Abstract
Loss of heterozygosity affecting chromosome 7q is common in acute myeloid leukemia and myelodysplastic syndromes, pointing toward the essential role of this region in disease phenotype and clonal evolution. The higher resolution offered by recently developed genomic platforms may be used to establish more precise clinical correlations and identify specific target genes. We analyzed a series of patients with myeloid disorders using recent genomic technologies (1458 by single-nucleotide polymorphism arrays [SNP-A], 226 by next-generation sequencing, and 183 by expression microarrays). Using SNP-A, we identified chromosome 7q loss of heterozygosity segments in 161 of 1458 patients (11%); 26% of chronic myelomonocytic leukemia patients harbored 7q uniparental disomy, of which 41% had a homozygous EZH2 mutation. In addition, we describe an SNP-A–isolated deletion 7 hypocellular myelodysplastic syndrome subset, with a high rate of progression. Using direct and parallel sequencing, we found no recurrent mutations in typically large deletion 7q and monosomy 7 patients. In contrast, we detected a markedly decreased expression of genes included in our SNP-A defined minimally deleted regions. Although a 2-hit model is present in most patients with 7q uniparental disomy and a myeloproliferative phenotype, haplodeficient expression of defined regions of 7q may underlie pathogenesis in patients with deletions and predominant dysplastic features.
Introduction
Complete loss of chromosome 7 (monosomy 7) or partial deletion involving its long arm [del(7q)] are highly recurrent chromosomal aberrations in myeloid disorders, including myelodysplastic syndrome (MDS), acute myeloid leukemia (AML), and juvenile myelomonocytic leukemia (JMML).1,2 The International Prognostic Scoring System (IPSS), the most validated score for predicting the evolution of patients with MDS, does not discriminate among chromosome 7 anomalies, uniformly assigning these patients to the poor-risk karyotype group.3 Other metaphase cytogenetic (MC) studies have consistently associated lesions involving the long arm of chromosome 7 with inferior survival in AML cases.1,4 However, there is a contention that monosomy 7 and del(7q) are not equivalent in prognosis and disease phenotype spectrum.5,6
In the traditional genetic view, loss of heterozygosity (LOH) for 1 tumor suppressor gene (TSG) allele increases the chance of inactivation of the remaining allele and total loss of function for a cancer-protective locus. In accordance to this 2-hit model, we and other groups found loss-of-function hypomorphic homo- and hemizygous mutations in a variety of genes, including TP53, CBL, or TET2.7-10 However, there is growing evidence that haploinsufficient TSGs also lead to hastened tumorigenesis, showing dramatic phenotypes with loss of only a single allele.11,12 The haploinsufficient model is supported by recent studies in the context of myeloid disorders harboring a deletion of the long arms of chromosome 5 or chromosome 20,13-15 and it is possible that monosomy 7/del(7q) cases are associated with a similar mechanism.
To better address the genomic and clinical complexity of myeloid malignancies associated with 7q abnormalities, we analyzed a large series of cases with single nucleotide polymorphism array (SNP-A)–based karyotyping, direct and next-generation sequencing (NGS), and microarray expression platforms to (1) examine the association of different SNP-A 7q lesions with certain clinical features and other genomic aberrations, (2) define a commonly deleted region or regions (CDRs) and search for recurrent tumor suppressor mutations, and (3) test the haploinsufficiency hypothesis.
Methods
Patients
Informed consent was obtained following the Declaration of Helsinki according to protocols approved by the review boards and ethics committees of the participating institutions. Presentation bone marrow (BM) aspirates from 1458 patients with myeloid malignancies were studied using SNP-A, including 200 AML cases analyzed by SNP-A and NGS 300 through The Cancer Genome Atlas project (TCGA; http://tcga-data.nci.nih.gov/tcga/tcgaHome2.jsp). Microarray expression data were available on a cohort of 183 patients with MDS and 17 healthy controls.16
Diagnosis of hypocellular myelodysplastic syndrome (hMDS) was made based on the presence of dysplastic features and the overall clinical presentation, including the presence of cytopenias, the absence of an excess of blasts (5% in BM or 2% in blood), and a decreased cellularity of the marrow of less than or equal to 20%. When indicated based on clinical suspicion, immunohistochemical staining for CD34 was performed to rule out, or find, collections of immature cells.
Metaphase cytogenetics
Chromosome preparations were G-banded using trypsin and Giemsa, and karyotypes were described according to the International System for Human Cytogenetic Nomenclature.17
SNP-A analysis
Genome-Wide Human SNP 6.0 and GeneChip Human Mapping 250K arrays (Affymetrix) were used for SNP-A analysis of bone marrow DNA as described previously.18 Germ-line encoded copy number variants and nonclonal areas of uniparental disomy (UPD) were excluded from further analysis by a bioanalytic algorithm, based on lesions identified by SNP-A karyotyping in an internal control series (n = 1003) and reported in the Database of Genomic Variants (http://projects.tcag.ca/variation/). Size and location criteria (telomeric > 8.7 Mb and interstitial, > 25 Mb) were used for identification of somatic UPD. In 11 patients, a 7q microdeletion (median size, 0.3 Mb; range, 0.1-0.7 Mb) was detected but testing of germ line DNA was not possible because of lack of appropriate samples. These patients were excluded from analysis as the pathophysiologic significance of such small lesions is not clear. None of those 7q microdeletions was included in any of the CDRs described subsequently.
Direct sequencing
Sanger technique was used for sequencing all exons of 36 candidate genes, included in our SNP-A–defined CDRs, screening a subset of 50 7q LOH patients [UPD(7q), n = 7; del(7q), n = 31; monosomy 7, n = 12]. Samples from this cohort were not used for NGS.
Next-generation sequencing
Two NGS approaches were used in this study. We generated exome chromosome 7 libraries that were enriched for the content of chromosome 7 coding sequences using the SureSelect capture synthetic biotinylated RNA probes from Agilent Technologies, tiling all the coding regions from chromosome 7. Libraries were subjected to high-throughput sequencing on a Genome Analyzer IIx (Illumina) and applied to 11 7q LOH patients [del(7), n = 6; del(7q), n = 2; UPD(7q), n = 3].
The second approach involved the sequencing of 15 paired bone marrow mononuclear cells and CD3+ lymphocytes (used as germ line controls) from 15 patients with different myeloid disorders and SNP-A findings. Among them, we included 2 patients with 7q LOH [UPD(7q) and del(7q)]. A rational bioanalytic algorithm was applied to identify candidate nonsynonymous alterations. First, nonredundantly mapped reads were used for whole exome assembly using the reference genome hg19. Next, the software algorithm called all the positions that vary from the reference genome. The candidate alterations were subtracted by the results of CD3+ lymphocyte-derived DNA (double-checked by direct and simultaneous visualization using DNAnexus Site) and subsequently validated using Sanger sequencing. Moreover, gene mutations affecting 7q LOH were screened using whole exome sequencing results available through TCGA.
Microarray data analysis
Previously published microarray expression data were obtained on a cohort of 183 MDS patients [monosomy 7/del(7q), n = 9].16 Cell intensity calculation and scaling was performed using GeneChip Version 1.40 operating software (Affymetrix). Affymetrix CEL files were preprocessed using robust multiarray average. Data from 17 healthy controls were used to obtain patient and control expression ratios.
Statistical analysis
Comparisons of proportions and ranks of variables between groups were performed by the χ2 test, Fisher exact test, Student t test or Mann-Whitney U test, as appropriate. We used the Kaplan-Meier and the Cox method to analyze overall survival (OS) and progression-free survival, with a 2-sided P less than or equal to .05 determining significance. In Cox models, examination of log (−log) survival plots and partial residuals was performed to assess that the underlying assumption of proportional hazards was met.
Results
Patient cohorts
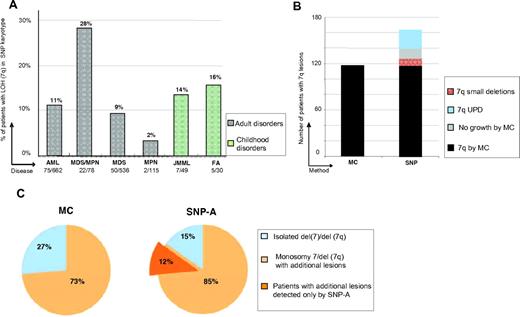
Using SNP-A karyotyping, LOH segments involving 7q were identified in 161 of 1458 patients (11%), consisting of 9% MDS, 28% MDS/myeloproliferative neoplasms (MDS/MPN), 11% AML, 14% JMML, and 16% Fanconi anemia subsets (Figure 1A). MC identified 7q LOH in each of the cases detected by SNP-A except for 26 UPD cases and 11 patients in whom no interpretable metaphases were obtained (Figure 1B). In addition, in 7 cases a balanced translocation with 7q material was noted by MC; in all instances, SNP-A analyses detected a small deletion (> 1 and < 5 Mb) affecting the boundaries of the translocation. In 16 of 67 monosomy 7 cases by MC, SNP-A detected retained chromosome 7 material, probably contributing to marker chromosomes found by MC analysis. With increased resolution, there was a shift toward identification of more complex karyotypes and of additional lesions among the patients with isolated MC 7q aberrations (Figure 1C). By SNP-A, previously cryptic lesions were identified in 45% of the patients who otherwise showed a singular 7q LOH lesion by MC.
Frequency of detection of 7q and additional abnormalities by SNP-A. (A) Distribution of 7q LOH among the 1458 SNP-A–tested patients with myeloid malignancies, according to World Health Organization disease classification. (B) Number of patients with 7q LOH seen on MC and SNP-A. Lesions were observed in 117 of 1458 and 161 of 1458 patients when using MC and SNP-A, respectively. The additional 7q lesions found by SNP-A included those found in patients with no growth of MC cultures, small deletions affecting balanced translocation boundaries,11 and UPD undetectable by MC.26 (C) Percentage of patients with a sole 7q lesion versus accompanied by other abnormalities as identified by MC and SNP-A. SNP-A indicates single nucleotide polymorphism array;MDS/MPN, myelodysplastic syndrome/myeloproliferative neoplasm; AML, acute myeloid leukemia; MPN myeloproliferative neoplasms; JMML, juvenile myelomonocytic leukemia; FA, Fanconi anemia; UPD, uniparental disomy; MC, metaphase cytogenetics; monosomy 7, deletion of whole chromosome 7; and del(7q), partial deltion involving 7q.
Frequency of detection of 7q and additional abnormalities by SNP-A. (A) Distribution of 7q LOH among the 1458 SNP-A–tested patients with myeloid malignancies, according to World Health Organization disease classification. (B) Number of patients with 7q LOH seen on MC and SNP-A. Lesions were observed in 117 of 1458 and 161 of 1458 patients when using MC and SNP-A, respectively. The additional 7q lesions found by SNP-A included those found in patients with no growth of MC cultures, small deletions affecting balanced translocation boundaries,11 and UPD undetectable by MC.26 (C) Percentage of patients with a sole 7q lesion versus accompanied by other abnormalities as identified by MC and SNP-A. SNP-A indicates single nucleotide polymorphism array;MDS/MPN, myelodysplastic syndrome/myeloproliferative neoplasm; AML, acute myeloid leukemia; MPN myeloproliferative neoplasms; JMML, juvenile myelomonocytic leukemia; FA, Fanconi anemia; UPD, uniparental disomy; MC, metaphase cytogenetics; monosomy 7, deletion of whole chromosome 7; and del(7q), partial deltion involving 7q.
The 7q LOH cohort included men (57%) and women (43%) with a median age of 65 years (interquartile range, 59-73 years). The distribution of disease subsets and associated genomic lesions among the 3 classes of chromosome 7 lesions [UPD7q, del(7q) and monosomy 7] is shown in Figure 2, and Table 1 shows clinical characteristics at baseline.
Distribution of disease subsets and associated genomic lesions among the 3 classes of chromosome 7 lesions. (Top) Distribution of LOH detected by SNP-A in the cohort, separated according to the nature of the lesion ([UPD(7q), del(7q), monosomy 7]. Patients have been grouped as follows: red, AML + high risk and intermediate-2 MDS; gray, low risk and intermediate-1 MDS; blue, hypocellular MDS; black, MDS/MPN; and green, Fanconi anemia and JMML. (Middle) Distribution of disease status in patients with 7 LOH separated according the nature of the lesion. (Bottom) Additional SNP-A–detected genomic lesions separated according the same criteria as stated herein. MDS indicates myelodysplastic syndrome; AML, acute myeloid leukemia; UPD, uniparental disomy; monosomy 7, deletion of whole chromosome 7; del(7q), partial deletion involving 7q; and MDS/MPN, myelodysplastic syndrome/myeloproliferative neoplasm.
Distribution of disease subsets and associated genomic lesions among the 3 classes of chromosome 7 lesions. (Top) Distribution of LOH detected by SNP-A in the cohort, separated according to the nature of the lesion ([UPD(7q), del(7q), monosomy 7]. Patients have been grouped as follows: red, AML + high risk and intermediate-2 MDS; gray, low risk and intermediate-1 MDS; blue, hypocellular MDS; black, MDS/MPN; and green, Fanconi anemia and JMML. (Middle) Distribution of disease status in patients with 7 LOH separated according the nature of the lesion. (Bottom) Additional SNP-A–detected genomic lesions separated according the same criteria as stated herein. MDS indicates myelodysplastic syndrome; AML, acute myeloid leukemia; UPD, uniparental disomy; monosomy 7, deletion of whole chromosome 7; del(7q), partial deletion involving 7q; and MDS/MPN, myelodysplastic syndrome/myeloproliferative neoplasm.
Comparative of clinical characteristics of patients at baseline according to the SNP-A–detected lesion nature
| . | Deletion(7), n = 63 (A) . | Deletion(7q), n = 72 (B) . | UPD(7q), n = 26 (C) . | P (only significant comparisons reported) . |
|---|---|---|---|---|
| Median age, y (range) | 58 (27-70) | 64 (56-72) | 68 (63.2-77) | A vs B (P = .01) A vs C (P = .009) |
| Sex | ||||
| Male, % | 59 | 56 | 57 | |
| Female, % | 41 | 44 | 43 | |
| White blood cell count, × 109/L, median (IQR) | 5.5 (2.7-25.7) | 4.3 (2.1-13.5) | 10.5 (8.1-37.9) | C vs A (P = .028) C vs B (P = .01) |
| Hemoglobin, g/dL, mean ± SD | 9.1 ± 1.9 | 9.2 (8.9-10) | 9.1 ± 2.1 | |
| Mean corpuscular volume, median (IQR) | 91 (85-99) | 90 (86-104.8) | 90 (85-103) | |
| Platelets, × 109/L, median (IQR) | 47 (22-71) | 50 (24-86) | 46 (20.2-146.2) | |
| BM cellularity, median (%) | 35 (22-61) | 75 (45-90) | 76(55-100) | A vs B (P = .03) A vs C (P = .036) |
| . | Deletion(7), n = 63 (A) . | Deletion(7q), n = 72 (B) . | UPD(7q), n = 26 (C) . | P (only significant comparisons reported) . |
|---|---|---|---|---|
| Median age, y (range) | 58 (27-70) | 64 (56-72) | 68 (63.2-77) | A vs B (P = .01) A vs C (P = .009) |
| Sex | ||||
| Male, % | 59 | 56 | 57 | |
| Female, % | 41 | 44 | 43 | |
| White blood cell count, × 109/L, median (IQR) | 5.5 (2.7-25.7) | 4.3 (2.1-13.5) | 10.5 (8.1-37.9) | C vs A (P = .028) C vs B (P = .01) |
| Hemoglobin, g/dL, mean ± SD | 9.1 ± 1.9 | 9.2 (8.9-10) | 9.1 ± 2.1 | |
| Mean corpuscular volume, median (IQR) | 91 (85-99) | 90 (86-104.8) | 90 (85-103) | |
| Platelets, × 109/L, median (IQR) | 47 (22-71) | 50 (24-86) | 46 (20.2-146.2) | |
| BM cellularity, median (%) | 35 (22-61) | 75 (45-90) | 76(55-100) | A vs B (P = .03) A vs C (P = .036) |
IQR indicates interquartile range; BM, bone marrow; UPD, uniparental disomy; and CMML, chronic myeloid leukemia..
Clinical and genomic correlates of monosomy 7/del(7q) patients
Compared with cases of partial deletions, those patients with del(7) were characterized by a lower number of genomic lesions per patient (1.2 vs 4.8; P < .001); the most remarkable the absence of 17p LOH cases among MDS patients.
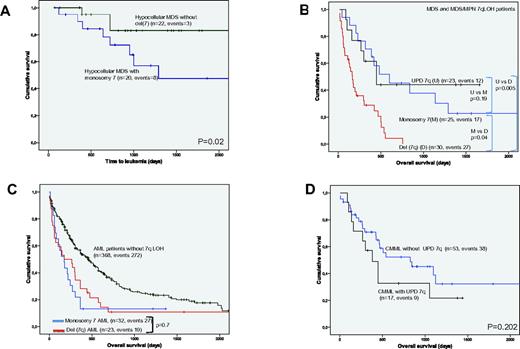
Of 26 patients with monosomy 7 by SNP-A and a diagnosis of MDS, 20 (77%) fulfilled the diagnostic criteria of hMDS. Of note, these 20 patients had no other lesion detectable by SNP-A (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). That strong correlation between hMDS and the presence of an isolated monosomy 7 could not be established by MC, because no-growth was obtained in 8 hMDS-MC analyses and 4 high-risk MDS patients were described to harbor an isolated monosomy 7 by MC, whereas SNP-A found additional lesions in all of them. When patients with hMDS with or without monosomy 7 were compared, those with monosomy 7 showed a worse prognosis, with a higher transformation to leukemia (P = .02; hazard ratio, 3.4, 95% confidence interval, 1.2%-9.7%; Figure 3A). Interestingly, 3 cases of monosomy 7 AML had a history of antecedent aplastic anemia. All the chromosome 7 lesions detected in 79 pediatric patients were monosomies; monosomy 7 was detected in 14% and 16% of patients with JMML and FA, respectively.
Differences in survival outcomes and progression-free survival of 7q LOH patients.P values presented correspond to the Cox regression between the groups indicated. AML indicates acute myeloid leukemia; Chr, chromosome, MDS/MPN, myelodysplastic syndrome/myeloproliferative neoplasm; UPD, uniparental disomy; monosomy 7, deletion of whole chromosome 7; del(7q), partial deletion involving 7q; and CMML, chronic myeloid leukemia.
Differences in survival outcomes and progression-free survival of 7q LOH patients.P values presented correspond to the Cox regression between the groups indicated. AML indicates acute myeloid leukemia; Chr, chromosome, MDS/MPN, myelodysplastic syndrome/myeloproliferative neoplasm; UPD, uniparental disomy; monosomy 7, deletion of whole chromosome 7; del(7q), partial deletion involving 7q; and CMML, chronic myeloid leukemia.
The subset of patients with del(7q) presented with an elevated percentage of high-risk disease (88% had AML or high-risk/intermediate-2 IPSS MDS) and a higher number of associated genomic lesions per patient. Del(5q) was the most common del(7q)-associated lesion found, occurring in 35 of 72 patients, all of which had AML or higher risk MDS. Of note, all del(5q) segments, except in 4 cases, involved either the centromeric or the telomeric extremes of the long arm of chromosome 5. The high frequency of del(5q) was followed closely by LOH 17p, seen in 14 of 72 patients, of which 5 of 14 were UPDs. Similar to patients with 5q, all patients had advanced stages of MDS or AML at diagnosis. All patients with LOH 17p spanned TP53; somatic mutations were present in 77% of cases tested.
The del(7q) MDS and MDS/MPN cohort had a shorter OS and time to leukemia transformation compared with patients with UPD(7q) or monosomy 7 (Figure 3B). In contrast, OS was similar in AML cases with monosomy 7 and del(7q) (Figure 3C), with both showing a significantly worse survival than in those patients with AML but without 7q LOH.
Clinical and genomic correlates of UPD(7q) patients
The UPD(7q) subset consisted of 26 patients, of which 17 were diagnosed with chronic myelomonocytic leukemia (CMML). Interestingly, 2 cases of AML and UPD(7q) also had history of antecedent CMML. The number of associated genomic lesions in the UPD(7q) cohort was lower than in the monosomy 7 and del(7q) subsets (P = .03 and P < .001, respectively), with a predominant presence of other regions of somatic copy neutral LOH rather than unbalanced defects. UPD(7q) was not associated with 5q or 17p LOH segments.
Comparing the 17 CMML patients to 55 CMML patients without UPD(7q) by SNP-A analysis, we found a trend toward worse survival. Those CMML patients with UPD(7q) showed a trend toward a shorter median OS (460 vs 730 days (P = .2; Figure 3D) and a higher rate of transformation to leukemia; whereas 26% of UPD(7q) patients progressed to higher-risk MDS or AML, advanced disease was observed in 13% of CMML patients without UPD(7q) (P = .001).
To test the prognostic validity and independence from known clinical variables of chromosome 7 SNP-A findings in patients' MDS and CMML, we developed a multivariate model for each cohort (Table 2). In the MDS model, the absence or presence of del(7q) or monosomy 7 kept the independent prognostic value when analyzed controlling for the clinical variables from the IPSS, ie, bone marrow blast percentage and number of cytopenias retained, whereas in the CMML model, the presence or absence of UPD 7q showed a trend toward statistical significance (P = .1) when tested together with the variables included in the score described by Onida et al,19 that is, hemoglobin level below 12 g/dL, presence of circulating immature myeloid cells, absolute lymphocyte count greater than 2.5 × 10/L,9 and marrow blasts greater than 10%.
Multivariate Cox proportional hazards regression models testing the prognostic value of SNP-A chromosome 7 findings in MDS and CMML
| MDS multivariate Cox model (n = 274) . | CMML multivariate Cox model (n = 70) . | ||||
|---|---|---|---|---|---|
| . | P . | HR (95% CI) . | . | P . | HR (95% CI) . |
| BM blasts* | ≤ .001 | 1.8 (1.3-2.4) | BM blasts > 10% | .01 | 10.4 (2.6-41.4) |
| Presence of blasts in PB | .01 | 10.3 (2.5-42.2) | |||
| No. of cytopenias† | .13 | 1.7 (0.8-3.4) | Lymphocyte count > 2.5 × 109/L | .6 | 0.7 (0.2-2.7) |
| 7q LOH SNP-A category‡ | ≤ .001 | 4.5 (3.1-6.7) | Hemoglobin level < 12 g/dL | .4 | 1.7 (0.4-8) |
| Presence of a UPD(7q) | .1 | 4.4 (0.8-16) | |||
| MDS multivariate Cox model (n = 274) . | CMML multivariate Cox model (n = 70) . | ||||
|---|---|---|---|---|---|
| . | P . | HR (95% CI) . | . | P . | HR (95% CI) . |
| BM blasts* | ≤ .001 | 1.8 (1.3-2.4) | BM blasts > 10% | .01 | 10.4 (2.6-41.4) |
| Presence of blasts in PB | .01 | 10.3 (2.5-42.2) | |||
| No. of cytopenias† | .13 | 1.7 (0.8-3.4) | Lymphocyte count > 2.5 × 109/L | .6 | 0.7 (0.2-2.7) |
| 7q LOH SNP-A category‡ | ≤ .001 | 4.5 (3.1-6.7) | Hemoglobin level < 12 g/dL | .4 | 1.7 (0.4-8) |
| Presence of a UPD(7q) | .1 | 4.4 (0.8-16) | |||
HR indicates hazard ratio; BM, bone marrow; PB, peripheral blood; SNP-A, single nucleotide polymorphism array; and CI, confidence interval.
Three BM blasts categories according to the percentage described: < 5; 5-10; and 11-20.
Number of cytopenias categories defined as good (0-1) and poor (2-3). Cytopenias defined as hemoglobin less than 10 g/dL, absolute neutrophil count less than 1.8 × 109/L, and platelets less than 100 × 109/L.
7q LOH SNP-A category defined as good, no deletion; intermediate, monosomy 7; and poor, partial deletion involving 7q.
Comparative analysis of SNP-A and MC
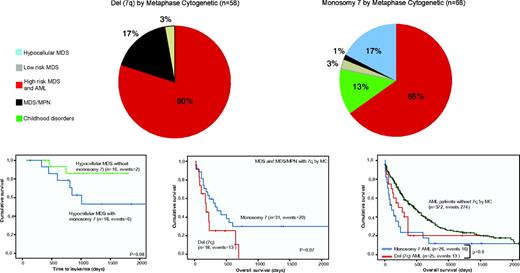
To provide a more detailed analysis about the added information that these karyotyping techniques could offer, Figure 4 illustrates how the distribution of disease subsets and outcome associations would be according to the lesion found by MC. Leaving aside those 44 patients with 7q LOH not detected by MC (26 UPD, 11 no growth, 7 small deletions in balanced translocations), the allocation of entities among MC del(7q) and monosomy 7 did not show a significant change. In fact, when only MC informative cases were considered, a multivariate model including both SNP-A and MC 7q lesions in MDS resulted in the variables cancelling each other (P = .7). No SNP-A–defined monosomy 7 was defined as a partial deletion by MC, because of which the strong association among monosomy 7 and hMDS and its high rate of transformation compared with hMDS without monosomy 7 remains unaltered. However, we must remark that this subgroup of MDS patients showed a higher frequency of no-growth MC analysis (in 8 hMDS patients, half of them harbored a monosomy 7 by SNP-A).
Illustration of how the distribution of disease subsets and outcome associations would be according to the lesion found by MC. (Top) Distribution of patients detected separated according to lesion detected by metaphase cytogenetics. Patients have been grouped as follows: red, AML + high risk and intermediate-2 MDS; gray, low risk and intermediate-1 MDS; blue, hypocellular MDS; black, MDS/MPN; and green, Fanconi anemia and JMML. (Bottom) Differences in survival outcomes and progression-free survival according to MC findings. P values presented correspond to the Cox regression between the groups indicated. AML indicates acute myeloid leukemia; Chr, chromosome, MDS/MPN, myelodysplastic syndrome/myeloproliferative neoplasm; UPD, uniparental disomy; monosomy 7, deletion of whole chromosome 7; del(7q), partial deletion involving 7q; and CMML, chronic myeloid leukemia.
Illustration of how the distribution of disease subsets and outcome associations would be according to the lesion found by MC. (Top) Distribution of patients detected separated according to lesion detected by metaphase cytogenetics. Patients have been grouped as follows: red, AML + high risk and intermediate-2 MDS; gray, low risk and intermediate-1 MDS; blue, hypocellular MDS; black, MDS/MPN; and green, Fanconi anemia and JMML. (Bottom) Differences in survival outcomes and progression-free survival according to MC findings. P values presented correspond to the Cox regression between the groups indicated. AML indicates acute myeloid leukemia; Chr, chromosome, MDS/MPN, myelodysplastic syndrome/myeloproliferative neoplasm; UPD, uniparental disomy; monosomy 7, deletion of whole chromosome 7; del(7q), partial deletion involving 7q; and CMML, chronic myeloid leukemia.
As a result of SNP findings, 16 patients were erroneously assigned to the monosomy 7 group by MC. SNP-A revealed that these samples instead had partial deletions and thus had been misclassified as high-risk patients. Probably, “relocation” of patients led to the lack of statistical difference noted in survival between MC-defined del(7q) and monosomy 7 patients (although a trend is still noted, P = .07).
There are 2 main limitations of SNP-A relative to MC: SNP-A does not detect balanced translocations and SNP-A cannot distinguish whether multiple abnormalities exist in a single clone. We tested whether both techniques could complement each other to solve these shortcomings. We grouped patients based on the presence of monosomy 7/del(7q) in less than 100% or in 100% of the metaphases analyzed. Patients with a clone burden of 100% had a lower median OS (175 vs 235 days; P = .150), probably because of the presence of more patients with MDS-derived AML (12 vs 6 cases; P = .103), although neither of these findings reached statistical significance. Regarding balanced rearrangements, they were present in 28% of patients, included in complex karyotypes with 5 or more abnormalities in all cases except for 1 patient with a del(7q) and an inv(3)(q21q26). The latter was the only recurrent balanced rearrangement, present in a second patient. The accumulation of this balanced aberrations patients with complex karyotypes and more than 10% of blasts explains partly why the presence of balanced rearrangements did not add independent prognostic value when 1 of those 2 variables was tested simultaneously
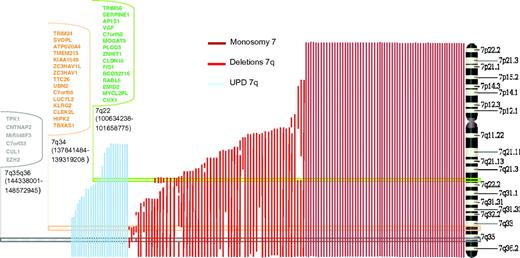
Exploring the 2-hit model: SNP-A definition of CDRs and NGS approach
To determine the location of genes on 7q that may be involved in clonal hematopoiesis, we analyzed the SNP-A karyotyping results from 161 patients and defined 3 CDRs, localized in bands 7q22 (100634238-101658775), 7q34 (137841484-139319208), and between bands 7q35 and 7q36.1 (144338001-148572945, Figure 4A). Genomic annotation of the CDRs was performed, and several candidate genes mapping within the CDR were noted (Figure 5); these genes were Sanger sequenced in a cohort of 50 cases with 7q LOH. The third CDR was defined by a single patient with a small deletion containing 6 genes. We sequenced all exons of these genes and detected a mutation in EZH2, located in exon 19 involving position Ile715, that produced a frameshift mutation. We found no other somatic mutations by this strategy. EZH2 proved to be recurrent in patients with a myelodysplastic/myeloproliferative component and UPD(7q). Supplemental Table 1 summarizes EZH2 mutations found in our 7q LOH patients that we reported previously in part.20
Identification of candidate genes on 7q by mapping CDRs by SNP-A. Three distinct CDRs, indicated by horizontal rectangles, were identified on 7q by mapping of SNP-A karyotyping. The connected keys show the candidate genes contained in each CDR; those genes sequenced in a test cohort of 50 patients with LOH 7q are in bold. CDR indicates commonly deleted region; SNP-A, single nucleotide polymorphism array; and LOH, loss of heterozygosity.
Identification of candidate genes on 7q by mapping CDRs by SNP-A. Three distinct CDRs, indicated by horizontal rectangles, were identified on 7q by mapping of SNP-A karyotyping. The connected keys show the candidate genes contained in each CDR; those genes sequenced in a test cohort of 50 patients with LOH 7q are in bold. CDR indicates commonly deleted region; SNP-A, single nucleotide polymorphism array; and LOH, loss of heterozygosity.
In an effort to overcome the limitations inherent to the classic screening method, which was limited to the genes located in the CDRs, we applied 2 NGS approaches. First, we generated exome chromosome 7 libraries from 11 cases with LOH 7q (monosomy 7, n = 6; del(7q), n = 2; UPD(7q), n = 3) and subjected them to high-throughput sequencing on a Genome Analyzer (Illumina). Second, paired (bone marrow and CD3+) samples from 15 myeloid neoplasms were subjected to whole exome sequencing using HiSeq 2000 (Illumina), including 3 patients with 7q LOH (UPD, deletion, and monosomy). Finally, we used publically available NGS data from TCGA for 74 AML patients. Supplemental Figure 2 shows the somatic mutations found in regions of 7q LOH: NRCAM (Q1040K) in a patient with refractory cytopenia with multilineage dysplasia with monosomy 7, LMTK2 (A1147T) in a refractory cytopenia with multilineage dysplasia patient with del(7q), and EZH2 (R690H) in a third MDS/MPN patient with UPD(7q). Of note, only mutations of EZH2 proved to be recurrent [10/19 CMML patients with UPD(7q)], when sequencing by Sanger technique a confirmatory cohort of 50 cases with 7q LOH. Table 3 shows somatic mutations, localized on chromosome 7q, in patients with 7q LOH found using NGS in our patients and in the TCGA project.
Somatic mutations found in regions of 7q LOH patients by NGS
| Diagnosis . | SNP-A LOH on chromosome 7 . | Gene . | Mutation . |
|---|---|---|---|
| MDS/MPN (CMML) | UPD 7q11.21-qter | EZH2 | R690H |
| MDS/MPN (aCML) | UPD 7q32.1-qter | EZH2 | R690H |
| MDS (RCMD) | Del 7q21.3-qter | LMTK2A | A1147T |
| MDS (RCMD) | Monosomy 7 | NRCAM | Q1040K |
| AML | Del 7q21.12q36.3 | ZAN | N1098Del |
| AML | Monosomy 7 | GRM8 | A686V |
| AML | Monosomy 7 | ENSG00000133375 | R68Q |
| AML | Del 7q31.31-qter | LOC641808 | V162fs |
| AML | Monosomy 7 | SEMA3A | R613Q |
| AML | Del 7q31.1q36.3 | DYNC1I1 | R239W |
| AML | Monosomy 7 | HYAL4 | N253K |
| AML | Monosomy 7 | FAM40B | C182R |
| AML | Monosomy 7 | LOC100128744 | P354L |
| AML | Monosomy 7 | LUC7L2 | R252fs |
| AML | Del 7q21.11q36.3 | CTAGE6 | T288M |
| AML | Monosomy 7 | FAM115A | F193S |
| AML | Del 7q35-qter | CUL1 | E241D |
| AML | Monosomy 7 | EZH2 | E745fs |
| AML | Del 7q11.21q36.3 | EZH2 | R690H |
| AML | Monosomy 7 | SSPO | T426R |
| Diagnosis . | SNP-A LOH on chromosome 7 . | Gene . | Mutation . |
|---|---|---|---|
| MDS/MPN (CMML) | UPD 7q11.21-qter | EZH2 | R690H |
| MDS/MPN (aCML) | UPD 7q32.1-qter | EZH2 | R690H |
| MDS (RCMD) | Del 7q21.3-qter | LMTK2A | A1147T |
| MDS (RCMD) | Monosomy 7 | NRCAM | Q1040K |
| AML | Del 7q21.12q36.3 | ZAN | N1098Del |
| AML | Monosomy 7 | GRM8 | A686V |
| AML | Monosomy 7 | ENSG00000133375 | R68Q |
| AML | Del 7q31.31-qter | LOC641808 | V162fs |
| AML | Monosomy 7 | SEMA3A | R613Q |
| AML | Del 7q31.1q36.3 | DYNC1I1 | R239W |
| AML | Monosomy 7 | HYAL4 | N253K |
| AML | Monosomy 7 | FAM40B | C182R |
| AML | Monosomy 7 | LOC100128744 | P354L |
| AML | Monosomy 7 | LUC7L2 | R252fs |
| AML | Del 7q21.11q36.3 | CTAGE6 | T288M |
| AML | Monosomy 7 | FAM115A | F193S |
| AML | Del 7q35-qter | CUL1 | E241D |
| AML | Monosomy 7 | EZH2 | E745fs |
| AML | Del 7q11.21q36.3 | EZH2 | R690H |
| AML | Monosomy 7 | SSPO | T426R |
SNP-A indicates single nucleotide polymorphism array; LOH, loss of heterozygosity; Del, deletion; MDS/MPN, myelodysplastic syndrome/myeloproliferative neoplasm; CMML, chronic myelomonocytic leukemia; UPD, uniparental disomy; RCMD, refractory cytopenia with multilineage dysplasia; AML, acute myeloid leukemia; and aCML, atypical chronic myelogenous leukemia.
Testing the haploinsufficiency hypothesis: microarray expression data
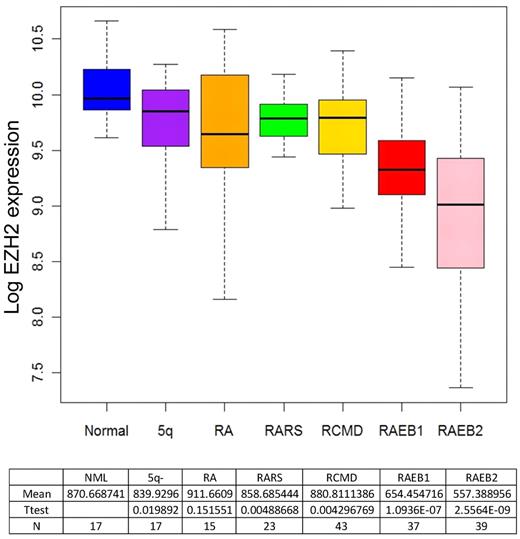
We examined the gene expression profiles of the CD34+ cells of 183 MDS patients, of which 9 cases had monosomy 7 or del(7q). We found that expression of 40% of the genes included in our SNP-A–defined CDRs were significantly reduced in those monosomy 7/del(7q) patients. These genes included LUC7L2, ZNHIT1, TTC26, RABL5, TRIM24, EZH2, ZC3HAV1L, CNTNAP2, TRIM24, CUX1, FIS1, RABL5, ZC3HAV1, and TBXAS1 (supplemental Figure 3). The mean decrease in expression levels was 42% to 33% of that in healthy controls. We also determined the expression of these genes in the 174 cases of MDS that did not have any chromosome 7 deletions, and most interestingly, we found that EZH2 and RABL5 were significantly down-regulated even in samples diploid for chromosome 7. Of note, we found that down-regulation of EZH2 was significantly reduced in patients with excess of blasts (Figures 5–6 and supplemental Figure 4).
Box plots showing the EZH2 expression ratios obtained in CD34+ cells of 174 MDS cases without 7q LOH and 17 healthy controls. A significant down-regulation of expression was identified for EZH2 in excess of blasts subgroups. NML indicates normal controls; 5q-, 5q-syndrome; RA, refractory anemia; RARS, refractory anemia with ring sideroblasts; RCMD, refractory anemia with multilineage dysplasia; and RAEB, refractory anemia with excess of blasts.
Box plots showing the EZH2 expression ratios obtained in CD34+ cells of 174 MDS cases without 7q LOH and 17 healthy controls. A significant down-regulation of expression was identified for EZH2 in excess of blasts subgroups. NML indicates normal controls; 5q-, 5q-syndrome; RA, refractory anemia; RARS, refractory anemia with ring sideroblasts; RCMD, refractory anemia with multilineage dysplasia; and RAEB, refractory anemia with excess of blasts.
Discussion
Unlike myeloid disorders harboring an isolated chromosome 5q deletion, a clear genotype-phenotype relationship has not been described in cases with 7q LOH, and underlying pathogenetic mechanisms remain unclear. Here, we applied high-resolution genomic technologies to accurately define the extent and nature of chromosomal lesions and to explore relevant clinical associations of inactivating mutations or insufficient gene dosage in a large cohort of patients with myeloid malignancies involving LOH of the long arm of chromosome 7. Our analyses demonstrate that in those subsets with isolated 7q LOH or accompanied by a very low number of additional lesions, the genotype-phenotype relation is clearly discernible. We found a correlation between an isolated deletion of the long arm of chromosome 7 and MDS with hypoplastic features, and between the presence of UPD(7q) in diploid MDS/MPN patients. In the latter group, the predominant driving genomic event was the presence of inactivating mutations involving EZH2, a finding supported by previous studies,21 whereas gene dosage effect seems to be paramount in typically large monosomy 7/del(7q) cases.
As expected, the spectrum of entities in each 7q LOH subgroup was relatively heterogeneous. Nevertheless, we found strong associations of SNP-A abnormalities with particular clinical entities that merit emphasis. We found a significant association of large MC-cryptic UPD(7q) segments among CMML patients (26%). In our experience, CMML shows a strikingly elevated frequency of somatic UPD compared with other myeloid disorders (data not shown). Particularly high frequencies of somatic UPD have been described for some neoplasms, suggesting that this specific type of chromosomal instability may be related to pathologic pathways that are common in some malignancies but absent in others.22 In addition, we observed a trend toward worse median survival of CMML patients harboring UPD(7q). The lack of MC lesions in a significant proportion of CMML patients and their controversial impact on survival,19,23 increases the value of a possible prognostic significance of UPD(7q).
Hypocellular MDS is a relatively uncommon entity among myeloid disorders, with a possible immune-related pathogenetic component, and a diagnosis that frequently overlaps with aplastic anemia.24 Within our cohort, 8% of patients had hypocellular MDS, a slightly lower percentage than what has been described in other studies.25,26 Of note, half of our hMDS cohort harbored monosomy 7 as the sole SNP-A lesion detected, conferring on them a higher rate of leukemia transformation. This finding may be helpful for distinguishing hMDS from other MDS.
The risk group assignment of MDS patients with monosomy 7 has been investigated in several studies.5,6 These studies reported dissimilar results that could be driven by the difficulty of dissecting, in a highly precise and reproducible way, the karyotype defects by conventional chromosome banding techniques. We and others, using more accurate karyotyping means, showed discrepancies in the context of 5q lesions.27,28 In our cohort, MDS patients harboring monosomy 7 presented a longer median OS than patients with partial deletions, more closely approximating that reported for the intermediate cytogenetic group in the IPSS.3 We also showed that the wrong assignment to the monosomy 7 subgroup by MC of a significant number of cases with partial deletions by SNP-A seems to be the responsible of a loss in the prognostic value of the conventional karyotyping technique. The better survival of those patients with a wider loss of genes in chromosome 7 (monosomy 7) than those with partial deletions could be presented as paradoxical. The frequent presence of monosomy 7 either in childhood disorders and not accompanied by other chromosomal lesions on one hand, and the common association of partial deletions of 7 with other chromosomal abnormalities shown to be early events in the genesis of dysplasia [del(5q)] on the other hand,29,30 prompt us to speculate that monosomy 7 might be a founding genomic aberration and that partial deletions of 7 might represent a secondary event in the context of preexisting genomic instability and therefore within a more aggressive clone.
The large size of the typical chromosomal LOH involving the long arm of chromosome 7 in myeloid disorders has complicated the search for a mutated TSG in this region. In this study, we used 2 approaches: (1) a classic approach with the definition of commonly deleted regions and direct sequencing of candidate genes and (2) a next-generation whole exome strategy. Three SNP-A–defined CDRs were described encompassing, with slight differences, those described previously.31,32 NGS technology allowed us to cover all coding exons, and because no recurrent mutation other than EZH2 was found, led us to conclude that the absence of recurrent somatic mutations in patients with monosomy 7/del(7q) is a hallmark of the disease pathogenesis in this unique category of myeloid neoplasms
The lack of recurrent mutations in any of the genes mapping to the segment of LOH in most of the patients with large monosomy 7/del(7q) prompted us to test the haploinsufficiency hypothesis by analyzing the expression profiles of patients with that kind of lesions. The dosage effect resulting from the loss of the whole q arm of chromosome 7 particularly affected genes localized in our 3 CDRs; 14 genes included in our SNP-A–defined minimally deleted regions had a mean decreased expression between 42% and 33%. In addition, 2 of these genes, EZH2 and RABL5, were significantly down-regulated even in samples that did not have monosomy 7/del(7q). This current study showed that down-regulation of EZH2 in the absence of LOH is common in advanced MDS. These results point to the importance of haploinsufficiency of the genes located in the 7q CDRs in the pathobiology of MDS and suggests that other genetic or epigenetic mechanisms may silence these genes in cases without 7q LOH.
In summary, the present study of 7q disorders, gathering data from a large series of patients using recent genomics technologies, shows that SNP-A complements traditional MC not only by detection of cryptic abnormalities but also by precisely defining the extent and nature of the lesions with strong clinical associations. Although a 2-hit model is supported for most patients with UPD(7q) and a overlapping MDS/MPN phenotype, our results suggest that haploinsufficient expression of select regions of 7q is the driving pathogenetic mechanism in those patients with predominant dysplastic features and loss of chromosome 7 material.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported in part by National Institutes of Health grants R01 HL082983 (J.P.M.), U54 RR019391 (J.P.M., M.A.S.), and K24 HL077522 (J.P.M.); Department of Defense grant MPO48018 (M.A.M.); funds from Leukaemia & Lymphoma Research of the United Kingdom (A.P. and J.B.) and Fundacion Caja Madrid (A.J.); and a charitable donation from Robert Duggan Cancer Research Foundation.
The results published here were partly based on data generated by The Cancer Genome Atlas pilot project established by the National Cancer Institute and National Human Genome Research Institute. Information about TCGA and the investigators and institutions that constitute the TCGA research network can be found at http://cancergenome.nih.gov.
National Institutes of Health
Authorship
Contribution: A.J., Y.S., and J.P.M. were responsible for overall design, data collection, analysis, and interpretation, statistical analysis, manuscript preparation, and writing and completion of the manuscript; H.M., A.V., A.M.J., B.P., V.V., R.V.T., C.L.O., A.M.M., and A.P. analyzed data and edited the manuscript; A.G.K., K.M., H.M., A.R.M., M.A.S., M.A.M., S.K., A.L., J.B., and G.J.M. gathered data and edited the manuscript; and all authors approved the final version of the manuscript and its submission.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jaroslaw P. Maciejewski, Taussig Cancer Institution/R40, 9500 Euclid Ave, Cleveland, OH; e-mail: maciejj@ccf.org.

![Figure 2. Distribution of disease subsets and associated genomic lesions among the 3 classes of chromosome 7 lesions. (Top) Distribution of LOH detected by SNP-A in the cohort, separated according to the nature of the lesion ([UPD(7q), del(7q), monosomy 7]. Patients have been grouped as follows: red, AML + high risk and intermediate-2 MDS; gray, low risk and intermediate-1 MDS; blue, hypocellular MDS; black, MDS/MPN; and green, Fanconi anemia and JMML. (Middle) Distribution of disease status in patients with 7 LOH separated according the nature of the lesion. (Bottom) Additional SNP-A–detected genomic lesions separated according the same criteria as stated herein. MDS indicates myelodysplastic syndrome; AML, acute myeloid leukemia; UPD, uniparental disomy; monosomy 7, deletion of whole chromosome 7; del(7q), partial deletion involving 7q; and MDS/MPN, myelodysplastic syndrome/myeloproliferative neoplasm.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/119/25/10.1182_blood-2011-12-397620/4/m_zh89991292640002.jpeg?Expires=1769157004&Signature=xK8vBsLgCCFHuGRHGi85SB523wNVEkZ2nNQ-fIrgyF3q-4hVD89h7Ch7k3XfmqRNe5jh57ehftBP2F~lgBrn9fr6mRMo8H7Gb1uVl~eC-lYyurVgNJcJlclC5CEzh6C3nhGUB7RiWxKLyWlxCfiw2GryJvYkerI20q0poZAXMgamr6umJAMK~zCbko43jYPPRVogmsQv6bTzBWD5oO~ZsRTUMOGu~BKTA-A8xgWWKTQyrmqKfp9PIaNSBfviRlabXj6h60bPACkpVA4lyYEi2Onn~~rCSJJzVaDVrFFyk7IuNKNpia7w-Sn6bPgrrUnPfUI7q9zzR9kqow1tJ2QA1A__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal