095-1455 and MBPFANCA1095-1200 lacking the motif were able to interact with FAAP20. Bottom panel: immunoblot analysis using anti-MBP or anti-FAAP20 confirmed the identity of the bands as FANCA or FAAP20, respectively.
FAAP20 interact with FANCA in vivo and in vitro. (A) Schematic of FANCA deletion constructs expressed in HSC72 cells. Nuclear localization signal (NLS), leucine-zipper (LZ) motif and C-terminal FLAG tag are shown. (B) Immunoblot of FLAG IP showing that full-length FANCA and the N-terminal deletion interact with FAAP20, but the C-terminal deletion shows weak or no interaction. (C) Immunoblot showing FANCD2 mono-ubiquitination in MMC treated (MMC) or untreated (UT) HSC72 cells expressing various FANCA deletion fragments. Although full-length FANCA corrected the FANCD2 monoubiquitination defect in HSC72 cells, all 3 deletion constructs failed to do so. (D) Schematic of FANCA deletion constructs expressed in E coli cells. MBP is fused to N-terminus. (E) Schematic of FAAP20 full-length construct expressed in E coli. (F) Top panel: Coomassie-stained gel showing FAAP20 and FANCA purified from E coli cell lysate expressing His6-FLAGFAAP20 or MBPFANCA fragment or both using M2 agarose (FAAP20 target) or amylose (FANCA target) beads. MBPFANCA1065-1101 construct with the leucine-zipper motif failed to interact with FAAP20. MBPFANCA1095-1455 and MBPFANCA1095-1200 lacking the motif were able to interact with FAAP20. Bottom panel: immunoblot analysis using anti-MBP or anti-FAAP20 confirmed the identity of the bands as FANCA or FAAP20, respectively.
Because leucine-zipper motifs mediate protein-protein interactions, we hypothesized that the C-terminus region of FANCA, containing a leucine-zipper motif between aa residues 1069 and 1090, may interact with FAAP20.17 We therefore made a series of MBP-fused FANCA deletion constructs with and without the leucine-zipper motif (Figure 3D) to test a direct interaction between full-length His6 and FLAG-tagged FAAP20 (Figure 3E). We first tested interaction between FAAP20 and a FANCA fragment representing aa 1065 to 1455 that harbors the leucine-zipper motif and the entire C-terminus (MBPFANCA1065-1455). Lysates of E coli cells expressing MBPFANCA1065-1455, His6-FLAGFAAP20, or both were immunoprecipitated with M2-agarose (FAAP20 target) or amylose (FANCA target). Coomassie staining of both immunoprecipitates revealed interaction between FAAP20 and FANCA (supplemental Figure 3), and immunoblot analysis with anti-FANCA and anti-FAAP20 antibodies confirmed the identity of the 2 bands (supplemental Figure 3 bottom panel).
We next wished to test whether the leucine-zipper motif is essential for interaction with FAAP20, and to localize the region of FANCA that interacts with FAAP20. To do this, we made 3 additional FANCA fragments: MBPFANCA1065-1101 that harbors only the leucine-zipper motif, as well as MBPFANCA1095-1455 and the smaller MBPFANCA1095-1200 that both lack the motif (Figure 3D). Lysates of E coli cells expressing these 3 constructs, with or without His6-FLAGFAAP20, were subjected to coprecipitation with M2-agarose or amylose. Coomassie staining of coprecipitates showed that although the FANCA construct with the leucine-zipper motif failed to interact with FAAP20 (Figure 3F lanes 9-12), both of the constructs lacking the motif were able to interact with FAAP20 (Figure 3F lanes 10,11,13,14). Therefore, the leucine-zipper motif of FANCA is not required for interaction with FAAP20, and the region of FANCA that interacts with FAAP20 lies between residues 1095 and 1200. Immunoblot analysis using anti-MBP or anti-FAAP20 confirmed the identity of the bands as FANCA or FAAP20, respectively (Figure 3F bottom panel).
FAAP20 contains a putative Zn-finger domain of UBZ type and is conserved across all vertebrates
BLAST analysis identified probable orthologs of FAAP20 in all vertebrates examined, but not in invertebrates or other lower organisms (supplemental Figure 4A). Although a database search did not reveal homology to any known domains, a secondary structure prediction identified a conserved C-terminal region with cysteines and histidine arranged in a CysCysHisCys (CCHC)–type Zn-finger motif; the CCHC motif was conserved across all species examined (supplemental Figure 4B). Alignment of this presumptive Zn-finger domain with Zn-finger domains of other proteins revealed a high similarity with ubiquitin-binding Zn-finger (UBZ) domain (supplemental Figure 4B). Several proteins involved in the FA pathway (eg, FAN1, SLX4, and Rad18) contain UBZ domains that are essential for their function.13,18-20
FAAP20 UBZ domain binds ubiquitin in vitro
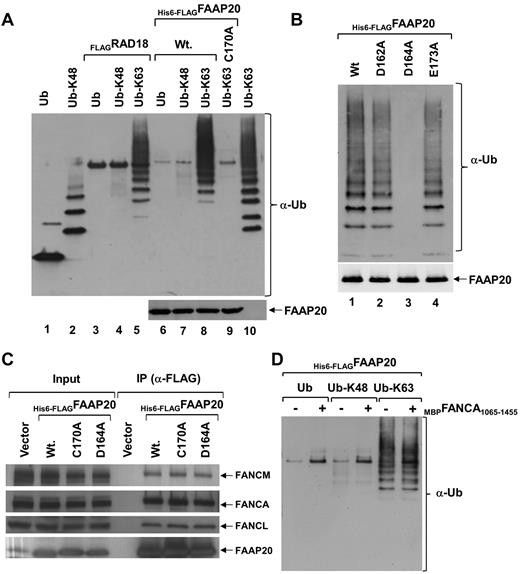
Ubiquitin plays a major role in DNA damage sensing and repair.18,19 It has been shown that UBZ domains in DNA repair proteins bind ubiquitin, and that this binding is required for these proteins to regulate their targets.13,18-21 To determine whether FAAP20 binds ubiquitin noncovalently, we performed an ubiquitin-binding assay by expressing His6-FLAGFAAP20 in E coli, immobilized it to M2-agarose beads, and incubated it with mono or poly-ubiquitin chains linked through either Lys-48 or Lys-63. Immunoblot analysis showed that FAAP20 bound only K-63–linked chains, not K-48–linked chains (Figure 4A lanes 6-8), and interaction with poly-ubiquitin chains was robust (Figure 4A lane 8). On the other hand, interaction with mono or di-ubiquitin was not detected, and interaction with tri-ubiquitin was weak (Figure 4A lane 8). E3 ubiquitin protein ligase (RAD18), which binds K-63–linked ubiquitin chains, served as a positive control (Figure 4A lanes 3-5).21 To determine whether binding of FAAP20 and K-63–linked poly-ubiquitin chains is mediated via the UBZ domain, we mutated Cys-170 in the domain, and then studied binding, as previously described. Immunoblot analysis revealed that the Cys-170 mutant failed to bind K-63–linked poly-ubiquitin chains (Figure 4A lane 9), suggesting that the UBZ domain is required for binding.
FAAP20 binds ubiquitin in vitro. (A) Immunoblot showing mono-ubiquitin (Ub), K-48 and K-63–linked ubiquitin chains. Interaction of M2-agarose bound His6-FLAGFAAP20 was tested with mono-ubiquitin, K-48 and K-63–linked ubiquitin chains. FAAP20 binds K-63 linked ubiquitin chains (lane 8), but not mono-ubiquitin (lane 6) or K-48 linked (lane 7). Mutation in the UBZ domain abolished binding (lane 9). RAD18 served as a positive control (lanes 3-5). (B) Immunoblot showing K-63- linked ubiquitin chains. Interaction of M2-agarose bound His6-FLAGFAAP20 WT (Wt) and various point mutants was tested with K-63–linked ubiquitin chains. FAAP20 constructs with single residue mutations in the UBZ domain, D164A mutation (lane 3) abolishes binding of FAAP20, whereas D162A (lane 2) and E173A (lane 4) bind K-63–linked chains. (C) Immunoblot showing no apparent difference between WT FAAP20 and UBZ mutant (C170A or D164A) binding to core-complex proteins. (D) Interaction of M2-agarose bound His6-FLAGFAAP20 alone (−) or in complex with MBPFANCA1065-1455 (+) was tested with mono-ubiquitin, K-48 and K-63–linked ubiquitin chains. No apparent difference was found in binding of FAAP20 with K-63–linked chains in the absence or presence of FANCA.
FAAP20 binds ubiquitin in vitro. (A) Immunoblot showing mono-ubiquitin (Ub), K-48 and K-63–linked ubiquitin chains. Interaction of M2-agarose bound His6-FLAGFAAP20 was tested with mono-ubiquitin, K-48 and K-63–linked ubiquitin chains. FAAP20 binds K-63 linked ubiquitin chains (lane 8), but not mono-ubiquitin (lane 6) or K-48 linked (lane 7). Mutation in the UBZ domain abolished binding (lane 9). RAD18 served as a positive control (lanes 3-5). (B) Immunoblot showing K-63- linked ubiquitin chains. Interaction of M2-agarose bound His6-FLAGFAAP20 WT (Wt) and various point mutants was tested with K-63–linked ubiquitin chains. FAAP20 constructs with single residue mutations in the UBZ domain, D164A mutation (lane 3) abolishes binding of FAAP20, whereas D162A (lane 2) and E173A (lane 4) bind K-63–linked chains. (C) Immunoblot showing no apparent difference between WT FAAP20 and UBZ mutant (C170A or D164A) binding to core-complex proteins. (D) Interaction of M2-agarose bound His6-FLAGFAAP20 alone (−) or in complex with MBPFANCA1065-1455 (+) was tested with mono-ubiquitin, K-48 and K-63–linked ubiquitin chains. No apparent difference was found in binding of FAAP20 with K-63–linked chains in the absence or presence of FANCA.
Multiple sequence alignment of UBZ domains of other proteins revealed a conserved aspartate at position +2, with respect to the histidine of the CCHC motif (supplemental Figure 4B light gray shading), which is functionally required.21 In one of these proteins, WRNIP1, an aspartate to alanine mutation abolishes the ubiquitin-binding activity of the UBZ domain in vitro.21 Surprisingly, the +2 position in the UBZ domain of FAAP20 was occupied by an alanine, not by an aspartate (supplemental Figure 4B). Because our in vitro ubiquitin-binding experiments suggest that the FAAP20 UBZ domain binds ubiquitin, we hypothesized that an aspartate or glutamate at some other position in the FAAP20 UBZ domain performs the function of the aspartate shown to be conserved in UBZ domains of all other proteins tested. An inspection of the amino acids around the conserved histidine of the CCHC motif identified 2 aspartate residues at positions −4 and −2 and a glutamate residue at position +7, which were conserved across all FAAP20 orthologs (supplemental Figure 4B; the absolute positions of −4, −2, and +7 are 162, 164, and 173, respectively). We mutated each of the 2 aspartates and a glutamate to alanine (D162A, D164A, and E173A) in FAAP20, and tested their K-63–linked ubiquitin-binding activity (Figure 4B). Immunoblot analysis showed that D164A abolished the in vitro ubiquitin-binding activity of FAAP20 UBZ domain (Figure 4B lane 3), but neither D162A nor E173A had an effect. This suggests that aspartate-164 in FAAP20 is essential for the function of the UBZ domain, and may be functionally equivalent to the aspartate at position +2 in other UBZ domains.
To determine whether FAAP20 interaction with FANCA requires the UBZ domain, we used M2-agarose to purify His6-FLAGFAAP20, His6-FLAGFAAP20D164A, and His6-FLAGFAAP20C170A from HeLa cells. Immunoblot analysis of purified complex revealed FANCA and other core-complex proteins, suggesting that the UBZ domain is not required for interaction of FAAP20 with FANCA (Figure 4C). To determine whether FANCA binding to FAAP20 affects the ability of FAAP20 to bind ubiquitin in vitro, we purified His6-FLAGFAAP20 alone or in complex with MBPFANCA1065-1455, a FANCA fragment (supplemental Figure 5), and performed the ubiquitin-binding assay. Immunoblot analysis showed no apparent difference between FAAP20 ubiquitin binding with or without FANCA, suggesting that FANCA binding to FAAP20 has no effect on the ability of FAAP20 to bind ubiquitin in vitro (Figure 4D). It is not known if full-length FANCA has any effect.
FAAP20 is required for FA pathway
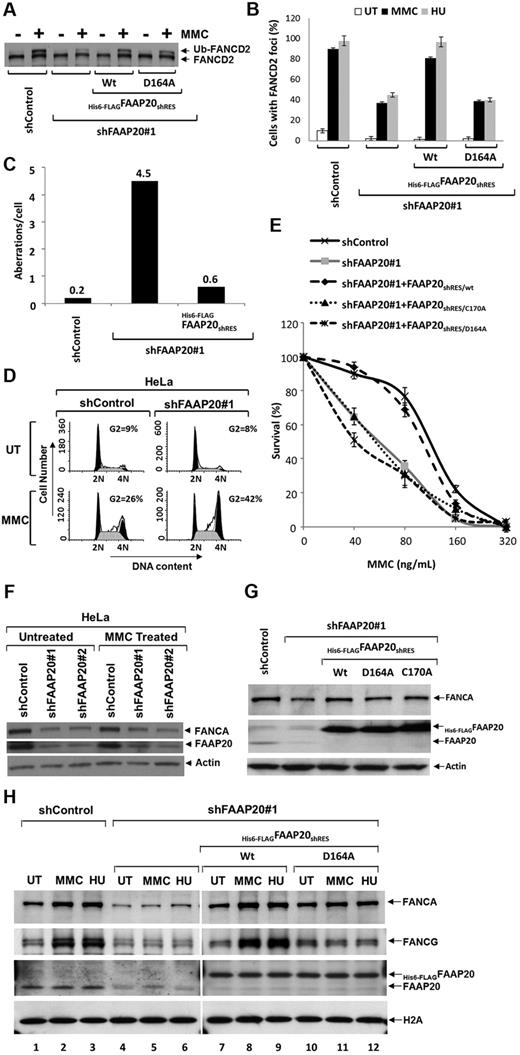
Cells derived from FA patients, which are defective in core-complex proteins, exhibit “FA phenotypes” when treated with chemicals, such as MMC or HU: impaired FANCD2 or FANCI mono-ubiquitination, defective FANCD2 foci formation, chromosomal breaks, higher sensitivity in a clonogeneic cell survival assay, and higher G2/M fraction in cell-cycle analysis. Similar FA-phenotypes are found in cells in which any of the core-complex proteins are knocked down with siRNA or shRNA.5-7,9 Because FAAP20 is an integral component of the core complex, we wished to determine whether FAAP20 knockdown also result in “FA-phenotypes.” We screened several siRNA oligos targeted against FAAP20 mRNA and found 2, siFAAP20#1 and siFAAP20#2, that were able to knockdown FAAP20 (supplemental Figure 6). We next wished to use these siRNAs to determine whether the FA phenotype FANCD2 mono-ubiquitination is dependent on FAAP20. Cells transfected with siFAAP20#1 or siFAAP20#2 and treated with MMC exhibited impaired FANCD2 mono-ubiquitination (supplemental Figure 6 lanes 3-6); cells transfected with control siRNA (siControl) and treated with MMC had normal FANCD2 mono-ubiquitination (supplemental Figure 6 lanes 1-2). We then made shRNA constructs using the siFAAP20#1 or siFAAP20#2 sequences, and generated a cell line stably expressing shRNA against FAAP20. FAAP20 was knocked down in the shRNA-transduced cells and FANCD2 mono-ubiquitination impaired after MMC treatment, compared with shControl-transduced cells (Figure 5A). Mono-ubiquitination of FANCD2 is known to target FANCD2 to DNA repair foci.9 Indeed, whereas MMC treatment resulted in dramatic induction in FANCD2 focus formation in control cells, FAAP20 knockdown compromised focus formation (Figure 5B). A higher number of chromosomal abberations, including breaks and radials, per cell were observed in FAAP20 knockdown cells (Figure 5C, supplemental Figure 7). In addition, propidium iodide- and RNase-stained MMC-treated FAAP20-knockdown cells revealed an increased number of G2/M-phase cells, suggestive of G2/M arrest (Figure 5D); these cells also showed higher sensitivity to MMC in a clonogeneic cell survival assay (Figure 5E). To determine whether these observed FA phenotypes are a direct result of FAAP20 knockdown, and not an off-target effect of shRNA, we stably expressed shRNA-resistant FAAP20 (FAAP20shRES) in cells stably expressing shFAAP20#1; indeed, in these cells FANCD2 mono-ubiquitination and focus formation, chromosome breaks, and MMC sensitivity were rescued (Figure 5A-C,E). Therefore, the observed FA-phenotypes were specific to FAAP20 depletion, and not an off-target effect of the shRNA.
FAAP20 is required for activation of the FA pathway. (A) Immunoblot showing levels of FANCD2 mono-ubiquitination and FAAP20 in HeLa cells stably expressing shControl or shFAAP20#1 siRNAs. Knockdown of FAAP20 expression reduced levels of mono-ubiquitinated FANCD2 in cells treated with MMC or HU compared with UT cells. (B) Immunofluorescence analysis of FANCD2 nuclear foci. HeLa cells stably expressing shControl showed an induction of FANCD2 foci on MMC and HU treatment; knockdown of FAAP20 by shFAAP20#1 resulted in decreased foci formation. Expression of WT His6-FLAGFAAP20shRES/Wt rescues capacity to form foci but mutant His6-FLAGFAAP20shRES/D164A does not. The percentage of cells with 5 or more foci was determined by examining at least 150 cells. Data are presented as the average of 3 independent experiments with standard deviations. (C) Bar diagram showing chromosome aberrations analysis data. Human HSC93 lymphoblast cells stably expressing control shRNA (shControl), shFAAP20 or shFAAP20, and His6-FLAGFAAP20shRES together were analyzed for diepoxybutane-induced chromosomal aberrations like breaks, gaps, and radials. Compared with shControl cells, shFAAP20 cells showed higher number of aberrations per cell and this phenotype was rescued by expressing wildtype FAAP20 resistant to shRNA. Fifty metaphase spreads were prepared and scored for chromosomal aberrations as described in supplemental Methods. (D) Cell-cycle analysis of PI and RNase-stained HeLa:shControl and HeLa:shFAAP20#1 cells that were left UT or treated with MMC for 24 hours. HeLa:shFAAP20#1 MMC treated cells showed an increased number of cells in G2 phase compared with HeLa:shControl MMC treated cells. (E) MMC survival curve showing that reduced FAAP20 expression results in increased sensitivity to MMC; control levels of MMC sensitivity are restored by expressing FAAP20shRES/wt, but not FAAP20shRES/C170A or FAAP20shRES/D164A mutants. Data represent percent survival, compared with untreated, MMC-naive cells. Each experiment was performed in triplicate, and mean values are shown with standard deviations, derived by comparing each dose to no MMC (0 value on the x-axis). (F) Immunoblot showing that FANCA is reduced when FAAP20 is knocked down using either of 2 shRNAs. (G) Immunoblot showing FANCA is reduced when FAAP20 is knockdown and the reduced levels can be rescued by expressing either WT (WT) or 1 of 2 FAAP20 mutants (D164A or C170A). (H) Immunoblot showing chromatin association of FANCA, FANCG, and FAAP20. HeLa:shControl cells treated with MMC or HU exhibited increased association of both FANCA and FANCG with chromatin, compared with UT cells (lanes 1-3). In contrast, HeLa cells depleted of FAAP20 showed reduced FANCA-chromatin association (lanes 4-6). Induction of chromatin association of FANCA in FAAP20 depleted cells can be rescued by ectopic expression of WT FAAP20 (lanes 7-9), but not by the FAAP20D164A mutant (lanes 10-12) suggesting FAAP20-ubiquitin binding activity is required for chromatin association of FANCA and FANCG on DNA damage.
FAAP20 is required for activation of the FA pathway. (A) Immunoblot showing levels of FANCD2 mono-ubiquitination and FAAP20 in HeLa cells stably expressing shControl or shFAAP20#1 siRNAs. Knockdown of FAAP20 expression reduced levels of mono-ubiquitinated FANCD2 in cells treated with MMC or HU compared with UT cells. (B) Immunofluorescence analysis of FANCD2 nuclear foci. HeLa cells stably expressing shControl showed an induction of FANCD2 foci on MMC and HU treatment; knockdown of FAAP20 by shFAAP20#1 resulted in decreased foci formation. Expression of WT His6-FLAGFAAP20shRES/Wt rescues capacity to form foci but mutant His6-FLAGFAAP20shRES/D164A does not. The percentage of cells with 5 or more foci was determined by examining at least 150 cells. Data are presented as the average of 3 independent experiments with standard deviations. (C) Bar diagram showing chromosome aberrations analysis data. Human HSC93 lymphoblast cells stably expressing control shRNA (shControl), shFAAP20 or shFAAP20, and His6-FLAGFAAP20shRES together were analyzed for diepoxybutane-induced chromosomal aberrations like breaks, gaps, and radials. Compared with shControl cells, shFAAP20 cells showed higher number of aberrations per cell and this phenotype was rescued by expressing wildtype FAAP20 resistant to shRNA. Fifty metaphase spreads were prepared and scored for chromosomal aberrations as described in supplemental Methods. (D) Cell-cycle analysis of PI and RNase-stained HeLa:shControl and HeLa:shFAAP20#1 cells that were left UT or treated with MMC for 24 hours. HeLa:shFAAP20#1 MMC treated cells showed an increased number of cells in G2 phase compared with HeLa:shControl MMC treated cells. (E) MMC survival curve showing that reduced FAAP20 expression results in increased sensitivity to MMC; control levels of MMC sensitivity are restored by expressing FAAP20shRES/wt, but not FAAP20shRES/C170A or FAAP20shRES/D164A mutants. Data represent percent survival, compared with untreated, MMC-naive cells. Each experiment was performed in triplicate, and mean values are shown with standard deviations, derived by comparing each dose to no MMC (0 value on the x-axis). (F) Immunoblot showing that FANCA is reduced when FAAP20 is knocked down using either of 2 shRNAs. (G) Immunoblot showing FANCA is reduced when FAAP20 is knockdown and the reduced levels can be rescued by expressing either WT (WT) or 1 of 2 FAAP20 mutants (D164A or C170A). (H) Immunoblot showing chromatin association of FANCA, FANCG, and FAAP20. HeLa:shControl cells treated with MMC or HU exhibited increased association of both FANCA and FANCG with chromatin, compared with UT cells (lanes 1-3). In contrast, HeLa cells depleted of FAAP20 showed reduced FANCA-chromatin association (lanes 4-6). Induction of chromatin association of FANCA in FAAP20 depleted cells can be rescued by ectopic expression of WT FAAP20 (lanes 7-9), but not by the FAAP20D164A mutant (lanes 10-12) suggesting FAAP20-ubiquitin binding activity is required for chromatin association of FANCA and FANCG on DNA damage.
Interestingly, FANCA levels were also reduced in FAAP20 knockdown cells expressing either shFAAP20#1 or shFAAP20#2 (Figure 5F); levels of FANCA were restored by expressing FAAP20shRES, suggesting that reduced FANCA levels were because of reduced FAAP20 (Figure 5G). This observation raised the question whether the observed FA-phenotypes in FAAP20-knockdown cells were a direct result of FAAP20 knockdown or because of reduced levels of FANCA. We also wished to determine whether the FAAP20 UBZ domain is required for expression of FA-phenotypes. To address both of these questions, we stably expressed FAAP20shRES with 1 of 2 mutations in the UBZ domain that render the domain nonfunctional, either Cys170 (FAAP20shRES/C170A) or Asp164 (FAAP20shRES/D164A; Figure 5G). Although both mutations rescued the reduced levels of FANCA (Figure 5G), they failed to correct the observed FA-phenotypes (Figure 5B-D), thereby suggesting that the FAAP20 UBZ domain is required for expression of the FA-phenotypes, and that they are not because of reduced FANCA levels. Taken together, these data indicate that FAAP20 plays a role in ICL DNA repair.
FAAP20 is required for DNA-damage–induced chromatin loading of FANCA
It has been reported that an increased amount of FANCA is associated with chromatin in response to DNA damage.22,23 Because FAAP20 interacts with FANCA, we wished to determine whether FAAP20 is required for an increased association of FANCA with chromatin in response to DNA damage. HeLa cells stably expressing shControl exhibited increased association of both FANCA and FANCG with chromatin, compared with untreated cells (Figure 5H lanes 1-3). In contrast, untreated HeLa cells depleted of FAAP20 showed reduced FANCA-chromatin association (Figure 5H lanes 4-6), which could be because of reduced stability of FANCA in the absence of FAAP20 (Figure 5F). Interestingly, we failed to see an induction in chromatin association of FANCA in FAAP20-knockdown cells treated with MMC or HU, suggesting that FAAP20 is, in fact, required for FANCA-chromatin association (Figure 5H); this FANCA-chromatin association defect was rescued by ectopic expression of WT FAAP20 but not by the FAAP20D164A mutant, despite restoration of FANCA levels (Figure 5H). Thus, ubiquitin binding via the UBZ domain of FAAP20 is required for increased chromatin loading of FANCA. Interestingly, unlike FANCA, FAAP20-chromatin association does not increase in response to DNA damage (Figure 5H).
Discussion
Earlier attempts to purify the FA nuclear core complex relied on affinity/IP of individual protein components of the complex,8,9 which resulted in isolation of core complex, as well as subcomplexes associated with the target protein compounding the analysis. To purify the core complex to homogeneity, we generated a cell line stably expressing 2 different protein components of the core complex, each with a different tag: His6-tagged FANCA and FLAG-tagged FAAP100. MS analysis of the core complex purified from these cells revealed a near stoichiometry of all components, a significant improvement over purifications performed with 2 tags on a single protein. Using this 2-protein approach, we identified a novel uncharacterized protein, which we named FAAP20. A reciprocal purification, done using ectopically expressed or endogenous FAAP20 as target, revealed the presence of other core-complex proteins. We also observed interaction between FAAP20 and core-complex proteins, in the presence or absence of DNA damage. These data all suggest that FAAP20 is a constitutive member of the FA nuclear core complex. FAAP20 may also exist in subcomplexes with FANCA and FANCG, because these proteins have an identical gel filtration profile, and FAAP20 IPs revealed higher amounts of FANCA and FANCG compared with other core-complex proteins. These presumptive subcomplexes may represent an early stage in the sequential assembly of the core complex, or they may have additional functions in the FA pathway.24,25
Expression levels of individual components of multiprotein complexes are often coregulated to ensure proper assembly and function.5-9,24 Studies of FA subcomplexes suggest coreliance of members for their stability and function. For example, FANCG and FANCL are unstable in the absence of FANCA, FAAP100 is unstable in the absence of FANCB or FANCL, and FANCM and MHF2 stability is dependent on MHF1.5-9,24 In this study, we observed that FANCA and FAAP20 are dependent on each other for their stability. Absence of FANCA results in significant reduction of FAAP20, and this effect can be reversed by blocking the proteasome pathway, which suggests that, in the absence of FANCA, FAAP20 is degraded via a proteasome-mediated protein-degradation pathway. Absence of FAAP20 resulted in partial destabilization of FANCA. Also overexpression of FANCA resulted in dramatic stabilization of FAAP20. Consistent with our data suggesting that FANCA and FAAP20 coexist in subcomplexes and are required for each other's stability, we present evidence for a direct, physical interaction between FANCA and FAAP20, and demonstrate that an approximately 96-aa-long region of FANCA, representing aa 1095 to 1200, is sufficient for interaction with FAAP20 in vitro.
Approximately 60%-80% of FA patients have mutations in the FANCA gene, and a majority of these mutations result in loss of FANCA stability.26-29 Genotype-phenotype correlation studies suggest that null mutations resulting in complete loss of the FANCA protein are the most severe, with earlier onset of anemia and a higher incidence of leukemia.26,30 We show here that absence of FANCA results in negligible amounts of FAAP20. This led us to hypothesize that the severe phenotype observed in FANCA-null patients could be partly because of loss of FAAP20 protein. In light of our observations, it will be interesting to make a genotype-phenotype correlation of FA-A patients with respect to FAAP20 protein levels. Given the role of FAAP20 in regulating FANCA, it would not be surprising to find that patients lacking both FAAP20 and FANCA exhibit more severe phenotypes.
UBZ domain-containing proteins and ubiquitination play major roles in the FA pathway.13,18-20 Two FA proteins, FANCD2 and FANCI, are mono-ubiquitinated, and this ubiquitination event is essential for the integrity of the FA pathway. Loss of any one of the core-complex proteins results in loss of FANCD2-FANCI mono-ubiquitination. One core-complex protein, FANCL, is an E3-ubiquitin ligase that, along with UBE2T, mono-ubiquitinates FANCD2-FANCI dimer. The mono-ubiquitinated FANCD2-FANCI dimer interacts with FAN1, a nuclease containing a UBZ domain, and recruits it to the site of DNA damage.31-35 FANCP/SLX4, also a member of the FA pathway, contains 2 UBZ domains shown to be important for FA pathway function.13,36-38 Recently, RAD18, a conserved UBZ domain-containing E3 ligase was shown to be a critical regulator of the FA pathway.20,39,40 In this study, we show that FAAP20 contains a highly conserved UBZ-type zinc-finger domain that binds K-63–linked chains but not K-48–linked chains in vitro, consistent with its function in DNA repair. We show that the FAAP20-UBZ domain is essential for FAAP20 function, and plays a role in recruitment of FANCA and other core-complex proteins to chromatin, probably to DNA-damaged sites. We hypothesize that the FAAP20-UBZ domain may bind K-63–linked poly-ubiquitinated DNA-damage sensor protein(s), thereby recruiting the core complex to the DNA repair site. In support of this hypothesis, we found several ubiquitin peptides in MS analyses of core complex immunopurified using FAAP20 antibody (data not shown). Further studies are required to identify the probable cellular target(s) of the FAAP20-UBZ domain. While this paper was under review, another group independently identified Rev1 as the probable target of FAAP20 in vivo.41 Kim et al show that FAAP20 binds mono-ubiquitinated Rev1 and this binding stabilizes Rev1 nuclear foci and promotes interaction of the FA core with PCNA-Rev1 DNA damage bypass complexes, thus providing a critical role of FAAP20 in linking the FA pathway to translesion synthesis (TLS).41
One interesting observation we made regarding the FAAP20-UBZ domain is the absence of a conserved aspartate at the +2 position (with respect to histidine) that is required for function of UBZ domains in other proteins.13,21 We show that an aspartate at the −2 position probably complements the function of aspartate at +2 position. FAAP20 interaction with FANCA is not dependent on the UBZ domain function, because FAAP20-UBZ domain mutants, which fail to bind ubiquitin in vitro, were still able to bind FANCA. In addition, FANCA is not required for function of the FAAP20-UBZ domain, because the ubiquitin-binding activity of the domain is unaltered in the presence or absence of FANCA.
Similar to other core-complex proteins, FAAP20 is essential for proper function of the FA pathway. Ablation of FAAP20 expression, using siRNA or shRNA, resulted in phenotypes that were characteristic of FA cells. We also show that the UBZ domain and ubiquitin-binding activity of FAAP20 are essential for FA pathway function, because UBZ-domain mutants failed to correct the cellular FA phenotypes.
There are currently 15 complementation groups of FA, and the genes mutated in those complementation groups have been described. Some FA patients do not exhibit mutations in any of the known FA genes. Using complementation analysis, we screened several human FA cell lines for mutations in FAAP20, but failed to find one that could be complemented by FAAP20 (data not shown). This apparent lack of FAAP20 patients could be that FA patients with FAAP20 mutations are rare and so not represented in current repositories. Indeed, several FA complementation groups (eg, L, M, O, and P) have extremely few patients.6,7,13,42
In summary, we identified FAAP20, a novel component of the FA nuclear-core complex that is required for FA pathway-mediated repair of ICLs. FAAP20 stability is controlled by FANCA, and these 2 core-complex proteins directly interact via the C-terminal region of FANCA. FAAP20 is highly conserved, found only in vertebrates, and contains a UBZ-type zinc-finger domain. The UBZ domain binds K-63–linked poly-ubiquitin chains in vitro, and is required for proper functioning of the FA pathway. The ubiquitin-binding property of FAAP20 is required for DNA-damage induced chromatin loading of FANCA.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the Viral Vector Core, DNA Sequencing, and Fluorescent Activated Cell Analyzing and Sorting facility of the Cincinnati Children's Research Foundation for their excellent service. They also thank the Fanconi Anemia Research Fund for antibodies and cell lines.
This work was supported by National Institutes of Health research grants HL084082 and HL084082-03S1 to A.R.M., and HL076712 to Q.P. A.R.M was also supported by grants from the Ohio Cancer Research Associates and the Fanconi Anemia Research Fund.
National Institutes of Health
Authorship
Contribution: A.M.A. designed and performed research, analyzed data, and wrote the paper; A.P., C.D., T.R.S., J.L., and K.W. performed research; E.G., A.A., and Q.P. provided reagents and designed research; and A.R.M. designed and performed research, analyzed and controlled data, and edited the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Amom Ruhikanta Meetei, Division of Experimental Hematology and Cancer Biology, Cincinnati Children's Hospital Medical Center, 240 Albert Sabin Way, Cincinnati, OH 45229; e-mail: ruhikanta.meetei@cchmc.org.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal