Abstract
The proto-oncogene EVI1 (ecotropic viral integration site-1), located on chromosome band 3q26, is aberrantly expressed in human acute myeloid leukemia (AML) with 3q26 rearrangements. In the current study, we showed, in a large AML cohort carrying 11q23 translocations, that ∼ 43% of all mixed lineage leukemia (MLL)–rearranged leukemias are EVI1pos. High EVI1 expression occurs in AMLs expressing the MLL-AF6, -AF9, -AF10, -ENL, or -ELL fusion genes. In addition, we present evidence that EVI1pos MLL-rearranged AMLs differ molecularly, morphologically, and immunophenotypically from EVI1neg MLL-rearranged leukemias. In mouse bone marrow cells transduced with MLL-AF9, we show that MLL-AF9 fusion protein maintains Evi1 expression on transformation of Evi1pos HSCs. MLL-AF9 does not activate Evi1 expression in MLL-AF9–transformed granulocyte macrophage progenitors (GMPs) that were initially Evi1neg. Moreover, shRNA-mediated knockdown of Evi1 in an Evi1pos MLL-AF9 mouse model inhibits leukemia growth both in vitro and in vivo, suggesting that Evi1 provides a growth-promoting signal. Using the Evi1pos MLL-AF9 mouse leukemia model, we demonstrate increased sensitivity to chemotherapeutic agents on reduction of Evi1 expression. We conclude that EVI1 is a critical player in tumor growth in a subset of MLL-rearranged AMLs.
Introduction
Acute myeloid leukemia (AML) is a heterogeneous disease, which can be classified based on cytogenetic aberrations, molecular abnormalities, and gene expression and methylation signatures.1-3 One group of AML patients is particularly characterized by the overexpression of the proto-oncogene EVI1 (ecotropic viral integration site-1), which was first identified as a common proviral integration site in retrovirally induced murine myeloid leukemias.4 EVI1 encodes a nuclear zinc finger protein, capable of DNA binding in a sequence-specific manner.5,6 EVI1 recruits a variety of transcriptional and epigenetic regulators, such as CTBP (C-terminal–binding protein), CBP (CREB-binding protein), P/CAF (P300/CBP-associated factor), HDAC (histone deacetylase), DNMT (DNA-methyltransferase), MBD3, or histone methyltransferases, suggesting a role in the control of gene expression.7-13 Human EVI1-mediated disease can partly be recapitulated using in vitro and in vivo models. Aberrant expression of EVI1 in mouse BM precursors in vivo causes a disease which resembles myelodysplastic syndrome (MDS).14,15 In vitro overexpression of EVI1 in transformed myeloid progenitors blocks myeloid differentiation, and affects survival and proliferation of these progenitors.16,17
Aberrant expression of EVI1 occurs in ∼ 8%-10% of human adult AML patients and is associated with a poor outcome.18-20 In approximately one-third of those, the EVI1 gene is highly expressed as the result of rearrangements of chromosome 3q26, the locus where the gene resides. However, high expression of EVI1 was also found in leukemias with chromosomal abnormalities, other than the ones affecting the EVI1 locus.19,21 Strikingly, ∼ 20% of EVI1-overexpressing AMLs have a concurrent mixed lineage leukemia (MLL) gene rearrangement,20 but the prevalence within the specific MLL rearrangements remains to be investigated. In addition, in pediatric AMLs, in 27% of MLL rearranged cases, EVI1 overexpression was detected.22 In a knockin MLL-AF9 mouse model mimicking human AML development, high Evi1 expression was detected in preleukemic stem and progenitor cells compared with corresponding wild-type cells.23 Importantly, among the various MLL-AF9 hematopoietic stem and progenitor cells, a direct correlation between Evi1 expression and the level of transformation was observed, where the Lin−/Sca1+/c-Kit+ (LSK) cells exhibiting highest Evi1 expression induced leukemias with the highest efficiency in recipients that received transplants.23 Furthermore, conditional ablation of Evi1 expression significantly reduces the colony-forming capacity of MLL-ENL–transformed BM progenitors.24 These studies suggest that, at least in murine model systems, a causal relationship between EVI1 and MLL rearrangements exists.
In the present study, we investigated the frequency and the prevalence of EVI1 overexpression in a large cohort of MLL-rearranged human AMLs. No prevalence was observed for high EVI1 expression in any of the most commonly detected MLL rearrangements in human AML, that is, MLL-AF6, MLL-AF9, and MLL-ENL. We also show that EVI1pos MLL-rearranged AMLs are different subtypes compared with EVIneg AMLs, based on their gene expression signature, morphology, and immunophenotype. The role of EVI1 in MLL-rearranged AMLs was studied in MLL-AF9–transformed mouse BM cells and in their human AML counterparts. We show that EVI1 expression is uncoupled from normal myeloid differentiation and is regulated by MLL-AF9, only in EVI1pos-transformed cells, in agreement with data reported by Arai and colleagues.25 The critical contribution of Evi1 in these model systems was shown by applying lentiviral knockdown strategies. Evi1 knockdown resulted in reduced survival in vitro and in vivo of MLL-AF9–transformed cells. Furthermore, Evi1 knockdown enhanced sensitivity of MLL-AF9–transformed cells to chemotherapeutic drugs. Our findings suggest a critical role for EVI1 in the pathogenesis of a subset of MLL-rearranged leukemias, and targeting of EVI1 could be beneficial for patients with EVI1pos MLL-rearranged AMLs.
Methods
For primers, Abs, culture conditions, and additional experimental methods, please refer to supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Patient material
Leukemic blast cells were purified from BM or blood of patients presenting with AML as previously reported.3,20 Patients were recruited from the Dutch-Belgian Cooperative Trial Group for Hematology (HOVON) and the AML Study Group (AMLSG) trials. All trails have been approved by the Institutional Review Board of the Erasmus University Medical Center and the University of Ulm.
Cytogenetic and molecular analysis
AML samples were routinely checked for cytogenetic abnormalities using a combination of standard chromosome-banding analysis and FISH. Additional RT-PCR was performed to verify the most common MLL fusions (primers are described in supplemental Table 1, and Balgobind et al26 and Jansen27 ). The karyotype of each patient according to the International System for Human Cytogenetic Nomenclature,28 French-American-British classification (FAB) type morphology, and WHO classifications, and if applicable, the MLL-fusion gene are depicted in supplemental Table 2.
Flow cytometric cell sorting
BM cells from healthy donors and of AML patients were isolated using Ficoll-Hypaque gradients and viably frozen in liquid nitrogen until use. The leukemic and normal CD34+/CD38− or CD34+/CD38+ subpopulations were obtained by flow cytometric cell sorting using double staining with anti-CD34 and anti-CD38 mAbs, with an exclusion of at least 500 channels between the CD38+ and CD38− subpopulations. In all cases, the purity of the preparation (99%) was verified by flow cytometry of separated cells. The sorting strategy is shown in supplemental Figure 2A.
Mononucleated murine normal BM cells were isolated on Ficoll-Hypaque gradients and stained with the BD Pharmingen Biotin-conjugated Mouse Lineage Panel (containing Abs against CD3e, CD11b, CD45R, Gr-1, and Ter119) and fluorophore-labeled Abs against c-kit (allophycocyanin), Sca-1 (PE-cy7), CD16/32 (PE), and CD34 (Pacific Blue). Biotin was targeted with streptavidin conjugated to allophycocyanin-Cy7. Subsequently, the BM cells were sorted for LSK (lineage marker negative [lin−], Sca1+, c-kit+), common myeloid precursors (CMPs; lin−/ckit+/CD34+/CD16/CD32low), granulocyte macrophage progenitors (GMPs; lin−/ckit+/CD34+/CD16/CD32high), and megakaryocyte-erythroid progenitor (MEP; lin−/ckit+/CD34−/CD16/CD32low) cells using a FACSAria flow cytometer (BD Biosciences). Dead cells were excluded by staining with 7-AAD. Subsequently, for the analysis of single MLL-AF9 colonies, a similar procedure was followed without the Ficoll separation. Cells were then analyzed on an LSR II (BD Biosciences).
Chromatin immunoprecipitation
ChIP experiments were performed on the knockin MLL-AF9 cell line 4166,29 MLL-AF9 mBM clones, and primary AML samples of patients with known MLL-AF9 rearrangements, according to the ChIP protocol (available on the Millipore Web site, http://www.millipore.com/userguides/tech1/mcproto407). Immunoprecipitation of cross-linked chromatin was performed with Abs against Histone H3 (trimethyl K4), Histone H3 (dimethyl K79; both Abcam; Ab ab8580, respectively, ab3594), and anti–trimethyl-Histone H3 (Lys27; Millipore) or an equal amount of isotype IgG (Cell Signaling Technology) as a background control. Three independent experiments were performed and the amount of immunoprecipitated DNA is represented as signal relative to cross-linked input DNA. Q-PCR was performed using primers (supplemental Table 1) directed against promoter regions of EVI1, HoxA9, and β-actin.
Transplantation experiments
For in vivo experiments, 4166 cells were transduced with shRNA-bearing or control vector at an MOI of 100. Transduced cells were grown in 4166 medium overnight without puromycin selection. Transduced or untreated 4166 cells combined with wild-type BM cells were transplanted via tail vein injection into 8-week-old lethally irradiated (8 Gy) C57BJ/6 mice (1 × 105 4166 cells + 5 × 105 wild-type normal BM cells/mouse; 9 mice in each of the Evi1 shRNA, control virus, and untreated groups). All animal experiments were approved by the Institutional Animal Care and Use Committee at the University of Minnesota.
Cell cycle, apoptosis, and in vitro drug-resistance assay after Evi1 knockdown
Evi1 knockdown in 4166 cells was performed as described in the previous paragraph, and cells were selected for 3 days on 1.5 μg/mL puromycin before conducting experiments. Analyses for cell cycle and apoptosis were performed as previously described.29
In vitro drug resistance was assessed with the MTT assay.30 Briefly, 96-well microculture plates contained 1.5 × 104 4166 cells suspended in 100 μL with 6 duplicate concentration ranges of each drug. The following drugs and range of concentrations were used: cytarabine (0-2μM); idarubicin (0-0.16μM). Untreated 4166 were used as a control. Cells were incubated for 3 days in the presence of each drug, and cell viability was measured by adding 10 μL of MTT solution (5 mg/mL) and culturing for an additional 6 hours, followed by addition of 100 μL of acidified 2-propanol. Optical density (OD) was measured at 562 nm on a microplate reader (Victor 3; PerkinElmer). Pilot assays were conducted to secure that the OD is linearly related to the number of viable cells within the settings of our experiments. Cell survival was calculated by the following formula: (mean OD drug-treated wells)/(mean OD control wells) × 100%. Dose-response curves and assessment of LD50 (lethal dose for 50%) was performed with GraphPad Prism software.
Statistical analysis and gene expression profiling
Statistical analyses were performed using Mathworks (Matlab) with the statistical and bioinformatics toolbox. Differences in FAB classification for the EVI1pos and EVI1neg AMLs were assessed using the Fisher exact test. Clustering analyses of the gene expression profiles (Gene Expression Omnibus GSE6891) were performed as previously described.31
Results
EVI1neg MLL-rearranged AMLs represents a distinct subtype with different gene expression profiles compared with EVI1pos AMLs
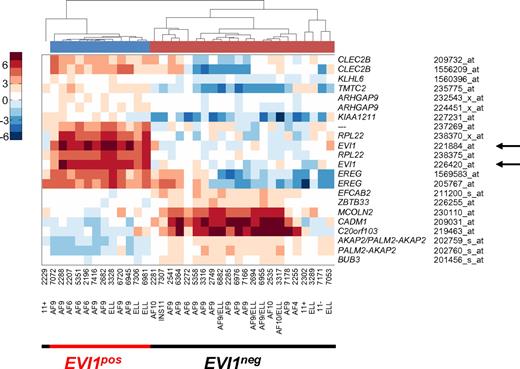
Previously, we found frequent EVI1 overexpression in AMLs with MLL rearrangements.19,20 To investigate whether EVI1pos and EVI1neg MLL-rearranged leukemias represent distinct molecular leukemia subtypes, we performed unsupervised clustering of gene expression profiling (GEP) data of 506 AMLs and identified a cluster that contains the majority of EVI1neg AMLs. Pearson correlation distances of the 35 MLL-rearranged AMLs illustrate EVI1pos being distinct from EVI1neg AMLs (supplemental Figure 1). Comparison of mRNA gene expression data of EVI1neg versus EVI1pos revealed 17 differentially expressed genes (22 probe sets), which strikingly contain 2 probe sets for EVI1 (Figure 1). EVI1 positivity was validated by real-time quantitative PCR (RQ-PCR) (Figure 2A; supplemental Table 2). Thus, EVI1pos MLL-rearranged AMLs represent a unique subtype that is different from EVI1neg MLL-rearranged myeloid leukemias.
Supervised gene expression profiling of MLL-rearranged AMLs uncovers 2 different subgroups. Pearson correlation clustering of 35 MLL-rearranged leukemias defined 2 subgroups, an EVI1pos group and an EVI1neg group (supplemental Figure 1). Supervised analysis revealed 22 probe sets that were differentially expressed between the 2 MLL-rearranged subgroups. Red color corresponds to high correlation, whereas blue color corresponds to low correlation of mRNA expression of genes in patient samples. Note that arrows (←) point to probe sets for EVI1 mRNA expression, which are part of the distinctive signature separating the 2 clusters.
Supervised gene expression profiling of MLL-rearranged AMLs uncovers 2 different subgroups. Pearson correlation clustering of 35 MLL-rearranged leukemias defined 2 subgroups, an EVI1pos group and an EVI1neg group (supplemental Figure 1). Supervised analysis revealed 22 probe sets that were differentially expressed between the 2 MLL-rearranged subgroups. Red color corresponds to high correlation, whereas blue color corresponds to low correlation of mRNA expression of genes in patient samples. Note that arrows (←) point to probe sets for EVI1 mRNA expression, which are part of the distinctive signature separating the 2 clusters.
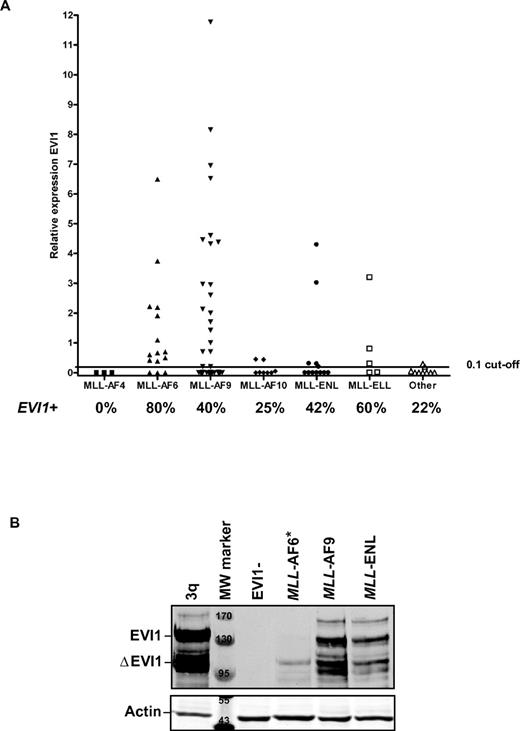
EVI1 is frequently expressed in MLL-rearranged leukemias. (A) Relative EVI1 expression of patients with different 11q23 aberrations, corresponding to MLL-AF4, MLL-AF6, MLL-AF9, MLL-AF10, MLL-ENL, and other MLL-fusions. A cutoff value of 0.1 relative to the calibrator ovarian carcinoma cell line SKOV3 was chosen to classify EVI1pos from EVI1neg cases.19 Per MLL translocation, the percentage (%) of EVI1pos patients is indicated. (B) EVI1 protein expression is shown in selected patient samples with MLL-rearranged leukemias. *For the MLL-AF6 sample, we observed EVI1 protein expression at longer exposure time. A 3q26/EVI1-rearranged AML and an EVI1-negative case served as, respectively, positive and negative control for EVI1 protein expression. Actin staining shows equal protein loading. MW marker indicates molecular weight marker; marking bands at, respectively 170, 130, 95 kDa on EVI1 Western and 55, 43 kDa on actin Western blot.
EVI1 is frequently expressed in MLL-rearranged leukemias. (A) Relative EVI1 expression of patients with different 11q23 aberrations, corresponding to MLL-AF4, MLL-AF6, MLL-AF9, MLL-AF10, MLL-ENL, and other MLL-fusions. A cutoff value of 0.1 relative to the calibrator ovarian carcinoma cell line SKOV3 was chosen to classify EVI1pos from EVI1neg cases.19 Per MLL translocation, the percentage (%) of EVI1pos patients is indicated. (B) EVI1 protein expression is shown in selected patient samples with MLL-rearranged leukemias. *For the MLL-AF6 sample, we observed EVI1 protein expression at longer exposure time. A 3q26/EVI1-rearranged AML and an EVI1-negative case served as, respectively, positive and negative control for EVI1 protein expression. Actin staining shows equal protein loading. MW marker indicates molecular weight marker; marking bands at, respectively 170, 130, 95 kDa on EVI1 Western and 55, 43 kDa on actin Western blot.
EVI1 is expressed in 11q23-rearranged AMLs with distinct MLL-fusion partners
To study the relationship between EVI1 expression and distinct MLL-fusion genes, we determined the relative EVI1 expression by RQ-PCR in a larger cohort of patients with MLL-rearranged leukemias (54 cases from the HOVON cohort and 48 AMLs from the AMLSG cohort). EVI1 expression (EVI1pos) was found in 44 of 102 MLL-rearranged cases. Patient characteristics and the relative EVI1 expression of the 102 MLL-rearranged AMLs included in this study are shown in supplemental Table 2. EVI1 positivity was found in 0 of 3 MLL-AF4, 12 of 15 MLL-AF6, 20 of 50 MLL-AF9, 2 of 8 MLL-AF10, 5 of 12 MLL-ENL and 3 of 5 MLL-ELL cases, and in 2 of 9 cases with other MLL rearrangements (Figure 2A). Western blot analysis on protein lysates of selected EVI1pos AML samples with MLL-AF6, MLL-AF9, and MLL-ENL rearrangements and a control AML with a 3q26 rearrangement (3q) revealed high EVI1 protein expression (Figure 2B). An AML sample that did not express EVI1 mRNA (EVI1neg) showed no EVI1 protein on the same Western blot (Figure 2B).
EVI1negMLL-AF9 AMLs are morphologically different from the EVI1pos cases
MLL-AF6, MLL-AF9, and MLL-ENL are among the most frequently occurring MLL rearrangements in AML.32,33 We found that 31 of 36 EVI1negMLL-AF6, MLL-AF9, and MLL-ENL cases for which morphologic data were available had FAB-M5 monoblastic morphology. On the other hand, only 5 of 28 EVI1pos MLL-AF6, MLL-AF9, and MLL-ENL cases were of the FAB-M5 subtype. In fact, EVI1posMLL-rearranged cases were found within all FAB classes (Fisher exact test P < .0001; supplemental Table 2). Because FAB-M5 cases mainly consist of monoblasts, we sought to determine whether the stem and progenitor cell–enriched CD34+/CD38− and CD34+/CD38+ populations in the EVI1neg AMLs were EVI1neg as well. CD34+/CD38− and CD34+/CD38+ fractions from 2 EVI1negMLL-AF9 cases were FACS-sorted (supplemental Figure 2a, supplemental Table 3) and studied for EVI1 expression. The phenotypically immature fractions in these AMLs did not show EVI1 expression (supplemental Figure 2b), emphasizing that these FAB-M5 MLL-AF9 AMLs are genuinely EVI1neg.
EVI1 expression pattern in EVI1posMLL-rearranged AMLs is aberrant
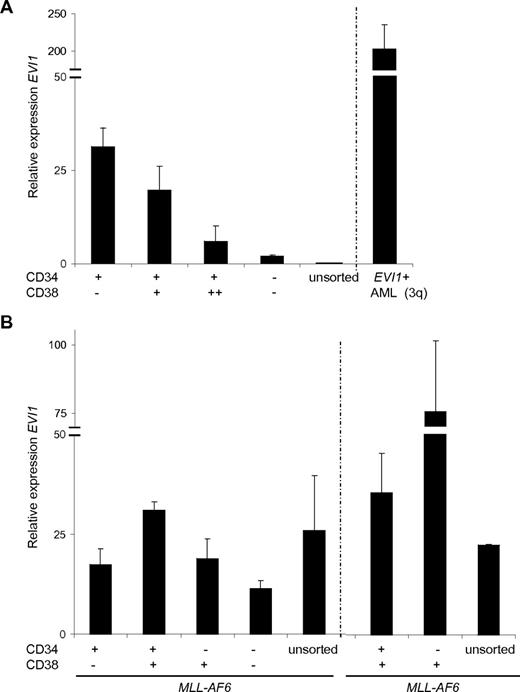
EVI1 mRNA expression is inversely correlated with differentiation in normal human BM samples, that is, EVI1 is high in primitive CD34+/CD38− fractions and markedly lower in the more differentiated CD34+/CD38+, CD34+/CD38++, or in CD34− cells (Figure 3A). To investigate whether the EVI1 expression patterns in defined subfractions of MLL-rearranged leukemias were abnormal, we determined EVI1 mRNA levels in sorted fractions of EVI1posMLL-rearranged AMLs. In the MLL-AF6 (n = 2) and MLL-AF9 (n = 2) cases, high EVI1 mRNA expression levels were observed in sorted CD34+/CD38−, CD34+/CD38+, CD34−/CD38+, and CD34−/CD38− fractions (Figure 3B, supplemental Figure 3). Thus, in contrast to normal human BM-sorted fractions, EVI1posMLL-rearranged AMLs show ubiquitous expression of EVI1 in the distinct stem and progenitor fractions.
Aberrant expression ofEVI1mRNA in sorted CD34/CD38 fractions of MLL-AF6–rearranged AMLs. BM cells from (B) 2 MLL-AF6 rearranged AMLs and (A) normal BM were FACS sorted for the distinct CD34/CD38 populations, with subsequent isolation of mRNA. Relative expression was normalized against the reference gene PBGD. Each measurement was carried out in triplicate and SD is shown per measurement.
Aberrant expression ofEVI1mRNA in sorted CD34/CD38 fractions of MLL-AF6–rearranged AMLs. BM cells from (B) 2 MLL-AF6 rearranged AMLs and (A) normal BM were FACS sorted for the distinct CD34/CD38 populations, with subsequent isolation of mRNA. Relative expression was normalized against the reference gene PBGD. Each measurement was carried out in triplicate and SD is shown per measurement.
Evi1 is expressed in MLL-AF9–transformed murine BM cells
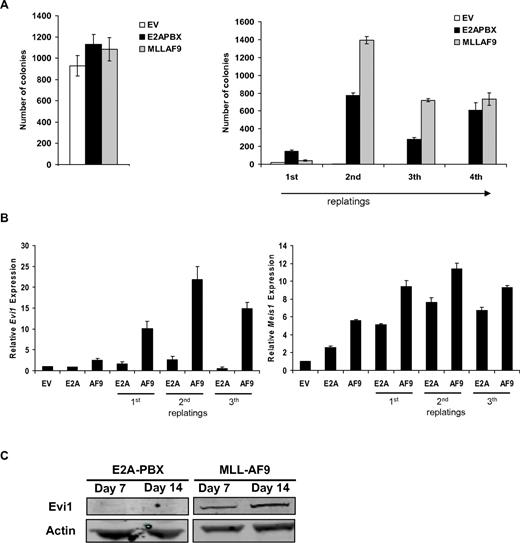
We next wished to investigate whether there was a causal relationship between the expression of MLL-fusion proteins and EVI1 in myeloid precursor cells. Because MLL-AF9 cases formed the major fraction of MLL-rearranged AMLs (49%) and EVI1posMLL-AF9–rearranged MLLs show a clinical behavior that is identical to the whole EVI1posMLL-rearranged AML cohort (Gröschel et al20 and Gröschel et al34 ), we focused for the remainder of our study on the effects of MLL-AF9. Ficoll-separated murine BM (mBM) cells were transduced with retrovirus containing MLL-AF9, E2A-PBX, or empty vector (EV) and subsequently cultured in an in vitro clonogenic assay (schematic outline of the procedure in supplemental Figure 4A). In agreement with the literature, MLL-AF9 and E2A-PBX–transformed cells provided colonies that could be serially replated, whereas EV control colonies could not (Figure 4A). Both MLL-AF9 and E2A-PBX–transformed mBM colony cells showed elevated Meis1 transcripts, whereas sustained, high Evi1 mRNA was primarily found in MLL-AF9–transformed colonies (Figure 4B). We also detected up-regulation of Evi1 mRNA in mBM cells transduced with other MLL-fusion constructs (supplemental Figure 4B-C). In accordance with the mRNA expression data, lysates of collected colonies of MLL-AF9–transduced mBM revealed EVI1 protein expression as detected by Western blotting, whereas no EVI1 protein was detected in E2A-PBX–transformed cells (Figure 4C). Together, these experiments demonstrate Evi1 expression in mBM cells on transformation by MLL fusion genes as observed in human MLL-rearranged AMLs.
High Evi1 expression in MLL-AF9–transformed mononucleated normal mouse BM. (A) Transduction of normal mBM with MLL-AF9 leads to colony formation and sustained replating capacity compared with empty vector control (EV). Normal mBM transduced with E2A-PBX served as a positive control for transformation. Experiments were performed in triplicate and presented as average number of CFUs with SD. (B) Evi1 and Meis1 relative mRNA expression was determined from colonies at day 7, 14, or 21. An average of 3 measurements is depicted with SD. (C) Detection of Evi1 protein by Western blot analysis in lysates of pooled colonies from day 7 or 14. Blots were reprobed with an α-actin Ab to show equal protein loading.
High Evi1 expression in MLL-AF9–transformed mononucleated normal mouse BM. (A) Transduction of normal mBM with MLL-AF9 leads to colony formation and sustained replating capacity compared with empty vector control (EV). Normal mBM transduced with E2A-PBX served as a positive control for transformation. Experiments were performed in triplicate and presented as average number of CFUs with SD. (B) Evi1 and Meis1 relative mRNA expression was determined from colonies at day 7, 14, or 21. An average of 3 measurements is depicted with SD. (C) Detection of Evi1 protein by Western blot analysis in lysates of pooled colonies from day 7 or 14. Blots were reprobed with an α-actin Ab to show equal protein loading.
Evi1pos and Evi1neg replatable colonies are generated on MLL-AF9 transformation of mBM cells
The above experiments show that Evi1 expression is high in pooled fractions of myeloid progenitors after transduction with MLL-AF9; however, they do not answer the question of whether MLL-AF9 transduction leads to Evi1 induction in all or in a subset of transformed progenitor cell. To address this question, we generated clonal MLL-AF9 mBM cell lines by picking and expanding single primary MLL-AF9–transduced colonies (strategy outlined in supplemental Figure 5A). Seventy-five of those picked colonies could be indefinitely replated. In 26 (35%) of 75 clones, Evi1 mRNA expression was detected, with half of them (13 of 75) expressing high Evi1 mRNA levels (supplemental Figure 5B). Evi1 mRNA expression levels remained stable after serial rounds of replating (supplemental Figure 5D). Evi1neg clones remained negative on replating. We did not observe a difference in growth capacity between Evi1pos or Evi1neg clones as monitored by colony or liquid cell cultures (supplemental Figure 5C,E).
Evi1 expression pattern in Evi1pos MLL-AF9–transformed mouse BM cells is abnormal
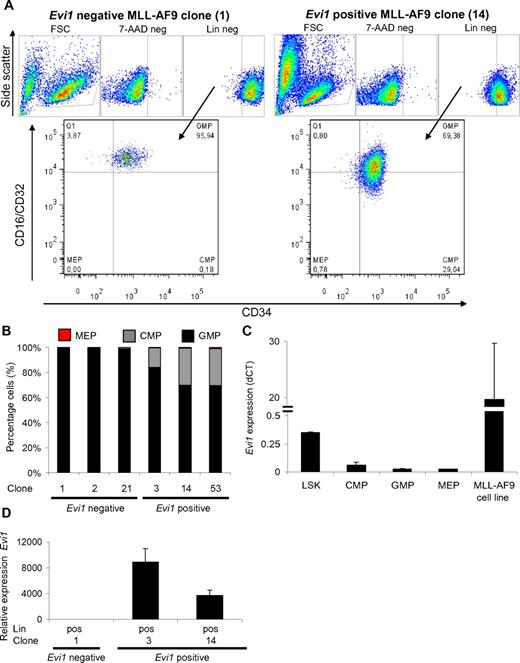
Flow cytometric analysis of serially replated MLL-AF9–transformed BM cells revealed a remarkable difference between Evi1pos versus Evi1neg MLL-AF9–transformed clones. Evi1pos MLL-AF9 cell fractions contained CMPs as well as GMPs, whereas in Evi1neg MLL-AF9–transformed cell fractions only GMPs were observed (Figure 5A-B). These data are in line with immunophenotyping and morphologic analysis of human MLL-rearranged AMLs which showed that Evi1neg cases were more mature than the Evi1pos leukemias.
Enrichment of CMPs in Evi1pos clonal MLL-AF9–transformed BM cell cultures. (A) Example of FACS-sorting strategy to select for CMP, GMP, and MEP derived from cultured Evi1pos and Evi1negMLL-AF9 clones. (B) Percentages of CMP, GMP, and MEP relative to the total number of progenitor cells are calculated for each clone. (C) Evi1 expression in LSK, CMP, GMP, and MEP subpopulations of normal mononucleated mBM cells. The MLL-AF9 cell line 4166 served as positive control for Evi1 expression. (D) Evi1 mRNA expression in lineage-positive subfractions of MLL-AF9 clones. The expression of Evi1 was calculated relative to the lineage-negative fraction of MLL-AF9 clone #1. Evi1 relative expression was normalized using Hprt as a reference gene. For the last 2 panels, the average of 3 experiments with its SD is shown.
Enrichment of CMPs in Evi1pos clonal MLL-AF9–transformed BM cell cultures. (A) Example of FACS-sorting strategy to select for CMP, GMP, and MEP derived from cultured Evi1pos and Evi1negMLL-AF9 clones. (B) Percentages of CMP, GMP, and MEP relative to the total number of progenitor cells are calculated for each clone. (C) Evi1 expression in LSK, CMP, GMP, and MEP subpopulations of normal mononucleated mBM cells. The MLL-AF9 cell line 4166 served as positive control for Evi1 expression. (D) Evi1 mRNA expression in lineage-positive subfractions of MLL-AF9 clones. The expression of Evi1 was calculated relative to the lineage-negative fraction of MLL-AF9 clone #1. Evi1 relative expression was normalized using Hprt as a reference gene. For the last 2 panels, the average of 3 experiments with its SD is shown.
Importantly, in normal mBM, Evi1 is expressed primarily in the HSC fraction while CMP, GMP, and MEP fractions show significantly reduced levels (Figure 5C). To address the question of whether the Evi1 expression pattern was aberrant in transformed mBM cells, we determined Evi1 mRNA levels in mature Lin+ cell fractions of Evi1pos clones. Q-PCR on lineage-positive cells sorted from Evi1pos MLL-AF9–transformed clones showed high Evi1 levels (Figure 5D) in the Lin+ cells. Because normal Lin+ cells are Evi1neg, these data point to an aberrant Evi1 expression pattern in these MLL-AF9–transformed clones. In fact, these data are in full agreement with the abnormal expression patterns observed in human EVI1posMLL-rearranged AMLs (Figure 3, supplemental Figure 3) and are highly suggestive of a role for MLL-AF9 in aberrant EVI1 expression in MLL-rearranged EVI1pos-transformed cells.
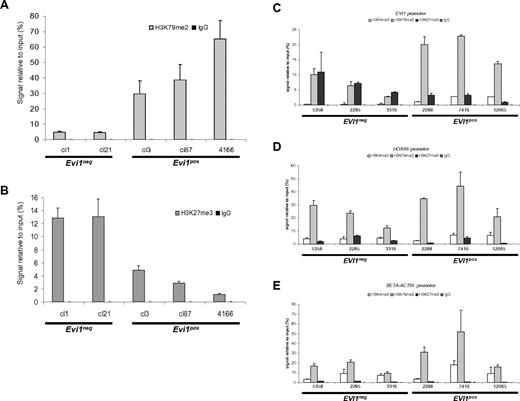
Enrichment of H3K79me2 on the EVI1 promoter in EVI1pos MLL-AF9–transformed cells
MLL-AF9 rearrangements result in loss of the H3K4 methyl transferase domain of MLL, but create a chimeric protein that recruits the H3K79 histone methyl transferase DOT1L, leading to H3K79 dimethylation (H3K79me2) at target promoter regions.35,36 To investigate whether the observed up-regulation of Evi1 is mediated by MLL-AF9, we used ChIP to assess H3K79me2 of the Evi1 promoter region. Clones with high Evi1 expression showed significant H37K79me2 enrichment on the Evi1 promoter (Figure 6A), whereas H3K27me3, a mark for repressed genes was low on the Evi1 promoter (Figure 6B). On the contrary, Evi1neg clones showed reduced levels of the H3K79me2 but high H3K27me3 at the Evi1 promoter (Figure 6A-B). Importantly, in the Evi1neg clones, we were able to detect expected enrichment of H3K79me2 on the putative MLL-AF9 target genes HoxA9 and Meis1 (supplemental Figure 6, and data not shown) indicating that the lack of H3K79me2 mark at the Evi1 locus was specific. H3K79me2 enrichment was also detected on the Evi1 promoter region in the MLL-AF9 knock-in cell line 4166, compared with wild-type mBM cells (supplemental Figure 7). Specificity of the H3K79me2 ChIP in 4166 cells was shown by the clear H3K79me2 mark on the promoter region of the MLL-AF9 target gene HoxA9, in contrast to the absence of an enrichment on the HoxC8 promoter, which is not a target gene of MLL-AF9 (supplemental Figure 7). Dot1L knockdown carried out on the Evi1-expressing MLL-AF9 knockin cell line 4166 showed a partial loss of Dot1L expression (supplemental Figure 8). Evi1 levels decreased significantly on Dot1L knockdown. Moreover, this decrease was comparable with that of HoxA9 and Meis1 (supplemental Figure 8). These data suggest that the H3K79me2 mark at the promoter of Evi1 is important for its expression in MLL-AF9–transformed cells.
Enrichment of H3K79me2 on the Evi1 promoter of MLL-AF9–transformed cells. ChIP using (A) H3K79me2 and (B) H3K27me3 Abs showing enrichment of the H3K79me2 mark on the Evi1 promoter of Evi1pos vs Evi1neg clones. Conversely, the H3K27me3 mark is enriched on the Evi1 promoter of Evi1neg clones compared with Evi1posMLL-AF9 clones. ChIP using H3K4me3, H3K79me2, and H3K27me3 Abs performed on 3 EVI1pos and 3 EVI1neg MLL-AF9 rearranged leukemias showed higher H3K79me2 marks present on the EVI1 promoter of EVI1pos MLL-AF9–rearranged human AMLs compared with EVI1neg cases. Q-PCRs were performed to monitor relative enrichment of each histone mark on the promoters of either (C) EVI1, (D) HoxA9 promoter, or (E) β-actin. Experiments show average relative enrichment with SD of either (A-B) 3 biologic replicates or (C-E) 2 independent ChIP pulldowns with triplicate Q-PCR measurements performed on the same cross-linked patient material.
Enrichment of H3K79me2 on the Evi1 promoter of MLL-AF9–transformed cells. ChIP using (A) H3K79me2 and (B) H3K27me3 Abs showing enrichment of the H3K79me2 mark on the Evi1 promoter of Evi1pos vs Evi1neg clones. Conversely, the H3K27me3 mark is enriched on the Evi1 promoter of Evi1neg clones compared with Evi1posMLL-AF9 clones. ChIP using H3K4me3, H3K79me2, and H3K27me3 Abs performed on 3 EVI1pos and 3 EVI1neg MLL-AF9 rearranged leukemias showed higher H3K79me2 marks present on the EVI1 promoter of EVI1pos MLL-AF9–rearranged human AMLs compared with EVI1neg cases. Q-PCRs were performed to monitor relative enrichment of each histone mark on the promoters of either (C) EVI1, (D) HoxA9 promoter, or (E) β-actin. Experiments show average relative enrichment with SD of either (A-B) 3 biologic replicates or (C-E) 2 independent ChIP pulldowns with triplicate Q-PCR measurements performed on the same cross-linked patient material.
We applied the same type of ChIP experiments for a panel of 6 human AML samples, that is, 3 with and 3 without EVI1 expression. Again, we observed enrichment of H3K79me2, with corresponding low levels of H3K27me3 levels on the EVI1 promoter of AML samples with high EVI1 expression. Notably, AML samples with no EVI1 expression exhibited lower levels of the H3K79me2 mark with a striking concomitant rise of equal levels of H3K27me3 on the EVI1 promoter region (Figure 6C). This phenomenon was only observed on the EVI1 promoter and not on the HoxA9 or β-actin promoter of EVI1neg human AML samples (Figure 6D-E)
MLL-AF9 maintains Evi1 expression in Evi1-expressing BM fractions
The flow cytometric difference between Evi1pos versus Evi1negMLL-AF9–transformed clones led us to hypothesize that MLL-AF9 may transform different progenitor fractions (Figure 5A). We therefore purified HSCs as Lin−CD34−Sca1+c-Kit+ and GMP as Lin−CD34+Sca1−c-Kit+CD16/32+ from mouse BM, which we subsequently transduced with MLL-AF9. Both HSC- and GMP-produced colonies were replatable in semisolid media supplemented with IL-3, IL-6, and SCF. MLL-AF9–expressing HSC-derived colonies expressed Evi1, while MLL-AF9–expressing GMP-derived colonies expressed no detectable levels of Evi1 mRNA (supplemental Figure 9). Thus, our data suggest that the difference between Evi1pos and Evi1neg MLL-AF9–transformed cells is the result of different progenitors that were initially transformed by the same MLL-fusion gene. In normal mouse BM fractions, Evi1 expression is high in the LSK fraction that contains the HSCs and low in CMP, GMP, or MEP compartments (Figure 5C). This difference was also observed in the different human stem cell and progenitor fractions. It is therefore suggestive that MLL-AF9 can transform Evi1-expressing primitive marrow precursors which subsequently maintain Evi1pos and the fusion gene is capable of transforming more mature Evi1neg progenitors that stay Evi1neg.
Evi1 knockdown inhibits the growth of MLL-AF9–transformed cells in vitro and in vivo
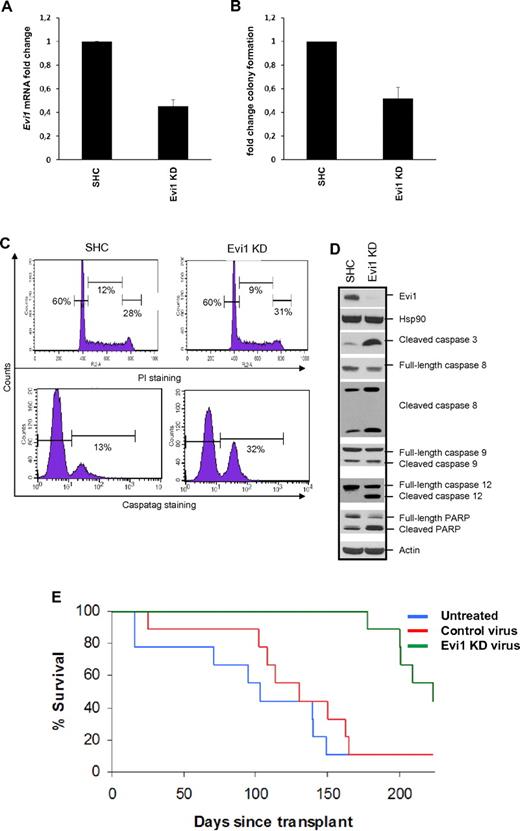
To investigate the role of EVI1 in MLL-AF9–directed leukomogenesis, we made use of lentiviral shRNA-mediated knockdown of Evi1 in experimental MLL-AF9 leukemia models. The MLL-AF9 knock-in leukemia cell line 4166 closely mimics human disease because each cell contains only one copy of wt-MLL and one copy of the MLL-AF9 fusion gene, under control of the endogeneous MLL promoter.29 This 4166 cell line expressed high levels of Evi1 mRNA and EVI1 protein, which could be down-regulated after Evi1 knockdown (Figure 7A,D). Evi1 knockdown resulted in reduced clonogenic survival in methylcellulose semisolid media (Figure 7B). Similarly, Evi1 knockdown also reduced colony formation in Evi1posMLL-AF9 mBM clones. Importantly, Evi1neg cells were unresponsive to Evi1 knockdown, showing that the observed effects on colony formation are not because of off-target effects of the applied shRNA for Evi1 (supplemental Figure 10).
Evi1 knockdown (KD) in MLL-AF9 cell line 4166 leads to reduced growth in vitro and in vivo. Transduction of 4166 with lentiviral vectors containing shRNA against Evi1 leads to reduced (A) Evi1 mRNA and (D top) protein expression. (B) Reduced Evi1 levels were accompanied with a significant reduction of colony formation. Fold change of 4 experiments (mean + SD) is shown for panels A and B. (C) Evi1 shRNA-mediated inhibition of cell growth was attributed to (bottom panel) an induction of apoptosis, (top panel) with no changes in cell-cycle profile. (D) Western blotting shows an increase of (pro)apoptotic markers after knockdown of Evi1. HSP90 and actin were used to show equal cell viability and loading of protein lysates. SHC indicates short hairpin control. (E) Survival curve of lethally irradiated mice transplanted with 1 × 105 4166 cells, transduced with either Evi1 shRNA or control virus supplemented with 5 × 105 normal mBM cells. As a control group, untreated 4166 cells were transplanted to monitor offset of normal 4166-mediated MLL-AF9 tumorigenesis. All 3 groups contained 9 mice each. By posttransplantation day 170, the majority of the mice in the untreated (8 of 9) or control virus (8 of 9) transduced group had succumbed to leukemia, while all mice receiving Evi1 shRNA-transduced cells were still alive. Leukemia development was significantly delayed in animals receiving Evi1 shRNA-transduced 4166 cells (log-rank P < .001) with 44% of the mice being long-term survivors.
Evi1 knockdown (KD) in MLL-AF9 cell line 4166 leads to reduced growth in vitro and in vivo. Transduction of 4166 with lentiviral vectors containing shRNA against Evi1 leads to reduced (A) Evi1 mRNA and (D top) protein expression. (B) Reduced Evi1 levels were accompanied with a significant reduction of colony formation. Fold change of 4 experiments (mean + SD) is shown for panels A and B. (C) Evi1 shRNA-mediated inhibition of cell growth was attributed to (bottom panel) an induction of apoptosis, (top panel) with no changes in cell-cycle profile. (D) Western blotting shows an increase of (pro)apoptotic markers after knockdown of Evi1. HSP90 and actin were used to show equal cell viability and loading of protein lysates. SHC indicates short hairpin control. (E) Survival curve of lethally irradiated mice transplanted with 1 × 105 4166 cells, transduced with either Evi1 shRNA or control virus supplemented with 5 × 105 normal mBM cells. As a control group, untreated 4166 cells were transplanted to monitor offset of normal 4166-mediated MLL-AF9 tumorigenesis. All 3 groups contained 9 mice each. By posttransplantation day 170, the majority of the mice in the untreated (8 of 9) or control virus (8 of 9) transduced group had succumbed to leukemia, while all mice receiving Evi1 shRNA-transduced cells were still alive. Leukemia development was significantly delayed in animals receiving Evi1 shRNA-transduced 4166 cells (log-rank P < .001) with 44% of the mice being long-term survivors.
To discern by which mechanism Evi1 knockdown leads to reduced cell growth, we performed cell cycle and apoptosis analysis on 4166 cells. Flow cytometric analysis showed that Evi1 knockdown did not lead to significant changes in cell-cycle profile (Figure 7C top panel). On the other hand, we observed a significant increase in apoptosis in 4166 cells after Evi1 knockdown compared with vector control transduced 4166 cells (Figure 7C bottom panel). The increase in apoptosis after Evi1 knockdown was characterized by an induction of the apoptotic markers: cleaved forms of Caspase -3, 8, and -12 and PARP (Figure 7D). These experiments demonstrate that down-regulation of EVI1 expression in MLL-AF9–transformed cells causes reduced cell growth through induction of apoptosis, without affecting cell-cycle progression.
We have shown that MLL-rearranged leukemia patients with high EVI1 expression have an adverse prognosis with a corresponding poor response to current treatment modalities.19,20 Therefore, we determined the cytotoxicity in 4166 cells of 2 cytostatic drugs, cytarabine and idarubicin, with a previously described in vitro toxicity test.30 We treated cells with Evi1 specific or control shRNA and observed that a reduction of Evi1 protein levels resulted in an increased sensitivity of 4166 cells to either idarubicin or cytarabine or the combination of the 2, as monitored by a left-shift of the dose-response curve and a reduced LD50 after Evi1 knockdown (supplemental Figure 11). These results show that Evi1pos MLL-AF9–transformed cells become more sensitive to chemotherapeutic agents on Evi1 down-regulation.
Previously, we have shown that transplantation of 4166 cells in irradiated mice recapitulates MLL-AF9 leukemia.29 To study the effect of Evi1 knockdown on in vivo tumor formation, lethally irradiated mice were transplanted with a mixture of wild-type donor cells plus 4166 cells that were either untreated or transduced with Evi1 shRNA or control vector. Mice were killed when becoming moribund. At necropsy, mice had signs of AML, displaying leukocytosis and splenomegaly. As depicted in Figure 7E, most of the control animals (transplanted with 4166 cells that were either untreated or transduced with control virus) died of leukemia within 170 days after transplantation. In contrast, leukemia development was significantly delayed in animals receiving 4166 cells transduced with Evi1 shRNA (P < .001, log-rank test), with 44% of the animals being long-term survivors (Figure 7E). Mice that did not develop leukemia were killed at the end of the experiments and showed no signs of leukemogenesis at autopsy.
Discussion
In previous studies, we found that a subset of AMLs overexpressed EVI1 and were associated with an adverse prognosis compared with EVI1neg AMLs that had similar molecular and karyotypic features. In particular, it was demonstrated that leukemias with MLL translocations showed frequent EVI1 expression.19,20 In the current study, we showed in a larger AML cohort carrying 11q23 translocations that ∼ 43% of all MLL-rearranged leukemias are EVI1pos, and that EVI1 expression was independent of the fusion partner involved in the translocation with MLL. In addition, we present evidence that, using MLL-AF9 AMLs as an example of the whole cohort, EVI1pos MLL-AF9 AMLs differ molecularly, morphologically, and immunophenotypically from EVI1neg MLL-AF9 leukemias.
Using mouse models for MLL-AF9 fusion leukemia, we provide evidence that on transformation of Evi1pos HSCs, the presence of the MLL-AF9 fusion protein causes Evi1 expression not to be turned off (supplemental Figure 9). Moreover, shRNA-mediated knockdown inhibits leukemia growth both in vitro and in vivo, suggesting that Evi1 is important for the growth of these EVI1pos MLL-fusion gene–transformed leukemias. These results are corroborated by recent reports showing that MLL-ENL up-regulates Evi1 expression in a mouse leukemia model, in which leukemic transformation was dependent on Evi1.24,25
We noted that although MLL-AF9 is readily able to transform normal mouse BM cells, only a fraction of colonies were Evi1pos (supplementary figure S5B). Similar to Arai et al, we found that in these retroviral models of MLL-fusion gene leukemia, Evi1 overexpression was predominantly associated with the stem cell–enriched LSK raction of normal BM compared with the more differentiated myeloid progenitors CMP, GMP, and MEP.25 Notably, several studies have shown that MLL-fusion gene–induced leukomogenesis is more efficient in LSKs compared with the more mature myeloid progenitors GMPs.23,37 Because Evi1 expression is normally high in these LSK cells, our results suggest that coexpression of Evi1 is at least partly responsible for the observed transformation of LSKs. This is in line with our observations in human AMLs, which show that EVI1neg MLL-rearranged leukemias are predominantly of the monoblastic subtype whereas the EVI1pos leukemias were not restricted to one morphologic subtype. Furthermore, we show that while EVI1 expression in normal hematopoiesis is primarily restricted to the stem cell–enriched CD34+/CD38− fraction, expression of EVI1 is aberrantly extended into the more differentiated CD34+/CD38+, CD34−/CD38+, and CD34−/CD38− fractions in EVI1pos MLL-rearranged leukemias. Recently, Eppert et al reported that EVI1 is part of the gene signature associated with HSCs and leukemia stem cells (LSCs).38 Moreover, they show that high expression of these HSC- and LSC-associated signatures is correlated with poor survival.37 Collectively, these results underline the prognostic significance of high EVI1 expression in AML and suggest that aberrant transcriptional regulation of EVI1 might impart aberrant self-renewal to LSCs. In EVI1neg MLL-rearranged leukemias, defects in other genes may have taken over this role.26,39
We further investigated what could be the mechanism leading to up-regulation of Evi1 in a subset of MLL-AF9–transformed normal mBM cells. It is well established that the Evi1 locus contains a hot spot for retroviral insertions.4 Strikingly, a significant number of retroviral-induced leukemias contain mutually exclusive retroviral insertions in Evi1 or its family member Prdm16.40 Retroviral insertions in Evi1 locus are predominantly between the Mds and Evi1 gene leading to activation of Evi1 and not MdsEvi1.4,40 Because we detected concomitant up-regulation of MdsEvi1 and Evi1 in our Evi1pos MLL-AF9–transduced mBM cells, and no up-regulation of Prdm16 transcript in Evi1neg MLL-AF9–transduced cells (supplemental Figure 12), we postulated that Evi1 up-regulation is unlikely the result of retroviral insertion into the Evi1 locus.
MLL fusions can transactivate the EVI1 promoter in luciferase reporter assays (supplemental Figure 13).25 Although others reported binding of MLL-ENL to the Evi1 promoter, we were not able to show direct interaction between MLL-AF9 and Evi1, possibly because of the lack of suitable ChIP-grade Abs. However, we observed clear differences between Evi1pos and Evi1neg MLL-AF9–transformed cells (both human or mouse) with regard to enrichments of H3K79/H3K4 versus H3K27 on the EVI1 promoter. We hypothesize, based on our ChIP data, that MLL-AF9 up-regulates EVI1 transcription via H3K79 methylation, which is known to be a major gene regulatory mechanism used by some MLL-fusion proteins in leukemia.35,36 In accordance, we witnessed that knockdown of Dot1L knockdown in 4166 resulted in decreased mRNA expression levels of Evi1 (supplemental Figure 8). The absence of increased H3K79 methylation at the EVI1 promoter of EVI1neg MLL-AF9 tumors was locus specific, because HoxA9, a known MLL-AF9 target gene, displayed a clear enrichment of the H3K79 mark in those cells. Thus, our findings may be explained by direct regulation of the Evi1 promoter by MLL-AF9 recruiting the DOT1L enzyme to the locus. ChIP experiments using Abs that recognize MLL-fusion proteins will be essential to adequately address this issue.
We suspect that the relatively silent Evi1 gene in GMPs was inaccessible for the MLL-AF9 protein complex and hence was not up-regulated. Our findings are in line with a recently described epigenetic profiling of L-GMPs, which showed no H3K79 methylation on the Evi1 locus in these transformed cells.41 The MLL-AF9 protein complex contains components of the PAFc and super elongation complex required for MLL-fusion protein function.42-46 At least for PAFc it is well established that its expression decreases during normal hematopoietic differentiation47 providing a potential additional layer of regulation of EVI1 expression by MLL-AF9. The above might explain why we detect MLL-fusion leukemias that are EVI1neg, because they might originate in cells that are more differentiated (GMP-like, with low EVI1 expression). In contrast, we propose that the EVI1pos MLL–rearranged leukemias might arise from cells that are more immature (HSC-like, with higher EVI1 expression).
Two recent reports describe the development of therapeutic interventions in MLL-fusion leukemias. They showed efficacy of 2 small molecule inhibitors, I-BET151 or EPZ004777, in which the first leads to displacement of the MLL-fusion protein complex from chromatin and the second selectively inhibits DOT1L.48,49 Because we have shown evidence for qualitative differences between EVI1pos and EVI1neg MLL-rearranged AMLs, we suggest that future research is needed to investigate whether EVI1 expression status would predict treatment response with inhibitors for MLL-fusion leukemias.
Here we showed evidence that MLL-rearranged leukemias can be classified on the basis of their relative EVI1 expression, separating these leukemias into EVI1pos and EVI1neg leukemias with clear distinct morphologic, molecular, and mechanistic differences. We provide indirect evidence for a role of MLL-fusion proteins in the regulation of EVI1 expression in those EVI1pos MLL-rearranged AMLs. Furthermore, we suggest a critical role for EVI1 in the pathogenesis of a subset of MLL-rearranged leukemias, and that targeting of EVI1 in combination with chemotherapeutic agents could be beneficial for patients with EVI1pos MLL-rearranged AMLs.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank their colleagues of the BM transplantation group and the Molecular Diagnostics Laboratory of the Department of Hematology (Erasmus University Medical Center, Rotterdam, The Netherlands) and the Cytogenetics and Molecular Genetics Laboratory (Department of Internal Medicine III, University of Ulm, Ulm, Germany) for storage and molecular analysis of primary leukemia cells. They thank Chi Wai Eric So for providing retroviral constructs containing MLL-AF9 MLL-AF6, MLL-ENL, and E2A-PBX. They thank Trui Visser and Martijn Schoester for technical assistance.
This work was supported by grants from the Dutch Cancer Society (R.D., M.H., and S.L.), Association for International Cancer Research (AICR) grant 118305-118297 (E.M.J.B.), Leukemia & Lymphoma Society grant 116196-118309 (E.M.J.B.), a European Hematology Association (EHA) research fellowship (S.L.), a ZonMW fellowship (S.L.), the National Institutes of Health (R01-CA087053, J.H.K.; K08-CA122191, A.R.K.), the Leukemia Research Fund (A.R.K.), and the Children's Cancer Research Fund (J.H.K., A.R.K.).
National Institutes of Health
Authorship
Contribution: E.M.J.B., M.H., S.L., C.E., E.W., A.V.K., E.R., E.T., J.R.H., and W.A.H. performed experiments; E.M.J.B., M.H., S.L., S.A.A., J.R.H., R.D., and A.R.K. analyzed the results and made the figures; E.M.J.B., S.L., J.H.K., R.D., and A.R.K. designed the research and wrote the manuscript; H.B.B. performed karyotyping and provided additional patient samples; and H.D. provided biologic materials.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ruud Delwel, Department of Hematology, Erasmus University Medical Center, Dr Molewaterplein 50, 3015 GE Rotterdam, The Netherlands; e-mail: h.delwel@erasmusmc.nl.