The choice of either induction or postremission therapy for adults with acute myeloid leukemia is still largely based on the “one size fits all” principle. Moreover, pretreatment prognostic parameters, especially chromosome and gene abnormalities, may fail in predicting individual patient outcome. Measurement of minimal residual disease (MRD) is nowadays recognized as a potential critical tool to assess the quality of response after chemotherapy and to plan postremission strategies that are, therefore, driven by the individual risk of relapse. PCR and multiparametric flow cytometry have become the most popular methods to investigate MRD because they have been established as sensitive and specific enough to allow MRD to be studied serially. In the present review, we examine the evidence supporting the appropriateness of incorporating MRD detection into the AML risk assessment process. A comprehensive prognostic algorithm, generated by combining pretreatment cytogenetics/genetics and posttreatment MRD determination, should promote advances in development of personalized therapeutic approaches.
Introduction
In adult patients with acute myeloid leukemia (AML), intensive chemotherapy achieves complete remission (CR) rates ranging from 50% to 80%. Despite these encouraging results, the majority of responding patients will eventually relapse, with only 30% to 40% of young and less than 20% of elderly patients being long-term survivors.1,,,–5 Advances in biologic characterization are expected to provide a more proper risk stratification of AML, allowing delivery of treatments proportional to the real aggressiveness of disease. In this view, cytogenetic abnormalities represent the most reliable prognosticator in adult AML.6,,,–10 Identification of specific gene abnormalities (eg, FLT3, NPM1, CEBPA, and DMNT3A) has further improved prognostic allocation of patients with AML, especially within homogeneous karyotypic groups (ie, intermediate karyotypes or favorable karyotypes), where the possible concomitant mutations of KIT, occurring in the context of core binding factor (CBF) translocations, confer a negative prognosis.11,,,–15 A proper assignment to low- or high-risk category is a critical step in the therapeutic decision-making process of patients with AML. Indeed, patients belonging to the low-risk category would not benefit from routine use of allogeneic stem cell transplantation (ASCT) in first CR because any advantages in terms of reduced relapse incidence will be counterbalanced by procedure-related morbidity and mortality.16 On the other hand, missing early identification of high-risk patients might hamper or delay the timely use of intensive treatments, such as ASCT.17 However, it is also well recognized that cytogenetic/genetic signature cannot always reliably predict outcome in individual patients; indeed, approximately 40% to 50% of those with favorable karyotype will eventually experience a relapse. In this view, measure of minimal residual disease (MRD) promises to be an efficient tool to establish on an individual basis the patient's leukemia's susceptibility to treatment, enhancing delivery of risk-adapted therapies.18,19 Despite these premises, the systematic applicability of MRD analysis in AML is not yet accomplished. For cases with a genetic signature that accounts for 60% to 70% of AML, the molecular approach still suffers from lack of standardized assays and cut-offs. Similarly, although flow cytometry can potentially investigate more than 85% of AML, its widespread applicability is complicated by lack of standardized procedure. The present review will focus on the technical issues and clinical relevance of MRD determination in non-M3 AML. In particular, we will try to outline how MRD assessment might improve risk stratification.
The case of outcome prediction
Clonal chromosome alterations are detected in AML in more than 50% of adults and are universally considered the strongest predictor of duration of response and overall survival (OS). Based on cytogenetics, patients are generally stratified into 3 risk groups: favorable, intermediate, and unfavorable.6,,,–10 Accordingly, patients falling into the category of favorable risk karyotype (F-RK) can expect an approximate 65% to 70% likelihood of cure, those who have an intermediate risk karyotype (I-RK) an approximate 40% chance of long-term disease-free survival (DFS), whereas patients belonging to the unfavorable risk karyotype (U-RK) category have a very dismal outcome, with less than 5% to 10% becoming long-term survivors.6,,,–10 However, it has become clear that cytogenetic risk allocation may help guide therapeutic decision, particularly for those who are at the extremes (F-RK or U-RK), whereas, because of the heterogeneity of this category, for patients within I-RK the therapeutic decision may be problematic. Indeed, 40% to 50% of patients categorized in this group lack any clonal abnormalities on standard cytogenetic analysis. Furthermore, together with cytogenetically normal AML (CN-AML), the I-RK also includes a miscellany of different structural and numerical changes that are too infrequent to be reliably assigned a prognostic significance and are not encompassed in the 2 other risk groups. In recent years, various molecular markers have been identified, allowing to dissect further cytogenetically defined subsets.20 This is critical to risk assessment of patients with CN-AML (Table 1). Although the expectations of using genetics to guide therapy were enormous, the delivery to the clinics has made slow progress and the therapeutic strategy for most patients still remains controversial. On one side, risk assessment based on pretreatment factors composes a large group of patients with broad differences in terms of risk of relapse. On the other side, fitting the increasing number of new gene abnormalities into a practical, prospective, decision-making algorithm is a very difficult task. Indeed, the excess of dissection in homogeneous genetic subsets may lead to a plethora of subgroups failing to provide physicians with an adequate statistical power (Figure 1). A reasonable way to improve outcome prediction in AML might be attained by combining pretreatment and posttreatment parameters into a common prognostic algorithm. Such a hypothesis found acceptance in a seminal manuscript published by Wheatley et al who generated a prognostic scoring system by integrating pretreatment cytogenetics and achievement of CR after 1 or 2 cycles of induction therapy.21 Patients with U-RK who entered CR after 2 induction courses had the worst outcome. The design of the analysis represented a first attempt to put together into a risk stratification algorithm 2 different classes of prognostic parameters: pretreatment factors and indicators of the quality of response after a chemotherapy-induced morphologic CR. Nowadays, such a working hypothesis can be developed further, thanks to the availability of techniques, such as PCR and multiparametric flow cytometry (MPFC). These techniques can reliably detect, at high sensitivity, leukemic cells at submicroscopic level, thus offering the opportunity to investigate MRD.18,19 Assessment of MRD promises to be a powerful and accurate tool to refine patients' risk category assignment as initially established on the basis of the sole cytogenetic/genetic findings.17,22,23
Gene mutations occurring in cytogenetically normal acute myeloid leukemia
| Gene . | Incidence . | Biologic features . | Impact on clinical outcome and targeted therapy opportunities . |
|---|---|---|---|
| FLT3 | |||
| ITD | Found in 28%-34% of CN-AML | Member of the class III receptor tyrosine kinase family | Consistently associated with inferior outcome in terms of OS, RFS, and EFS33,34 |
| In-frame mutations, mostly in exons 14 and 15 of the juxtamembrane domain | Constitutively active FLT3 molecules are targets for specific molecular inhibitors (phase 2 or 3 trials) | ||
| Level of mutant allele probably of importance | |||
| Homozygous FLT3 mutations as a result of mitotic recombination leading to partial UPD | |||
| TKD | Found in 11%-14% of CN-AML | In-frame point mutations of the TKD codon 835 and 836 | Meta-analysis suggesting a negative prognostic impact32 |
| Level of mutant allele may be of importance | More recently associated with better OS31 | ||
| High-level mutants associated with improved OS30 | |||
| NPM1 | Found in 25%-35% of AML(45%-62% of CN-AML) | The mutational event modifies specific nucleoli binding and nuclear export signal motifs coded by the exon 12 and determines an abnormal cytoplasmic localization of nucleophosmin | NPM1mut/FLT3-ITD− genotype associated with a favorable OS, RFS, and CR rate38,39–40 |
| Associated with presenting clinical and laboratory features, such as female sex, higher BM blast counts, and LDH levels, as well as high CD33 but low or absent CD34 levels | Patients with NPM1mut/FLT3-ITD− genotype may not benefit from MRD allogeneic transplantation in first CR11 | ||
| Associated with FLT3-ITD and TKD mutations (40% of cases) | Pharmacologic modulation of the RA-signaling pathway has been suggested as a therapeutic option11 | ||
| NPM1 acts as a corepressor in RA-associated transcriptional regulation41 | |||
| CEBPA | Found in 15%-20% of CN-AML | Transcription factor mediating lineage specification and differentiation of multipotent myeloid progenitors into mature neutrophils | Associated with higher CR rate and better RFS and OS45,46 |
| Concurrent mutations (ie, FLT3-ITD and NPM1) were virtually not present in cases with CEBPA–biallelic mutation compared with CEBPA–single allelic mutation44 | The presence of CEBPA biallelic (bm) but not single allelic mutation (sm) is an independent factor for a favorable outcome44 | ||
| MLL | PTD found in 5%-11% of CN-AML | Usually involves exons 5-11 or, less frequently, exons 5-12 | Associated with shorter CR duration or inferior RFS and EFS60 |
| MLL wild-type allele is silenced, probably as a result of differential DNA methylation and histone modifications | Allogeneic transplantation may improve outcome11 | ||
| Rationale for the use of DNA methyltransferase and/or histone deacetylase inhibitors based on in vitro data | |||
| IDH1/2 | Found in 15% of AML | Correlated with accumulation of the cancer-associated metabolite 2-hydroxyglutarate | IDH1 mutations identify patients at higher risk of relapse within the favorable risk category of NPM1 mutated AML50,,–53 |
| IDH2 gene encodes a mitochondrial protein homologous to IDH1 that also catalyzes isocytratecarboxylation | IDH2 mutations get along with specific pretreatment features (low WBC, lack of other mutations, and primary resistance to chemotherapy)50,,–53 | ||
| IDH1 and IDH2 mutations are mutually exclusive | |||
| DNMT3A | Found in 22% of AML. | Somatic mutation in DNMT3A, encoding a DNA methyltransferase | Independent determinant of dismal prognosis both in the overall study population and, significantly, within high-risk patients (FLT3-ITD, age > 60 years)54 |
| Highly enriched in the group of patients with an intermediate-risk cytogenetic profile (56/66, 33.7%) | Only ASCT provided a significant benefit54 | ||
| Absent in the subgroup of 79 patients carrying a favorable-risk cytogenetic | |||
| A perturbation in the methylation of specific DNA sequences has been postulated | |||
| BAALC overexpression | Rare | Expressed in neuroectoderm-derived tissues and hematopoietic precursors | Significantly worse CR rates and shorter DFS, EFS, and OS than patients with low expression55 |
| Detected in a subset of karyotypically normal adults < 60 years with de novo AML | SCT in first CR might overcome the adverse prognostic effect of high BAALC expression55 | ||
| ERG overexpression | Rare | Identified in CN-AML patients and patients with prognostically unfavorable complex karyotypes with cryptic amplification of chromosome | Significantly shorter OS and higher CIR56,57 |
| RAS | Found in 9%-13% of CN-AML | Constitutively activated by point mutations either in the GTP-binding domain (codon 12/13) or in the GTPase domain (codon 61) | A clear prognostic role could not be elucidated yet42 |
| Possible susceptibility blasts to FTIs | |||
| WT1 | Found in ∼ 10% of CN-AML | The majority of mutations in both studies were frameshift mutations of exon 7, whereas only one was identified in exon 9 | Initial studies on small patient cohorts suggest association with induction failure47,–49 |
| Gene . | Incidence . | Biologic features . | Impact on clinical outcome and targeted therapy opportunities . |
|---|---|---|---|
| FLT3 | |||
| ITD | Found in 28%-34% of CN-AML | Member of the class III receptor tyrosine kinase family | Consistently associated with inferior outcome in terms of OS, RFS, and EFS33,34 |
| In-frame mutations, mostly in exons 14 and 15 of the juxtamembrane domain | Constitutively active FLT3 molecules are targets for specific molecular inhibitors (phase 2 or 3 trials) | ||
| Level of mutant allele probably of importance | |||
| Homozygous FLT3 mutations as a result of mitotic recombination leading to partial UPD | |||
| TKD | Found in 11%-14% of CN-AML | In-frame point mutations of the TKD codon 835 and 836 | Meta-analysis suggesting a negative prognostic impact32 |
| Level of mutant allele may be of importance | More recently associated with better OS31 | ||
| High-level mutants associated with improved OS30 | |||
| NPM1 | Found in 25%-35% of AML(45%-62% of CN-AML) | The mutational event modifies specific nucleoli binding and nuclear export signal motifs coded by the exon 12 and determines an abnormal cytoplasmic localization of nucleophosmin | NPM1mut/FLT3-ITD− genotype associated with a favorable OS, RFS, and CR rate38,39–40 |
| Associated with presenting clinical and laboratory features, such as female sex, higher BM blast counts, and LDH levels, as well as high CD33 but low or absent CD34 levels | Patients with NPM1mut/FLT3-ITD− genotype may not benefit from MRD allogeneic transplantation in first CR11 | ||
| Associated with FLT3-ITD and TKD mutations (40% of cases) | Pharmacologic modulation of the RA-signaling pathway has been suggested as a therapeutic option11 | ||
| NPM1 acts as a corepressor in RA-associated transcriptional regulation41 | |||
| CEBPA | Found in 15%-20% of CN-AML | Transcription factor mediating lineage specification and differentiation of multipotent myeloid progenitors into mature neutrophils | Associated with higher CR rate and better RFS and OS45,46 |
| Concurrent mutations (ie, FLT3-ITD and NPM1) were virtually not present in cases with CEBPA–biallelic mutation compared with CEBPA–single allelic mutation44 | The presence of CEBPA biallelic (bm) but not single allelic mutation (sm) is an independent factor for a favorable outcome44 | ||
| MLL | PTD found in 5%-11% of CN-AML | Usually involves exons 5-11 or, less frequently, exons 5-12 | Associated with shorter CR duration or inferior RFS and EFS60 |
| MLL wild-type allele is silenced, probably as a result of differential DNA methylation and histone modifications | Allogeneic transplantation may improve outcome11 | ||
| Rationale for the use of DNA methyltransferase and/or histone deacetylase inhibitors based on in vitro data | |||
| IDH1/2 | Found in 15% of AML | Correlated with accumulation of the cancer-associated metabolite 2-hydroxyglutarate | IDH1 mutations identify patients at higher risk of relapse within the favorable risk category of NPM1 mutated AML50,,–53 |
| IDH2 gene encodes a mitochondrial protein homologous to IDH1 that also catalyzes isocytratecarboxylation | IDH2 mutations get along with specific pretreatment features (low WBC, lack of other mutations, and primary resistance to chemotherapy)50,,–53 | ||
| IDH1 and IDH2 mutations are mutually exclusive | |||
| DNMT3A | Found in 22% of AML. | Somatic mutation in DNMT3A, encoding a DNA methyltransferase | Independent determinant of dismal prognosis both in the overall study population and, significantly, within high-risk patients (FLT3-ITD, age > 60 years)54 |
| Highly enriched in the group of patients with an intermediate-risk cytogenetic profile (56/66, 33.7%) | Only ASCT provided a significant benefit54 | ||
| Absent in the subgroup of 79 patients carrying a favorable-risk cytogenetic | |||
| A perturbation in the methylation of specific DNA sequences has been postulated | |||
| BAALC overexpression | Rare | Expressed in neuroectoderm-derived tissues and hematopoietic precursors | Significantly worse CR rates and shorter DFS, EFS, and OS than patients with low expression55 |
| Detected in a subset of karyotypically normal adults < 60 years with de novo AML | SCT in first CR might overcome the adverse prognostic effect of high BAALC expression55 | ||
| ERG overexpression | Rare | Identified in CN-AML patients and patients with prognostically unfavorable complex karyotypes with cryptic amplification of chromosome | Significantly shorter OS and higher CIR56,57 |
| RAS | Found in 9%-13% of CN-AML | Constitutively activated by point mutations either in the GTP-binding domain (codon 12/13) or in the GTPase domain (codon 61) | A clear prognostic role could not be elucidated yet42 |
| Possible susceptibility blasts to FTIs | |||
| WT1 | Found in ∼ 10% of CN-AML | The majority of mutations in both studies were frameshift mutations of exon 7, whereas only one was identified in exon 9 | Initial studies on small patient cohorts suggest association with induction failure47,–49 |
LDH indicates lactate dehydrogenase; CN, cytogenetically normal; TKD, tyrosine kinase domain; MRD, matched related donor; JM, juxtamembrane domain; UPD, uniparental disomy; EFS, event-free survival; PTD, partial tandem duplication; NPM1, nucleophosmin (nucleolar phosphoprotein B23, numatrin); FLT3-ITD, internal tandem duplication of the fms-related tyrosine kinase 3 (FLT3) gene; RA, retinoic acid; DFS, disease-free survival; CEBPA, CCAAT/enhancer binding protein (C/EBP)–α; CRD, CR duration; MLL-PTD, partial tandem duplication of the myeloid/lymphoid or mixed-lineage leukemia (MLL) gene; IDH1/2, isocitrate dehydrogenase 1 and 2; DNMT3A, DNA methyltransferase 3A gene; BAALC, brain and acute leukemia gene, cytoplasmic; ERG, v-erythroblastosis virus E26 oncogene-like (avian); GTP, guanosine triphosphate; and FTIs, farnesyltransferase inhibitors.
Pie chart depicting the molecular heterogeneity of CN-AML. The analysis is based on mutations in the NPM1, CEBPA, MLL, and FLT3 (ITD and tyrosine kinase domain [TKD] mutations at codons D835 and I836), NRAS, and WT1 genes. Data are derived from mutational analysis of 485 younger adult patients with CN-AML from the German AML Study Group.90
Pie chart depicting the molecular heterogeneity of CN-AML. The analysis is based on mutations in the NPM1, CEBPA, MLL, and FLT3 (ITD and tyrosine kinase domain [TKD] mutations at codons D835 and I836), NRAS, and WT1 genes. Data are derived from mutational analysis of 485 younger adult patients with CN-AML from the German AML Study Group.90
The quest of residual leukemia
PCR and MPFC have proven sensitive and specific enough to allow MRD to be investigated and represent today the gold standard for MRD monitoring in AML.
MRD detection by PCR
Leukemic fusion genes.
The power of PCR relies on cloning of breakpoints of the chromosomal rearrangements and allows their detection in the postremission phase in at least 30% of the patients, using RT-PCR or real-time quantitative PCR. Common targets for PCR-based MRD detection are fusion transcripts of CBF-positive AM (eg, RUNX1-RUNX1T1, CBFB-MYH11, and MLL-gene fusions). RT-PCR in CBF-positive AML has a limited clinical applicability as persistent PCR positivity has been observed in long survivors even after ASCT.24 Real-time quantitative PCR has proven potentially more valuable because of its capability to anticipate impending relapse during follow-up monitoring.25 In a recent report, Corbacioglu et al established clinically relevant MRD cut-points at which persistence of CBFB-MYH11 transcript positivity singled out patients with significantly increased risk of relapse.26 The authors conclude that monitoring of CBFB-MYH11 transcript levels should be incorporated into future clinical trials to guide therapeutic decisions.26
Mutations.
The discovery of gene mutations in fusion gene-negative AML has potentially increased to 60% to 70% the percentage of cases suitable for PCR-based MRD monitoring (Table 1).27,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,,–59 The key question about the use of these genes as candidates for MRD monitoring regards their stability over the course of disease. The receptor tyrosine kinase FLT3 is mutated in a relevant proportion of CN-AML. Mutations resulting in the constitutive activation of FLT3 have been identified in 2 functional domains of the receptor: the juxtamembrane domain and the split tyrosine kinase domain.28,29 The juxtamembrane domain is disrupted by internal tandem duplications (ITDs) of various sizes in 28% to 34% of CN-AML, whereas point mutations of the tyrosine kinase domain codon 835 or 836 have been reported in 11% to 14% of CN-AML.30,–32 The role of FLT3-ITD in MRD monitoring is controversial: lack of stability of FLT3 mutations in paired samples from diagnosis and relapse raises concerns about the clinical usefulness of this marker.29,36 However, some authors claim that this lack of longitudinal stability might relate to lack of sensitivity. Using high-sensitivity RT-PCR, it was observed that, in all 25 cases under study, FLT3-ITD was detected at diagnosis and relapse. In 4 of them, relapse was predictable based on serially documented reappearance of FLT3-ITD. These results suggest that there is still room for revisiting the role of FLT3-ITD as a potential marker for MRD monitoring.60 Although there are only initial experiences dealing with the use of CEBPA and MLL-PTD in MRD monitoring,61,62 increasing knowledge is being accumulated on the role of NPM1 mutations. NPM1 mutations can be identified in 45% to 60% of patients with CN-AML, accounting for the most frequent genetic change in this subset. The mutational event modifies specific nucleoli binding and nuclear export signal motifs coded by exon 12 and determines an abnormal cytoplasmic localization of NPM1.37,38 Approximately 40% of patients with NPM1 mutations also carry FLT3-ITD. Some studies indicate that NPM1 mutations are very stable at relapse, and thus that they might have a role in MRD assessment.63 Schnittger et al developed a highly sensitive real-time quantitative PCR assay able to prime 17 different mutations of NPM1.64 In 252 NPM1-mutated AML, high levels of NPM1mut, quantified at 4 different time points, were significantly correlated with outcome at each tested time point. Multivariate analysis, including age, FLT3-ITD status, and the level of NPM1, demonstrated that the latter was the most relevant prognostic factor affecting event-free survival during first-line treatment, also in the subgroup of patients who underwent ASCT. Furthermore, the authors compared the kinetics of NPM1 and FLT3-ITD fluctuations and found that FLT3-ITD levels suffer from an unpredictable instability, making it unreliable for MRD purposes.29,36 In a further refinement of such an approach, Krönke et al demonstrated that the level of NPM1mut, measured after double induction and consolidation therapy, impacted on OS and cumulative incidence of relapse (CIR; P < .001 for all comparisons).65
Gene overexpression.
Techniques of real-time quantitative PCR intended to quantify genes overexpression theoretically provide the opportunity to study the whole population of AML. Various candidates have been proposed, with WT1 being the most reliable. Retrospective clinical data indicate that WT1 assessment after induction therapy is a predictor of outcome, with lower levels being associated with long-term remission.66 WT1 overexpression can be exploited as a marker to establish the presence, persistence, or reappearance of leukemic hematopoiesis. Cilloni et al have measured the levels of WT1 after chemotherapy-induced morphologic CR, in peripheral blood (PB), and BM of patients with AML.67 In multivariate analysis, they found that a more than or equal to 2-log MRD reduction in PB and/or BM was associated with a significantly lower CIR (P = .004). Although the manuscript is of outstanding relevance because of the provision of a common standardized protocol on the behalf of the LeukemiaNet, only in 46% and 13% of PB and BM samples, respectively, were the levels of WT1 sufficiently overexpressed, compared with normal samples, to allow a prognostic stratification. Based on this, concerns still remain about the confounding role of the physiologic background of WT1 in normal PB and BM.
MRD detection by flow cytometry
MRD monitoring by MPFC relies on the expression of “leukemia-associated immunophenotypes” (LAIPs) defined as the presence of a combination of antigens and/or flow cytometric physical abnormalities that are absent or very infrequent in normal BM. The growing interest surrounding this technique is because of its wide applicability (> 85% of AML), quickness, specificity, and ability to distinguish viable cells from BM debris and dead cells. Furthermore, the diffusion of devices equipped with multiple lasers allowed implementation of multiple color assays (> 4 or 5 monoclonal antibody combinations), thus favoring increment of sensitivity that, now, can be reasonably placed in between 10−4 and 10−5. An additional advantage of multiple color assays consists of a significant attenuation of concerns of phenotypic shifts that can be observed on recurrence.68 There is evidence that the use of an expanded 6-9 color polychromatic assay not only increases sensitivity69 but also results in improved qualitative information on the leukemic clone because of the superior definition of its phenotypic composition. In this context, MRD monitoring might not be compromised, especially if serial determinations are performed. The results from the literature demonstrate that detection of MRD by MPFC is technically sound and represents a powerful tool to segregate patients with AML into categories of risk (Table 2). Most studies do not deal with the MRD issue as a simple matter of a positive or negative finding; rather, they set a threshold below or above which the outcome can be significantly different. A common approach is to set up empirically the most significant level of MRD by choosing a logarithmic scale (eg, 101, 102, 103) or a quartile segregation that correlates with survival estimates and CIR. An alternative approach relies on the evaluation of the prognostic role of MRD in a continuous variable model. In the experience of Kern et al, such an analysis demonstrated a strong correlation with outcome when assessed on day 16 from induction and after consolidation.70,71 However, when it came to stratifying the patients in low- or high-risk categories, significant thresholds were identified. At least 4 manuscripts tried to address the issue of MRD “prognostically significant levels” applying specific statistical methods that would help selecting thresholds more appropriately.72,,–75 In the paper by Al-Mawali et al, to determine the optimal cut-off yielding the best segregation of AML patients into categories of risk, a receiver operating characteristic analysis was carried out.75 The threshold was established at the value of 0.15% residual leukemic cells; therefore, patients with MRD more than 0.15% qualified as MRD-positive, whereas those with MRD less than or equal to 0.15% as MRD-negative. We evaluated the trend of standardized log-rank statistics using relapse-free survival (RFS) and OS as dependent variables and the values of residual leukemic cells determined after induction and consolidation as independent variables (maximally selected log-rank statistics).76 Based on the results of this test, we currently use the value of 0.035% BM residual leukemic cells to discriminate MRD-negative from MRD-positive cases, both after induction and consolidation. Although the use of a dedicated statistical approach to establish the threshold for MRD negativity/positivity is recommended, the reproducibility of that specific threshold throughout different laboratories still remains an issue. Should we select a particular cut-off that different laboratories can refer to or should any laboratory set up its own? A universal cut-off would be desirable, but it might represent a very troublesome mission to accomplish because standardizing thresholds is affected by a number of technical (equipment, fluorochromes, sensitivity of LAIPs, procedures of acquisition and analysis) and clinical factors. Among clinical factors, the different intensity of chemotherapeutic regimens delivered may play a critical role. Indeed, the clinically relevant threshold of MRD may depend on the therapy used and might change with changes in therapeutic schedule. Divergence between our experience72,73 and that of San Miguel et al is an example of such a situation.77,78 In the study of San Miguel et al, the induction, consolidation, and intensification therapy consisted of the combination of an anthracycline and cytosine arabinoside.77,78 In our study, the patients were treated on 3 drug-based regimens, associating an anthracycline with cytosine arabinoside and etoposide.72,73 In addition, cytosine arabinoside administration was prolonged through 10 days instead of the conventional 7 during the induction phase. Thus, one may assume that, in the study of San Miguel et al, the less intensive therapy delivered was associated with a milder debulking effect that in turn may account for the higher level of MRD at which a significant influence on disease outcome was found.77,78 Based on this, we can also assume that each new protocol will most likely define a new threshold.
MRD by MPFC studies including more than 50 patients
| Reference . | No. of patients . | Time point . | Threshold postinduction, % . | Threshold postconsolidation, % . | Method . | Multivariate analysis . | Survival parameter . |
|---|---|---|---|---|---|---|---|
| San Miguel77 | 53 | I, C | 0.5 | 0.2 | NA | I,C | RFS |
| Venditti87 | 56 | I, C | 0.045 | 0.035 | empirical | C | OS, RFS |
| San Miguel78 | 126 | I | < 0.01 0.01-0.1, 0.1-1, > 1 | NA | NA | I | RFS |
| Kern70 | 62 | I, C | Continuous analysis log-difference | Continuous analysis log-difference | 75th percentiles of log difference | C | RFS |
| Kern71 | 106 | Day 16 after start of induction | Continuous analysis log-difference | NA | Median of log difference | Day 16 after start of induction | EFS, RFS |
| Buccisano72 | 100 | I, C | 0.035 | 0.035 | Maximally selected log-rank statistic | C | OS, RFS |
| Maurillo73 | 142 | I, C | 0.035 | 0.035 | Maximally selected log-rank statistic | C | OS, RFS |
| Al Mawali75 | 54 | I | 0.15 | 0.15 | ROC analysis | I | RFS, OS |
| Reference . | No. of patients . | Time point . | Threshold postinduction, % . | Threshold postconsolidation, % . | Method . | Multivariate analysis . | Survival parameter . |
|---|---|---|---|---|---|---|---|
| San Miguel77 | 53 | I, C | 0.5 | 0.2 | NA | I,C | RFS |
| Venditti87 | 56 | I, C | 0.045 | 0.035 | empirical | C | OS, RFS |
| San Miguel78 | 126 | I | < 0.01 0.01-0.1, 0.1-1, > 1 | NA | NA | I | RFS |
| Kern70 | 62 | I, C | Continuous analysis log-difference | Continuous analysis log-difference | 75th percentiles of log difference | C | RFS |
| Kern71 | 106 | Day 16 after start of induction | Continuous analysis log-difference | NA | Median of log difference | Day 16 after start of induction | EFS, RFS |
| Buccisano72 | 100 | I, C | 0.035 | 0.035 | Maximally selected log-rank statistic | C | OS, RFS |
| Maurillo73 | 142 | I, C | 0.035 | 0.035 | Maximally selected log-rank statistic | C | OS, RFS |
| Al Mawali75 | 54 | I | 0.15 | 0.15 | ROC analysis | I | RFS, OS |
I indicates induction; C, consolidation; NA, not available; EFS, event-free survival; and ROC, receiver operating characteristic.
PB versus BM MRD detection
PB may represent an alternative source of cells for the purpose of MRD studies. This is based on the assumption that the presence of circulating blasts at the time of CR might reflect the persistence of malignant cells in the BM. Studies using real-time quantitative PCR to monitor MRD in CBF-positive AML showed that the transcripts were detectable in PB and BM with a comparable sensitivity.79,80 The levels of WT1 transcript measured after consolidation in PB and BM were found to be equally associated with the risk of relapse, and the sensitivity of PB WT1 analysis resulted to be equivalent if not better than that of BM.66,67 We have demonstrated the feasibility of MRD detection in PB of 50 adult AML patients using MPFC.74 The levels of MRD after induction and consolidation in PB significantly reproduced those observed in BM (r = 0.86, P < .0001; and r = 0.82, P < .0001; respectively). A level of MRD more than 0.015% in PB after consolidation was associated with a significant likelihood of subsequent relapse and a shorter RFS. Our data suggest that PB may be a complementary source of cells for MRD studies in patients with AML. In addition, combined measurement of MRD in BM and PB might improve the risk stratification process. The clinical impact of MRD contamination of PB apheresis product is a further subject of research because it has been associated with a shorter RFS and OS.81 Further studies are warranted to clarify these issues.
When MRD assessment is clinically relevant
Timing of PCR assessment
As soon as experience was accumulated, it became clear that selection of the most appropriate time point to determine MRD was a key issue to have a risk-adapted approach transposed into the clinical reality. Ideally, such a time point should be the one havingthe power to provide the most informative prognostic indication, so that the choice of postremission therapy is driven by the actual risk of relapse. The paradigm of such a situation is represented by acute promyelocytic leukemia, where the persistence of PML-RARA fusion transcript at the end of consolidation therapy or subsequent recurrence of PCR positivity in patients previously in molecular remission was demonstrated to precede overt relapse.82,83 CBF-positive cases have been extensively investigated by real-time quantitative PCR for MRD persistence at specific time points. Perea et al found that MRD positivity was associated with an increased risk of relapse at any time of assessment but only MRD persistence at the end of treatment and during subsequent follow-up significantly anticipated relapse.24 Similar results have been reported in CBFB-MYH11 AML by Corbacioglu et al who identified in the postconsolidation and early follow-up phase (≤ 3 months) the critical time points allowing patients at higher risk of relapse to be recognized.26 Krönke et al observed that detection of high NPM1mut transcript levels after double induction and consolidation correlated significantly with an increased CIR.65 In accordance with what we have learned by assessing MRD in acute promyelocytic leukemia, experience with CBF-positive and NPM1mut AML appears to indicate that MRD levels as measured at delayed rather early time points are of superior prognostic relevance. At variance with this assumption, Cilloni et al suggest that risk stratification is indeed improved by early assessment.67 Indeed, they reported an increased risk of relapse when the level of WT1 failed to reduce more than or equal to 2-log after induction; the magnitude of WT1 reduction remained independently significant, even after adjusting for competitive covariates, such as age, white blood cell count, and cytogenetics. A final issue pertains to the hypothesis that the optimal sampling interval varies with molecular subgroups. Ommen et al have demonstrated that the kinetics of molecular relapse are remarkably different among NPM1, PML-RARA, RUNX1-RUNX1T1, and CBFB-MYH11 AML.84 The authors derived a mathematical model to investigate the molecular relapse and the time from molecular relapse to hematologic relapse. They found that CBFB-MYH11 AML displayed a slower clone regrowth than AML with other molecular signature. These results will potentially optimize MRD monitoring, allowing the identification of suitable sampling intervals for other molecular signatures.
Timing of MPFC assessment
With regard to the MPFC approach, the issue of the best time point is even more controversial. The German AML Cooperative group demonstrated that assessment of MRD persistence on day 16 from induction and the log-difference between MRD-positive cells on day 1 and 16 from induction represented an independent prognostic factor affecting CR, event-free survival, OS, and RFS.71 However, it is important to note that this analysis is something different from MRD assessment, for it pertains to the concept of “speed of blast clearance.” “Speed of blast clearance” is supposed to reflect the chemosensitivity of the leukemic clone, but it does not necessarily relate to the quality of response as determined, later on, on full hematopoietic reconstitution. To the best of our knowledge, no formal demonstration has been published on the correlation between “fast blast clearance” and achievement of MRD negativity.85,86 San Miguel et al demonstrated that a stratification according to levels of MRD less than 0.01%, 0.01% to 0.1%, 0.1% to 1%, and more than 1% after induction therapy, resulted in significant differences in OS.77,78 Al-Mawali et al found that a threshold of 0.15% residual leukemic cells discriminated MRD-negative from MRD-positive cases with optimal sensitivity and specificity, allowing impending relapse to be predicted at postinduction and postconsolidation time points.75 Multivariate analysis showed that the postinduction MRD level affected independently RFS and OS. However, also diverging opinions have been published supporting the hypothesis that delayed time points may be even more informative compared with earlier ones. We demonstrated that levels of MRD more than or equal to 0.035%, as measured after consolidation therapy, predicted a high frequency of relapse and a short duration of OS and RFS; the prognostic role of MRD positivity after consolidation was confirmed in multivariate analysis.72,73,87 In line with our experience, Kern et al reported that the 75th percentile of the MRD log-difference between the first day of induction and postconsolidation time point was the sole variable dividing the patients in 2 groups with significantly different OS.70 Selecting an early or delayed time point for MRD determination might entail the choice of different therapeutic options: the early time point option may prove useful to identify as soon as possible high-risk patients for whom a fast allocation to very intensive treatments is required. For these patients, approaches, such as dose-dense schedule88 and/or ASCT, could be incorporated into the upfront treatment strategy.89 On the other hand, opponents of this hypothesis raise concerns about situations of overtreatment for patients showing a slow blast clearance. In our experience, approximately 30% of patients who are MRD-positive after induction become negative at the end of consolidation and the clinical outcome of these “slow responders” was not significantly different from that of patients who tested MRD-negative soon after induction.72,73 Finally, we think that special consideration should be given to specific situations where a serial sampling may be recommended. In our experience, sequential MRD monitoring is required in the phase after autologous stem cell transplantation to enhance detection of impending relapse in patients who are MRD-negative after consolidation.90
Outcome prediction: cytogenetics/genetics, MRD, or both?
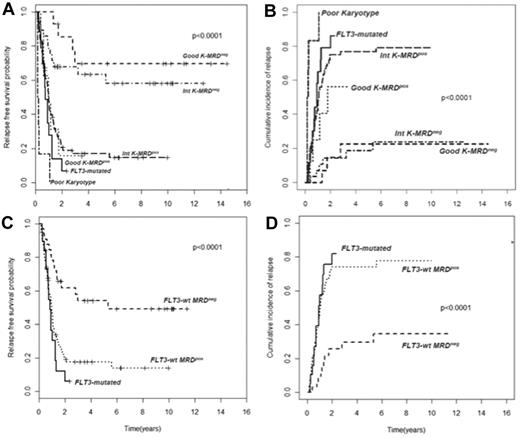
In AML, cytogenetic and molecular findings at diagnosis are critical determinants of outcome and allow stratification of approximately 40% of patients into “good risk” (based on the presence of mutated NPM1 without FLT3-ITD or F-RK) or “poor risk” (FLT3-ITD mutations or U-RK) categories.91 Good risk patients achieve high OS and DFS rates with standard treatments, whereas poor risk do unsuccessfully without intensified therapy with ASCT. On the other hand, there are no accepted criteria to direct the decision-making process after induction/consolidation for patients included in the I-RK (∼ 60%). For these patients, evaluation of MRD status appears appropriate to extrapolate those at high (MRD-positive) or low (MRD-negative) risk of relapse, for whom differentiated treatments may be adopted. Based on this assumption, we have tried to optimize risk assessment of patients with AML by integrating evaluation of pretreatment cytogenetics/genetics and MRD status at the postconsolidation time point.92 Of 143 adult patients, those with F-RK and I-RK who were MRD-negative had 4-year RFS of 70% and 63%, and OS of 84% and 67%, respectively. Patients with F-RK and I-RK who were MRD-positive had 4-year RFS of 15% and 17%, and OS of 38% and 23%, respectively (P < .001 for all comparisons; Figure 2A-B). Likewise, FLT3 wild-type patients achieving a MRD-negative status (Figure 2C-D) had a better outcome than those who remained MRD-positive after consolidation (4-year RFS 54% vs 17% P < .0001, OS 60% vs 23% P = .002). Therefore, patients with F-RK, I-RK, or FLT3 wild-type had a very different outcome depending on MRD status at the end of consolidation. The Children's Oncology Group has recently carried out the same analysis, based on which children with standard risk AML and MRD positivity after the first induction were reallocated to the high-risk category, whereas those MRD-negative were added to the favorable-risk cohort.93 This risk stratification system generated 2 new categories with a 3-year DFS of 20% and 68%, respectively (P < .001). The authors conclude that cytogenetics, molecular genotyping, and postinduction MPFC analysis provide a robust means of stratifying pediatric AML into 2 risk groups with significantly different outcomes.
Subgroup analysis of RFS and CIR of 143 AML patients stratified according to pretreatment karyotype or FLT3 status and levels of MRD after consolidation. (A-B) Those with a level of residual leukemic cells < 0.035% are referred to as intermediate karyotype-MRD−, favorable karyotype-MRD−, or FLT-wt MRD−, whereas those with levels ≥ 0.035% are categorized as intermediate karyotype-MRD+, favorable karyotype-MRD+, or FLT-wt MRD+. Survival outcomes of these subsets and of U-RK and FLT3-ITD category are shown (P < .001 for all comparisons). (C-D) FLT-wt patients achieving a MRD-negative status show a better outcome than those who remained MRD-positive after consolidation (P < .001).
Subgroup analysis of RFS and CIR of 143 AML patients stratified according to pretreatment karyotype or FLT3 status and levels of MRD after consolidation. (A-B) Those with a level of residual leukemic cells < 0.035% are referred to as intermediate karyotype-MRD−, favorable karyotype-MRD−, or FLT-wt MRD−, whereas those with levels ≥ 0.035% are categorized as intermediate karyotype-MRD+, favorable karyotype-MRD+, or FLT-wt MRD+. Survival outcomes of these subsets and of U-RK and FLT3-ITD category are shown (P < .001 for all comparisons). (C-D) FLT-wt patients achieving a MRD-negative status show a better outcome than those who remained MRD-positive after consolidation (P < .001).
The LSC residual disease
The role of the leukemic stem cell (LSC) in AML recurrence is now more than a simple hypothesis. Relapses derive from re-expansion of residual leukemic cells escaping the cytotoxic effect of chemotherapy; this leukemic cell population is thought to reside within the stem cell CD34+CD38− compartment. Experimental data indicate that LSC are more resistant to chemotherapy than the more mature CD34+CD38+ progeny and can be distinguished from their normal counterpart because of the expression of LSC-specific immunophenotype.94 Therefore, the concept of LAIP expression is something that applies, even to LSC on which aberrant overexpression of CD47,95 CD123, and CD44 has been described.94 Among markers of LCS aberrancy, C-type lectin-like molecule-1 (CLL-1) promises to be one of the most appealing because its expression is lacking on normal stem cells.96,97 Therefore, CLL-1 has the double role of LSC specific antigen and potential target for future LSC directed-therapy. Using a monoclonal antibody targeting CLL-1, Terwijn et al demonstrated that a LSC frequency more than 1 × 10−3 after induction and more than 2 × 10−4 after the second induction and consolidation predicted a short RFS (P = .00003 and .004, respectively).98 By combining the residual LSC and the “whole MRD” fractions, they came up with 4 different categories whose outcome was the best for the group with a negative residual LSC status.98 The persistence of residual LSC may explain why a certain proportion of MRD-negative patients experience disease recurrence: in our experience, such a proportion accounts for 20% to 25% of MRD-negative patients. Therefore, monitoring of LSC-LAIPs represents an additional tool capable to refine the MPFC MRD assessment, and the combination of LSC-LAIPs and “whole MRD blast” frequencies might prove useful to guide future therapeutic intervention.
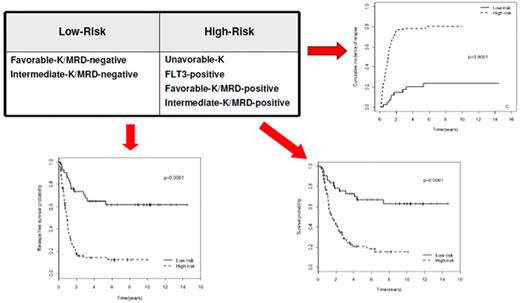
Implementing risk-adapted transplantation policy
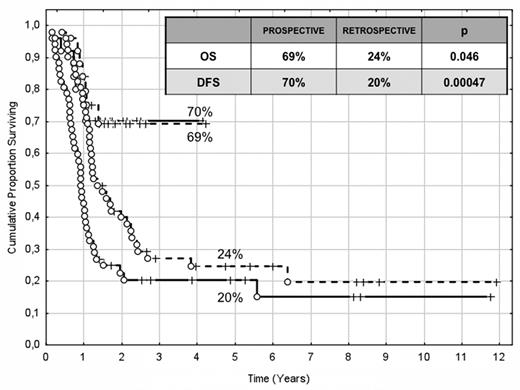
The choice of postremission therapy is too often driven by a “genetic randomization” resulting in a given patient being allocated to ASCT once the availability of an HLA-compatible sibling donor is documented. Meta-analysis of large prospective studies indicates that the beneficial effect of ASCT takes place as soon as the risk of relapse exceeds 35% to 40%99 ; when probabilities of relapse are below those percentages the risk of treatment-related mortality will attenuate the survival advantage of this procedure. Furthermore, the extensive use of ASCT is hampered by the paucity of candidates (25%-30%) with a fully matched family donor. For the remaining, even if a timely search of either international registries or cord blood banks is started, the probability of identifying a suitable donor is 46% and 73% at 3 and 6 months, respectively (W. Arcese, personal oral communication, June 30, 2011). In the meantime, almost 40% of candidates for transplant die of relapse or are relegated to less effective approaches, such as chemotherapy or autologous stem cell transplantation. We have demonstrated that an adjusted risk stratification based on pretreatment genetics/cytogenetics and MRD status at the end of consolidation refines the upfront genetic/cytogenetic risk classification.92 In this view, MRD assessment will greatly enhance selection procedures so that patients are assigned to ASCT not on the basis of donor availability but of the real risk of relapse.17 Applying this adjusted risk stratification, we distinguished 2 categories of patients: (1) low-risk: F-RK and I-RK, which were MRD-negative after consolidation; and (2) high-risk: U-RK, FLT3-ITD mutated cases, F-RK and I-RK, which were MRD-positive after consolidation (Figure 3). After these observations, we started a program for high-risk AML based on a prospective assignment to ASCT that should be delivered in the form of matched sibling donor, matched unrelated donor, umbilical cord blood, or haploidentical related donor transplant. We analyzed preliminarily a cohort of 21 high-risk patients who, according to the aforementioned policy, were assigned to ASCT (8 matched sibling donor, 7 matched unrelated donor/umbilical cord blood, and 6 haploidentical related donor). For comparative purposes, a matched historical cohort of 36 high-risk patients was analyzed: 12 were given matched sibling donor-ASCT and 24, lacking a matched sibling donor, received autologous stem cell transplantation. Survival estimates were significantly better for the prospective cohort compared with the control group (DFS 70% vs 20%, P = .000 47; OS 69% vs 24%, P = .046; Figure 4). The prospective cohort also showed a lower relapse rate (20% vs 52%, P = .003).
Risk assessment combining pretreatment and post-treatment prognosticators. Applying an adjusted risk stratification, combining genetics/cytogenetics and level of MRD at the end of consolidation therapy, we distinguished 2 categories of patients: (1) low-risk, including F-RK and I-RK that were MRD-negative; and (2) high-risk, including U-RK, FLT3-ITD cases, and F-RK/I-RK that were MRD-positive. The first group stands for significantly longer OS (73% vs 17%), RFS (58% vs 22%), and cumulative incidence of relapse (17% vs 77%; P < .001 for all comparisons).
Risk assessment combining pretreatment and post-treatment prognosticators. Applying an adjusted risk stratification, combining genetics/cytogenetics and level of MRD at the end of consolidation therapy, we distinguished 2 categories of patients: (1) low-risk, including F-RK and I-RK that were MRD-negative; and (2) high-risk, including U-RK, FLT3-ITD cases, and F-RK/I-RK that were MRD-positive. The first group stands for significantly longer OS (73% vs 17%), RFS (58% vs 22%), and cumulative incidence of relapse (17% vs 77%; P < .001 for all comparisons).
Comparison of clinical outcome in the “prospective” and “historical” cohort. The prospective cohort included 21 high-risk patients (4 MRD-positive favorable-karyotype, 9 MRD-positive intermediate-karyotype, 4 unfavorable-karyotype, and 4 FLT3-ITD) who underwent ASCT (8 matched sibling donor, 6 haploidentical related, and 7 matched unrelated/umbilical cord blood donor). The historical cohort accounted for 36 high-risk patients (6 MRD-positive favorable-karyotype, 23 MRD-positive intermediate-karyotype, 1 unfavorable-karyotype, and 6 FLT3-ITD). ASCT was offered to 12 patients with an available matched sibling donor, whereas those lacking this option were given autologous stem cell transplantation (n = 24). With a median follow-up of 18 months, survival estimates were significantly superior for the prospective cohort compared with the historical control (DFS 70% vs 20%, P = .000 47; OS 69% vs 24%, P = .046).
Comparison of clinical outcome in the “prospective” and “historical” cohort. The prospective cohort included 21 high-risk patients (4 MRD-positive favorable-karyotype, 9 MRD-positive intermediate-karyotype, 4 unfavorable-karyotype, and 4 FLT3-ITD) who underwent ASCT (8 matched sibling donor, 6 haploidentical related, and 7 matched unrelated/umbilical cord blood donor). The historical cohort accounted for 36 high-risk patients (6 MRD-positive favorable-karyotype, 23 MRD-positive intermediate-karyotype, 1 unfavorable-karyotype, and 6 FLT3-ITD). ASCT was offered to 12 patients with an available matched sibling donor, whereas those lacking this option were given autologous stem cell transplantation (n = 24). With a median follow-up of 18 months, survival estimates were significantly superior for the prospective cohort compared with the historical control (DFS 70% vs 20%, P = .000 47; OS 69% vs 24%, P = .046).
In conclusion, the paradigm of treatment for adult AML has largely been based on the “one size fits all” approach: in the short term, this has led to satisfactory rates of CR; but in the long term, survival estimates for young and elderly patients are disappointing. In acute promyelocytic leukemia and acute lymphoblastic leukemia, MRD positivity has consistently been shown to increase probabilities of relapse; therefore, attainment of MRD-negative remission represents a “gold standard,” leading to prolonged duration of CR.100 More recently, a trial of prospective MRD-driven therapy has also been initiated in childhood non-M3 AML, demonstrating an improvement of outcome in high-risk patients.101 Altogether, these observations suggest that, whatever the method, even in non-M3 AML measurement of MRD will potentially contribute refining risk assessment. In this view, a comprehensive risk stratification, generated by integrating the prognostic weight of pretreatment and posttreatment parameters (MRD),92 might help to allocate the majority of patients to a more realistic category of risk, thus favoring selection of more appropriate postremission strategies. In the context of such an integrated approach, pretreatment cytogenetic/genetic profile will dictate intensity and type of induction therapy, whereas posttreatment MRD status helps modulating intensity of postremission strategies, allowing a treatment proportional to the individual risk of relapse to be delivered. The final purpose is that patients will be assigned to ASCT not on the basis of donor availability (donor vs no donor approach) but on the basis of their adjusted (cytogenetic/genetic plus MRD) risk of relapse (transplant vs no transplant approach). This will also comply with the primary goal of saving additional lives, even without any major advances in chemotherapy or transplant technologies.17
Authorship
Contribution: All authors wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Adriano Venditti, Ematologia, Fondazione Policlinico Tor Vergata, Viale Oxford 81-00133, Rome, Italy; e-mail: adriano.venditti@uniroma2.it.

![Figure 1. Pie chart depicting the molecular heterogeneity of CN-AML. The analysis is based on mutations in the NPM1, CEBPA, MLL, and FLT3 (ITD and tyrosine kinase domain [TKD] mutations at codons D835 and I836), NRAS, and WT1 genes. Data are derived from mutational analysis of 485 younger adult patients with CN-AML from the German AML Study Group.90](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/119/2/10.1182_blood-2011-08-363291/5/m_zh89991183350001.jpeg?Expires=1767720350&Signature=FutLNwVhioa8pEDfswuAWKFVyC336Yv4Xbx4j7Ac5d1JC7RpFotwH89dDdE0iAq92XYo2Rr8L1NJpKWxx2IP0tTtcmu3pcoNNMnAkR44Tb65Tx2AwucsiIG-F0eVgFS0cbyC-e-HT57msy8btJybvho4gjmPEWhozX40TT1R1S31qJCmB62HDfkPzTkVBvr4hc7qxCyDdqKGqNarT~6~Hln~o7H767q~WSEewHKb~5do309O9hp13IAiPBS0A2Wg6JmdZ2GMRq7Fn-CFkup5cfxogMqp6d4JZ8NbEjd0VXouHJAmbcOz-nortuo6py8werRb8DYSu7mOaHQSUpmFdQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal