Abstract
Dendritic cells (DCs) are the most potent APCs. Whereas immature DCs down-regulate T-cell responses to induce/maintain immunologic tolerance, mature DCs promote immunity. To amplify their functions, DCs communicate with neighboring DCs through soluble mediators, cell-to-cell contact, and vesicle exchange. Transfer of nanovesicles (< 100 nm) derived from the endocytic pathway (termed exosomes) represents a novel mechanism of DC-to-DC communication. The facts that exosomes contain exosome-shuttle miRNAs and DC functions can be regulated by exogenous miRNAs, suggest that DC-to-DC interactions could be mediated through exosome-shuttle miRNAs, a hypothesis that remains to be tested. Importantly, the mechanism of transfer of exosome-shuttle miRNAs from the exosome lumen to the cytosol of target cells is unknown. Here, we demonstrate that DCs release exosomes with different miRNAs depending on the maturation of the DCs. By visualizing spontaneous transfer of exosomes between DCs, we demonstrate that exosomes fused with the target DCs, the latter followed by release of the exosome content into the DC cytosol. Importantly, exosome-shuttle miRNAs are functional, because they repress target mRNAs of acceptor DCs. Our findings unveil a mechanism of transfer of exosome-shuttle miRNAs between DCs and its role as a means of communication and posttranscriptional regulation between DCs.
Introduction
Cellular miRNAs are released membrane free1 or packaged inside microvesicles (0.1-1 μm) shed by the plasma membrane2,3 or within nanovesicles (< 100nm) derived from the endocytic pathway known as exosomes.4,5 Exosomes are generated as intraluminal vesicles by reverse budding of the membrane of multivesicular bodies (MVBs). Release of exosomes occurs when MVBs fuse their limiting membrane with the plasma membrane.6-9
Dendritic cells (DCs) are APCs with the ability to regulate adaptive immunity. Whereas immature DCs down-regulate T-cell responses, mature DCs promote activation, proliferation, and differentiation of effector T cells.10 Communication between DCs is essential to amplify their tolerogenic and immunogenic functions.11,12 This DC-to-DC interaction is mediated through cell-to-cell contact, soluble mediators, exchange of plasma membrane patches,13,14 nanotubules,15 and interaction with apoptotic cell–derived vesicles16 and exosomes.17,18
Although the mechanisms have not been elucidated, it has been reported that DCs acquire proteins/peptides from other cells via exosomes.17-19 Recently, it has been suggested that transfer of exosome-shuttle miRNAs might constitute a mechanism of cell-to-cell communication that regulates mRNA translation20 or, alternatively, a way to dispose of “unwanted” miRNAs.21 An important unanswered question in the field is how exosome-shuttle miRNAs, transported inside the vesicles, are delivered into the cytosol of the acceptor cells, a problem we have investigated in this study with the use of DCs. Addressing this point has been challenging because (1) the composition of DC exosomes depends on the maturation of the DC of origin22,23 ; (2) there is limited information on intercellular communication via “endogenous” (instead of exogenously added) exosomes22 ; (3) transfer of exosomes between cells probably occurs rapidly and below the limit of resolution of conventional microscopy; and (4) the function of exosome-shuttle miRNAs is difficult to test, because homologous cellular miRNAs can be present in the acceptor DCs. Our findings indicate that endogenously released exosomes constitute an effective means of communication between DCs and that such vesicles are capable of delivering their intraluminal content (including functional exosome-shuttle miRNAs) into the cytosol of the target DCs.
Methods
Generation of DCs
BM-derived DCs and splenic DCs were obtained as previously described (see supplemental Methods, available on the Blood Web site; see the Supplemental Materials link at the top of the online article).12 All mouse studies were approved by the University of Pittsburgh Institutional Animal Care and Use Committee.
Exosome purification
Exosomes were isolated from supernatants of B6 BMDCs maintained in medium with exosome-free FCS (overnight centrifugation, 100 000g) during the last 48 hours of culture. BMDCs (day 6) were incubated with 100nM ionomycin (1 hour), and culture supernatants were centrifuged at 300g (10 minutes), 1200g (20 minutes), 10 000g (30 minutes), and then ultrafiltered (2000g, 20 minutes) through a Vivacell 100 filter. The filtered supernatant was adjusted to 10 mL with PBS and ultracentrifuged (100 000g, 60 minutes) on top of 1.6 mL of 30% sucrose/D20 density cushion.24 The phase containing the exosomes was collected, adjusted to 10 mL of PBS, rinsed overnight (4°C), and centrifuged (100 000g, 60 minutes). Alternatively, flotation of exosomes on a continuous sucrose gradient was done as described by Raposo et al.7 In some experiments, B6 (IAb) BMDC-derived exosomes were loaded with the BALB/c IEα52-68 peptide (ASFEAQGALANIAVDKA) by incubating the B6 BMDCs with 50nM of the peptide the day before collecting the exosomes. The amount of exosome protein was assessed with a NanoDrop 2000c. The exosome size was measured with an LM10 NanoSight's instrument equipped with a high-sensitivity EMCCD camera and the NTA 2.0 software (NanoSight). For analysis of the stability of exosomal miRNAs, exosomes purified by 30% sucrose gradient were treated (37°C, 60 minutes) with 500 μg/mL proteinase K (Sigma-Aldrich) dissolved in RNAse-free water, followed by heat inactivation of the protease (60°C, 10 minutes), and incubation (37°C, 10 minutes) with 2 μg/mL protease-free RNase A (Sigma-Aldrich).
miRNA microarray analysis
BMDCs and BMDC-derived exosomes, purified by sucrose gradient, were treated with 2 μg/mL RNAse A (37°C, 10 minutes) and disrupted with Trizol. RNA isolation was performed with the miRNeasy Mini Kit (QIAGEN). Samples were quantified by picogreen assay and normalized to 27 ng/μL. The miRNA was amplified and hybridized to Illumina expression profiling microarrays according to the manufacturer's directions with minor modifications. Briefly, RNA was poly-A tailed and reverse transcribed from a polydT tailed primer to generate biotinylated cDNA. The cDNA underwent allele-specific extension with miRNA-specific oligos for all targeted sequences, followed by amplification with a cyanine 3 (Cy3) fluorophore–labeled primer. The double-stranded PCR product was bound to magnetic beads and denatured with 0.1N NaOH. The fluorophore-labeled strand was hybridized to the miRNA microarray containing 1536 IllumiCode sequences attached to 3-μm beads. Hybridization was performed overnight with a 60°C-45°C temperature ramp, washed to remove excess label, and dried in a vacuum dessicator before imaging by laser excitation of the fluorochrome-labeled sample. Fluorescence intensity was quantitatively imaged with the iScan system for expression analysis. Two total RNA samples normalized to 100 ng/μL were included to control for artifacts because of low sample concentration. All microarray data are available in the Gene Expression Omnibus under accession no. GSE33179.
Assay of exosome transfer
B6 BMDCs (1 × 106; CD45.2+) transduced with RAd-eGFP-tmFasLΔ or with control RAds (MOI = 100) were coincubated the following day at a 1:1 cell ratio with “acceptor” (CD45.1+) B6 BMDCs (at 37° or 4°C). After 3 hours, cells were rinsed with ice-cold 0.01M EDTA/PBS and labeled with PE-CD45.1 and CyC-CD45.2 Abs, fixed in 4% paraformaldehyde (PF) and analyzed with FACS. For assessment of exosome transfer between BMDCs and T cells, 106 B6 BMDCs, transduced with RAd-eGFP-tmFasLΔ or control RAds, were pulsed for 3 hours with IEα52-68 or OVA323-339 and cocultured 18 hours (37°C) with naive or in vitro–activated 1H3.1 and OT-II T cells (5 × 106 of each). At the end of the assay, cells were rinsed with cold 0.01M EDTA/PBS; labeled with PE-Thy1.2, CyC-Thy1.1, and APC-CD11c Abs; fixed in PF; and analyzed by FACS. Splenic T cells were isolated by negative selection (Dynabeads) and in vitro–activated with artificial APCs (Dynabeads).
Membrane fusion assay
Exosomes were labeled with 20μM R18 (1 hour, room temperature) and then purified by gel filtration (Sephadex G-75) to remove unbound dye. R18-labeled exosomes (10 μg) were added to 2 mL of 2 g/L glucose Ca/Mg HBSS (pH 7) in a magnetically stirred cuvette of a thermostated FluoroMax-3 spectrofluorometer (Horiba). After 3 minutes of equilibration, fusion was initiated by adding the unlabeled DCs (106 cells), and fluorescence was measured for 35 minutes on the spectrofluorometer set at 560-nm excitation and 590-nm emission (slits, 1.5 nm; integration time, 10 seconds). The assay was stopped by adding 0.3% Triton X-100. The fluorescence increase after the addition of DCs was expressed as the percentage of maximal fluorescence de-quenching (FD), as follows: FD (%) = [(F − Fi)/(Fmax − Fi)] × 100, where F is the fluorescence intensity at each point of the assay, Fi is the initial fluorescence of R18-exosomes, and Fmax is the maximum fluorescence of R18 after the addition of detergent.
Fluorescence time-lapse microscopy
BMDCs (150 000 cells; 150 μL of 2 g/L glucose Ca/Mg HBSS) were attached to poly-L-lysine–coated 35-mm2 coverslip bottom dishes (MatTek) and placed in an environmental chamber (37°C, 5% CO2). After the addition of 2.5 μg of R18-labeled exosomes, image acquisition was performed with an Nikon TE2000 inverted microscope with a 60×/1.40 NA oil objective and a QImaging Retiga SRV CCD camera. BMDCs were alternately imaged with differential interference contrast and DsRed filter sets every 30 seconds, for 30 minutes. Analysis was conducted with MetaMorph Version 7.7 software.
Content-mixing assay
Exosomes were loaded with DMNPE-caged-luciferin (10 μL of 10mM luciferin/200 μg of exosomes/200 μL PBS) for 1 hour, at 37°C, in the dark. Luciferin was released from the DMNPE group by UV-B (365 nm) photolysis (5 minutes, on ice). Nonincorporated luciferin was removed from the exosomes by gel filtration (Sephadex G-75).
BMDCs (1 × 106) transfected with a Rad-encoding luciferase (RAd-LUC) or a RAd without transgene (RAd-Empty; MOI = 50) were added to 2 mL of 2 g/L glucose Ca/Mg HBSS (pH 7) in a magnetically stirred cuvette of a thermostated FluoroMax-3 spectrofluorometer set at 0 nm excitation and 560 nm emission wavelengths (slits, 20 nm; integration time, 10 seconds). After 3 minutes of equilibration, fusion was initiated by adding 10 μg of luciferin (or unlabeled) exosomes. Light emission was measured for 35 minutes as percentage of increase in light emission according to the formula [(F × 100)/Fi) − 100, where F represents the light emitted at each time point and Fi is the initial background light emitted by the BMDCs before adding the exosomes.
Statistical analyses
Comparison between 2 groups was performed by Student t test. Graft survivals were compared by Kaplan-Meier analysis and the log-rank test. A P value < .05 was considered significant. For comparison of miRNAs, quantitative and qualitative analyses were conducted. For qualitative analysis, the efficiency analysis paradigm was applied to determine which among 315 methods for array-based expression analysis exhibited high internal consistency.25 For comparison between immature and mature exosomes, the J5 with threshold T = 1.644 method with Quantile99 normalization gave the highest internal consistency.25 For comparison between immature or mature exosomes and immature or mature BMDCs, the J5 test with z-transformation (within array) was optimal. For each comparison, the optimal method was applied with caGEDA.25 A quantitative analysis was conducted for each comparison with the use of intensity-rank plots. A threshold expression intensity value of 2500 was used to identify those miRNAs highly expressed only in immature or mature exosomes or shared by both. The same threshold was used to identify miRNAs highly expressed in immature or mature BMDCs or shared by their respective exosomes. mRNA targets of miRNAs were determined with TargetScan Version 5.2 software.26
Results
DCs at different stages of maturation release exosomes with different miRNA content
Exosomes released by DCs with synchronized maturation were purified from supernatants of maturation-resistant (vitamin D3-treated) B6 BMDCs (immature exosomes) or fully mature (LPS-treated) B6 BMDCs (mature exosomes), both treated with ionomycin during the last hour of culture. Immature BMDCs released more exosomes than LPS-matured BMDCs (13.60 ± 9.61 vs 9.52 μg ± 3.30 μg per 107 DCs). Immature and mature exosomes exhibited similar structure (supplemental Figure 1A) and size (103 ± 30nm vs 108 ± 36nm, respectively). Mature exosomes expressed more CD86 and CD54 and exhibited superior T-cell allostimulatory ability than immature exosomes (supplemental Figure 1B-D). As described in other transplantation models,23,27 only systemic administration donor-derived BMDC immature exosomes (intravenously, 7 days before transplantation) prolonged significantly (P < .001) survival of heart allografts in mice (supplemental Figure 1E).
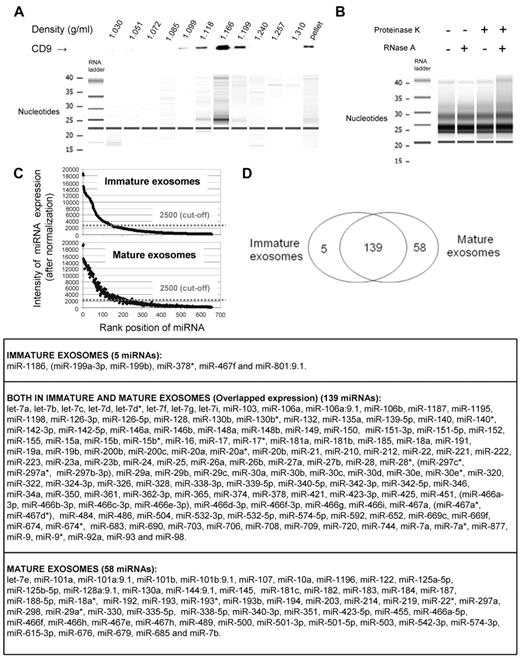
BMDCs release different exosomes depending on the stage of maturation of the parental DC.23 Therefore, we analyzed separately the miRNA content of exosomes secreted by maturation-resistant and LPS-matured BMDCs. The Bioanalyzer profiles indicated that the BMDC exosomes (pretreated with RNase A) contained small RNAs with minimal contamination with cellular RNAs (supplemental Figure 2). Purification of exosomes by continuous sucrose gradient confirmed that miRNAs were highly enriched at the characteristic density of exosomes (1.166 g/mL fraction), the latter identified by CD9 detection by Western blot analysis (Figure 1A). The amount of RNA recovered per microgram of exosomal protein did not differ significantly between immature (0.95 ± 0.28 ng of RNA) and mature exosomes (1.042 ± 0.38 ng of RNA). Exosomal miRNA was resistant to RNase A digestion, even after treatment with proteinase K (Figure 1B). This indicated that exosomal miRNA purified from BMDC culture supernatants was encapsulated inside the exosomes, instead of forming soluble ribonucloprotein complexes, as recently shown in a fraction of blood-borne miRNAs.28 Exosome RNA was used as template for miRNA profiling with the Illumina miRNA Expression Array, which targets 656 mature miRNAs. BMDC exosomes were positive for 202 miRNAs according to the intensity of miRNA expression above the threshold of data ranked by normalized intensity (n = 4 independent samples; Figure 1C). We detected 139 miRNAs in both exosomes, 5 only in immature vesicles and 58 exclusively in mature exosomes (Figure 1D). Most of the miRNAs detected in exosomes were also found in the parental cells (supplemental Figures 3-4). A percentage of the miRNAs detected in BMDCs were not found in their respective exosomes (supplemental Figures 3-4), indicating that in BMDCs, not all cellular miRNAs are sorted into exosomes. Other miRNAs were detected in the exosomes and not in the parental DCs, suggesting a preferential enrichment of some exosomal miRNAs (supplemental Figures 3-4).
miRNA in BMDC-derived immature and mature exosomes. (A) Western blot analysis (top) of 1-mL density fractions from a continuous sucrose gradient used to purify BMDC exosomes. Exosomes, detected by their CD9 expression, were enriched in those fractions with characteristic exosome density. The digital gel (bottom), obtained with Agilent 2100 Bioanalyzer, shows the presence of miRNAs in the density fractions in which exosomes were present. One of 2 experiments is shown. (B) Digital gel (Agilent 2100 Bioanalyzer) showing the presence of similar levels of miRNAs isolated from exosomes treated (or not) with proteinase K and then incubated (or not) with RNase A. Results are representative of 2 independent experiments. (C) Analysis of miRNAs from BMDC exosomes showing intensity of expression of miRNAs against their rank position. Signals above the 2500 cutoff were considered positive. (D) Venn diagram with the numbers of miRNAs detected in immature and mature exosomes released by BMDCs. List with the miRNAs detected by Illumina miRNA Expression Array. In those cases were the probe did not resolve miRNAs differing at only 1 or 2 positions, miRNAs were listed in parentheses. Results are based on the miRNA profiling of 4 different samples of each type of exosomes.
miRNA in BMDC-derived immature and mature exosomes. (A) Western blot analysis (top) of 1-mL density fractions from a continuous sucrose gradient used to purify BMDC exosomes. Exosomes, detected by their CD9 expression, were enriched in those fractions with characteristic exosome density. The digital gel (bottom), obtained with Agilent 2100 Bioanalyzer, shows the presence of miRNAs in the density fractions in which exosomes were present. One of 2 experiments is shown. (B) Digital gel (Agilent 2100 Bioanalyzer) showing the presence of similar levels of miRNAs isolated from exosomes treated (or not) with proteinase K and then incubated (or not) with RNase A. Results are representative of 2 independent experiments. (C) Analysis of miRNAs from BMDC exosomes showing intensity of expression of miRNAs against their rank position. Signals above the 2500 cutoff were considered positive. (D) Venn diagram with the numbers of miRNAs detected in immature and mature exosomes released by BMDCs. List with the miRNAs detected by Illumina miRNA Expression Array. In those cases were the probe did not resolve miRNAs differing at only 1 or 2 positions, miRNAs were listed in parentheses. Results are based on the miRNA profiling of 4 different samples of each type of exosomes.
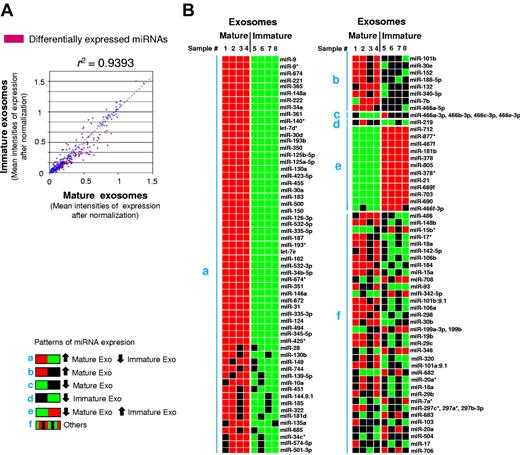
A semiquantitative analysis that was based on the number of individual samples of immature exosomes with intensity of expression of a specific miRNA in the upper/lower 95th percentile of those detected in mature exosomes (or vice versa) defined a pattern of differential miRNA expression between BMDC-derived immature and mature exosomes (Figure 2A-B; supplemental Table 1). Of note, a percentage of exosomal miRNAs from the presence/absence analysis represented in the Venn diagram (Figure 1D) are not listed in the semiquantitative analysis of the pattern grid (Figure 2B). Such miRNAs did not reach the level of statistical significance (in each of the 4 individual samples analyzed) required to be considered differentially expressed between immature and mature exosomes according to the statistical method used. The content of some of the most outstandingly differentially expressed exosomal miRNAs was confirmed by quantitative RT-PCR (supplemental Figure 5).
Differential miRNA content in BMDC-derived immature and mature exosomes. (A) Correlation plot of exosome miRNAs, indicating in red those miRNAs expressed differentially by immature and mature exosomes. (B) Expression pattern grid with miRNAs expressed differentially by immature and mature exosomes. Each box corresponds to a gene for a given sample of an experimental group. If the individual sample expression value for that miRNA is > 95 percentile of the same miRNA from the other sample group (considered as a group), it is colored red. If that individual miRNA expression value is < 5 percentile of the same miRNA from the other experimental group, it is painted green. If the expression value falls within the range of the 5th and 95th percentiles, it is represented as a black square. The groupings of miRNAs fall out logically according to the counts of the colored squares; the predominantly red and green group represents those miRNAs whereby the miRNA is overexpressed in all mature exosome samples (group a), or in all immature exosomes samples (group e). Group b represents miRNAs whereby the trend is toward overexpression in mature exosomes, and so on. The 2 main groups of most reliable differences are groups a and e. The black versus black (ie, group f) is actually miRNAs that have equal numbers of squares colored red, green, and black and are, therefore, not actually different; the measure of difference being high for such miRNAs may be because of outliers. Four independent samples of each type of exosome were analyzed.
Differential miRNA content in BMDC-derived immature and mature exosomes. (A) Correlation plot of exosome miRNAs, indicating in red those miRNAs expressed differentially by immature and mature exosomes. (B) Expression pattern grid with miRNAs expressed differentially by immature and mature exosomes. Each box corresponds to a gene for a given sample of an experimental group. If the individual sample expression value for that miRNA is > 95 percentile of the same miRNA from the other sample group (considered as a group), it is colored red. If that individual miRNA expression value is < 5 percentile of the same miRNA from the other experimental group, it is painted green. If the expression value falls within the range of the 5th and 95th percentiles, it is represented as a black square. The groupings of miRNAs fall out logically according to the counts of the colored squares; the predominantly red and green group represents those miRNAs whereby the miRNA is overexpressed in all mature exosome samples (group a), or in all immature exosomes samples (group e). Group b represents miRNAs whereby the trend is toward overexpression in mature exosomes, and so on. The 2 main groups of most reliable differences are groups a and e. The black versus black (ie, group f) is actually miRNAs that have equal numbers of squares colored red, green, and black and are, therefore, not actually different; the measure of difference being high for such miRNAs may be because of outliers. Four independent samples of each type of exosome were analyzed.
Combinatorial analysis of miRNA clusters detected predominantly in BMDC-derived immature or mature exosomes (Figure 2B groups a,e) showed that they target mRNAs involved in cytokine synthesis, cell survival, endocytosis, Ag cross-presentation, DC differentiation, TGF-β signaling, and for transcription factors (supplemental Tables 2-3). These findings indicate that the pattern of exosome-shuttle miRNAs depends on the maturation stage of the parental DCs and that the miRNAs target transcripts that regulate critical DC functions.
Efficiency of transfer of endogenous exosomes by DCs
If exosome-shuttle miRNAs constitute a mechanism of communication between DCs, the latter must capture exosomes released by other DCs. Accordingly, DCs take up exosomes added to the culture medium.18,22 However, the efficiency of spontaneous transfer of exosomes between DCs (or cells in general) is unknown. To address this, we tagged with encoding enhanced green fluorescent protein (eGFP) exosomes released by BMDCs, based on the sorting mechanism of Fas-ligand (FasL).29 BMDCs were transduced with a Rad-encoding eGFP linked to mouse FasL with a C-terminal deletion of its cytotoxic domain (RAd-eGFP-tmFasLΔ; supplemental Figure 6A). This resulted in eGFP attached to the proline and lysine residues of the intracellular domain of FasL required for sorting of eGFP into MVBs and exosomes. BMDCs transduced with RAd-eGFP-tmFasLΔ (eGFP-tmFasLΔ-BMDCs) concentrated eGFP in cytoplasmic vesicles, whereas BMDCs infected with control RAd-eGFP showed diffuse eGFP expression (supplemental Figure 6B). Quantitative colocalization analysis of confocal microscopy on eGFP-tmFasLΔ-BMDCs showed that a fraction of eGFP was sorted into LAMP-1+ compartments (Pearson correlation = 0.37 ± 0.08, with 1 being complete overlap between eGFP and LAMP-1 and −1 meaning no overlap; supplemental Figure 6C). The overlap between eGFP and LAMP-1 decreased when eGFP-tmFasLΔ-BMDCs were matured with LPS (Pearson correlation = 0.26 ± 0.10, P = .0006 compared with untreated controls; supplemental Figure 6C). This agrees with the findings of Buschow et al30 that a percentage of exosomes in MVBs is sorted for lysosomal degradation in immature DCs and that this phenomenon decreases in matured DCs. Ultrastructural analysis confirmed that the eGFP-containing domain of the chimeric protein was inside the vesicles (supplemental Figure 7A). eGFP-tmFasLΔ-BMDCs released eGFP-exosomes and exhibited similar viability to BMDCs nontreated or transduced with control RAds (supplemental Figure 7B). By FACS analysis, transgenic eGFP-tmFasLΔ was confined into MVBs without detectable leaking into the cell membrane (supplemental Figure 7C).
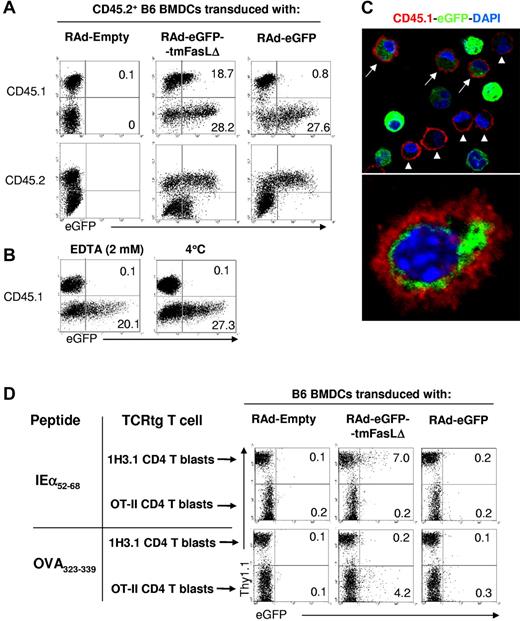
Next, we assessed the spontaneous transfer of eGFP-tagged exosomes between DCs. We cocultured CD45.2 congenic BMDCs, previously transduced with RAd-eGFP-tmFasLΔ (or with control RAd-eGFP or RAd-Empty), with equal numbers of “acceptor” CD45.1+ BMDCs. Quantification by flow cytometry showed that 15% ± 8% of CD45.1+ BMDCs acquired eGFP+ material from CD45.2+ eGFP-tmFasLΔ-BMDCs, compared with 0.1% in controls cocultured with RAd-Empty–infected BMDCs (Figure 3A). The fact that CD45.1+ BMDCs with eGFP+ material were CD45.2− excluded the possibility of doublets between acceptor and donor (CD45.2+) BMDCs (Figure 3A). The finding that only 0.9% ± 0.4% of the acceptor cells exposed to control RAd-eGFP–transduced BMDCs acquired eGFP, confirmed that the results were because of transfer of eGFP-tagged exosomes, instead of passage of other cell components, or apoptotic cell fragments (Figure 3A). Transfer of exosomes between BMDCs was abrogated in the presence of EDTA, indicating its requirement for extracellular Ca++, or at 4°C (Figure 3B). Analysis by confocal microscopy confirmed that the acquired eGFP+ material was inside the acceptor BMDCs (Figure 3C).
Transfer of endogenous exosomes between BMDCs. (A) FACS analysis of transfer of eGFP between 106 CD45.2+ BMDCs transfected with RAd-eGFP-tmFasLΔ (or control RAd-Empty or RAd-eGFP) and 106 acceptor CD45.1+ BMDCs. Numbers are percentages of cells. (B) Effect of EDTA and temperature on transfer of exosomes between CD45.2+ RAd-eGFP-tmFasLΔ-transfected BMDCs and CD45.1+ BMDCs. (C top) Confocal microscopy of CD45.1+ BMDCs (in red) incubated with CD45.2+ BMDCs transfected with RAd-eGFP-tmFasLΔ. Arrows indicate acceptor (CD45.1+) BMDCs with eGFP+ content. As control, arrowheads show some acceptor BMDCs without eGFP. (Bottom) eGFP inside an acceptor BMDC surface labeled with CD45.1 (in red; ×1000). (D) Detection by FACS of transfer of eGFP between BMDCs and TCR transgenic (tg) T cells in a 3-cell culture system. One million BMDCs transfected with RAd-eGFP-tmFasLΔ (or control RAd-Empty or RAd-eGFP) and pulsed with IEα52-68 or OVA323-339 peptide were cocultured with in vitro–activated (blasts) 1H3.1 (Thy1.1+) and OT-II (Thy1.2+) CD4 T cells (5 × 106 of each). CD11c+ cells (BMDCs) were gated out. Numbers are percentages of cells. One of 3 (A,D) or 2 (B-C) experiments is shown.
Transfer of endogenous exosomes between BMDCs. (A) FACS analysis of transfer of eGFP between 106 CD45.2+ BMDCs transfected with RAd-eGFP-tmFasLΔ (or control RAd-Empty or RAd-eGFP) and 106 acceptor CD45.1+ BMDCs. Numbers are percentages of cells. (B) Effect of EDTA and temperature on transfer of exosomes between CD45.2+ RAd-eGFP-tmFasLΔ-transfected BMDCs and CD45.1+ BMDCs. (C top) Confocal microscopy of CD45.1+ BMDCs (in red) incubated with CD45.2+ BMDCs transfected with RAd-eGFP-tmFasLΔ. Arrows indicate acceptor (CD45.1+) BMDCs with eGFP+ content. As control, arrowheads show some acceptor BMDCs without eGFP. (Bottom) eGFP inside an acceptor BMDC surface labeled with CD45.1 (in red; ×1000). (D) Detection by FACS of transfer of eGFP between BMDCs and TCR transgenic (tg) T cells in a 3-cell culture system. One million BMDCs transfected with RAd-eGFP-tmFasLΔ (or control RAd-Empty or RAd-eGFP) and pulsed with IEα52-68 or OVA323-339 peptide were cocultured with in vitro–activated (blasts) 1H3.1 (Thy1.1+) and OT-II (Thy1.2+) CD4 T cells (5 × 106 of each). CD11c+ cells (BMDCs) were gated out. Numbers are percentages of cells. One of 3 (A,D) or 2 (B-C) experiments is shown.
The capability of BMDCs to transfer exosomes to other leukocytes (ie, T cells) was investigated in a 3-cell Ag-specific system, whereby eGFP-tmFasLΔ-BMDCs (B6, IAb+) pulsed with IEα52-68 or OVA323-339 peptide were cocultured (37°C) with acceptor (naive or in vitro–activated) 1H3.1 (Thy1.1) and OT-II (Thy1.2) CD4 T cells, specific for IAb-IEα52-68 and IAb-OVA323-339, respectively. Eighteen hours later, the 1H3.1 and OT-II T cells were analyzed with FACS and identified by their Thy1 allele expression. BMDCs were excluded according to their CD11chi Thy1neg phenotype. After incubation with eGFP-tmFasLΔ-BMDCs pulsed with IEα52-68, 7.9% ± 3.9% of 1H3.1 blasts (but not naive cells; not shown) acquired eGFP (Figure 3D). By contrast, 0.2% ± 0.1% of 1H3.1 cells were eGFP+ after culture with control BMDCs transduced with RAd-eGFP or RAd-Empty (Figure 3D). As internal control, only 0.3% ± 0.1% of OT-II blasts became eGFP+, a percentage similar to that of OT-II cells exposed to BMDCs transduced with control RAds (Figure 3D). By contrast, when eGFP-tmFasLΔ-BMDCs pulsed with OVA323-339 were used as APCs, 6.1% ± 3.1% of the OT-II cells become eGFP+, compared with 0.2% ± 0.1% of the 1H3.1 cells (Figure 3D). Together, our findings indicate that BMDCs, and to a lesser extent activated (but not naive) Ag-specific CD4 T cells, acquire DC-derived exosomes released endogenously.
Mechanisms of transfer of exosomes between DCs
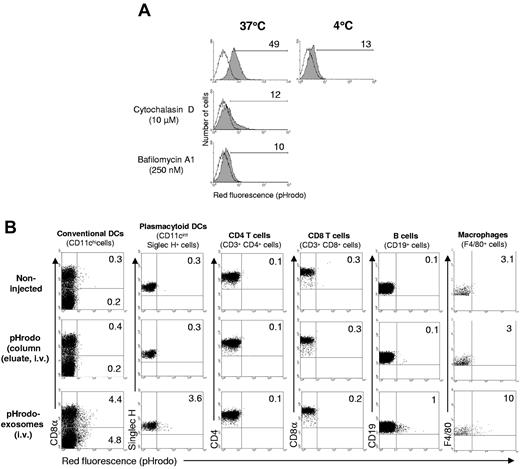
Delivery of exosome-shuttle miRNAs into the cytosol of DCs requires the following conditions: (1) endocytosis of exosomes followed by their back-fusion with the phagosome membrane and/or (2) fusion of the exogenous exosomes with the cell surface membrane. Exosomes can be phagocytosed or incorporated via a lipid-dependent fusion process to acceptor cells.18,22,31 However, the mechanism used to deliver the luminal content of the exogenous exosomes into target cells remains unknown. We quantified the ability of DCs to endocytose exosomes in vitro and in vivo by BMDC exosomes labeled with pHrodo, a dye that becomes fluorescent red at the phagosome pH. Acceptor BMDCs cocultured with pHrodo-exosomes purified by gel filtration emitted red fluorescence, confirming that DCs phagocytose exosomes. This phenomenon decreased substantially at 4°C, with the cytoskeleton inhibitor cytochalasin D, or with the blocker of the vacuolar proton ATPase, bafilomycin A1 (Figure 4A). Similar results were obtained in vivo after delivering intravenous Spephadex G75 column-purified pHrodo-exosomes in mice. The injected exosomes were internalized by splenic conventional (CD11chi) DCs and to a lesser extent by plasmacytoid DCs, B lymphocytes, and macrophages (Figure 4B). By contrast, splenic T cells did not become fluorescent red, which agrees with previous studies that reported that T cells do not internalize exosomes attached to their plasma membrane.30 The emission of red fluorescence was not because of internalization of free dye injected with the pHrodo-exosomes, because it was absent in splenocytes of control mice injected with eluates extracted from the Sephadex G75 columns after the exosome fraction, which contains the excess of free dye without vesicles.
Internalization of BMDC-derived exosomes. (A) In vitro assay of internalization of pHrodo-exosomes (5 μg, purified by gel filtration) by BMDCs (5 × 105 cells, 1 mL of final volume) assessed by FACS. Numbers are percentages of positive cells. One of 2 independent experiments is shown. (B) In vivo endocytosis of pHrodo-labeled exosomes (300 μg, intravenously, purified by gel filtration) by different splenic leukocytes, analyzed by FACS, 3 hours after exosome administration. Numbers are percentages of positive cells in the corresponding quadrant. Results are representative of 3 mice per group.
Internalization of BMDC-derived exosomes. (A) In vitro assay of internalization of pHrodo-exosomes (5 μg, purified by gel filtration) by BMDCs (5 × 105 cells, 1 mL of final volume) assessed by FACS. Numbers are percentages of positive cells. One of 2 independent experiments is shown. (B) In vivo endocytosis of pHrodo-labeled exosomes (300 μg, intravenously, purified by gel filtration) by different splenic leukocytes, analyzed by FACS, 3 hours after exosome administration. Numbers are percentages of positive cells in the corresponding quadrant. Results are representative of 3 mice per group.
Next, we sought whether BMDC exosomes fuse/hemifuse with membranes of acceptor DCs with the use of a fluorogenic de-quenching assay with the lipophilic dye R18, a method used to monitor fusion of enveloped viruses and lipid vesicles. On fusion/hemifusion of R18-labeled exosomes with lipid membranes, the surface density of the fluorophore incorporated into the outer leaflet of the exosome membrane decreases, resulting in relief of R18 quenching and the fluorescence increases proportionally to the extent of membrane fusion/hemifusion. Exosomes labeled with self-quenching concentrations of R18 and purified by gel filtration retained their membrane integrity (Figure 5A). Addition of (unlabeled) acceptor BMDCs to R18-exosomes resulted in a time-dependent fluorescence increase indicative of fusion/hemifusion of the exosomes with the BMDCs (Figure 5B). As controls, there was not spontaneous probe de-quenching in R18-exosomes incubated alone under similar conditions and before disruption with Triton X-100 (Figure 5B), and R18 de-quenching depended on the acceptor membrane concentration (Figure 5C). The findings that the rate and extent of fluorescence decreased drastically at 37°C when PF-fixed BMDCs were used as acceptor cells (Figure 5D), or at 4°C (Figure 5E), confirmed that R18 de-quenching was caused by fusion/hemifusion of R18-exosomes with BMDCs with minimum spontaneous transfer of R18 monomers.32 Fusion/hemifusion of R18-exosomes with BMDCs required cholesterol-rich microdomains, because R18 de-quenching decreased when the acceptor BMDCs were pretreated with the cholesterol-sequestering agent filipin (Figure 5F). Fusion/hemifusion of R18-exosomes with BMDCs was confirmed by time-lapse microscopy on the basis of the spread of R18 fluorescence (flashes) from the area of fusion/hemifusion of exosomes with the acceptor BMDC (Figure 5G; supplemental Video). We confirmed that either immature or mature exosomes fused/hemifused with BMDCs and with immature (freshly isolated) or mature (overnight-cultured) splenic DCs (supplemental Figure 8), suggesting that release and fusion/hemifusion of exosomes are a general mechanism of communication between conventional DCs.
Fusion/hemifusion of exosomes with BMDCs. (A) Structure of R18-labeled exosomes purified by gel filtration (×100 000). Bar = 100 nm. (B) Spectrofluorimetric analysis after addition of (unlabeled) BMDCs to R18-exosomes. When R18-exosomes were incubated alone (control) fluorescence increased only after their disruption with Triton X-100 (T X100). (C-F) Fusion assays of R18-exosomes with BMDCs at different conditions. (B-F) Results are expressed as percentages of maximal fluorescence de-quenching [FD (%)]. Spectrofluorimetric assays were done exposing 106 BMDCs to 10 μg of R18-exosomes in a final volume of 2 mL of 2 g/L glucose Ca/Mg HBSS. (G) Differential interference contrast (top) and fluorescence (bottom) images during fusion/hemifusion of R18-exosomes with BMDCs. Arrow indicates the fusion/hemifusion of one R18-exosome(s) with the BMDC on the right, detected when the area of diffusion of de-quenched R18 on the DC surface reached the limit of resolution of the objective. Images were pseudo-colored to indicate the intensity of light. Arrowhead points to another fusion of an R18-exosome(s) with a neighboring BMDC. The chart shows the quantitative analysis of the cells from the images (×400). For fluorescence time-lapse microscopy, 150 000 BMDCs were incubated with 2.5 μg of R18-exosomes in a final volume of 500 μL. Results (B-G) represent ≥ 3 experiments per condition.
Fusion/hemifusion of exosomes with BMDCs. (A) Structure of R18-labeled exosomes purified by gel filtration (×100 000). Bar = 100 nm. (B) Spectrofluorimetric analysis after addition of (unlabeled) BMDCs to R18-exosomes. When R18-exosomes were incubated alone (control) fluorescence increased only after their disruption with Triton X-100 (T X100). (C-F) Fusion assays of R18-exosomes with BMDCs at different conditions. (B-F) Results are expressed as percentages of maximal fluorescence de-quenching [FD (%)]. Spectrofluorimetric assays were done exposing 106 BMDCs to 10 μg of R18-exosomes in a final volume of 2 mL of 2 g/L glucose Ca/Mg HBSS. (G) Differential interference contrast (top) and fluorescence (bottom) images during fusion/hemifusion of R18-exosomes with BMDCs. Arrow indicates the fusion/hemifusion of one R18-exosome(s) with the BMDC on the right, detected when the area of diffusion of de-quenched R18 on the DC surface reached the limit of resolution of the objective. Images were pseudo-colored to indicate the intensity of light. Arrowhead points to another fusion of an R18-exosome(s) with a neighboring BMDC. The chart shows the quantitative analysis of the cells from the images (×400). For fluorescence time-lapse microscopy, 150 000 BMDCs were incubated with 2.5 μg of R18-exosomes in a final volume of 500 μL. Results (B-G) represent ≥ 3 experiments per condition.
Release of the luminal content of exosomes into the cytosol of DCs
In R18-assays, probe de-quenching still occurs if the membrane fusion is arrested at the hemifusion stage. At this phase, the outer leaflet of the exosome membrane coalesces with the external monolayer of the DC membrane without formation of the fusion pore.33 To test whether exosomes release their content into the DC cytosol, we set up a content-mixing assay with the use of luciferin-loaded exosomes and BMDCs expressing transgenic luciferase (LUC-BMDCs). Exosomes were loaded with membrane-permeable DMNPE-caged luciferin that was released inside the exosomes by UV-B photolysis. The nonencapsulated luciferin was removed from the vesicles by gel filtration. Because LUC-BMDCs retained the transgenic luciferase in the cytosol and the uncaged luciferin inside the exosomes is unable to cross lipid membranes, the assay measures content mixing by the emission of light produced by oxidation of the exosome-shuttle luciferin by the cytosolic luciferase. Addition of luciferin-exosomes to LUC-BMDCs was followed by emission of light that started 8.0 ± 0.6 minutes later and increased continuously until the end of the assay (4 independent experiments; Figure 6A). The average percentage of increase in light emission at the assay end point (35 minutes) was 17.25% ± 4.80% in LUC-BMDCs incubated with luciferin-exosomes, and 1.28% ± 1.14% in LUC-BMDCs exposed to control-exosomes (P = .0006). By contrast, LUC-BMDCs incubated with control exosomes (without luciferin) emitted light at background levels, comparable to LUC-BMDCs incubated alone (Figure 6A). As controls, addition of luciferin-exosomes or unlabeled (control) exosomes to control BMDCs (transduced with RAd-Empty) gave background light emissions (Figure 6B).
Release of luminal content of exosomes and acquisition of exosome-shuttle miRNAs by DCs. (A) Content-mixing assay between 106 BMDCs transduced with RAd-LUC (Luciferase BMDCs or with RAd-Empty, control BMDCs) and 10 μg exosomes loaded with luciferin (or not, Control-Exosomes) added to the DCs 3 minutes later. The assays were done in a final volume of 2 mL of 2 g/L glucose Ca/Mg HBSS. Luciferase BMDCs incubated alone were included as negative controls. (B) Content-mixing assay between 106 control BMDCs and 10 μg of exosomes loaded with luciferin (or not, Control-Exosomes) added to the DCs 3 minutes later. (C) Ultrastructural analysis of the interaction of exogenously added (BALB/c) BMDC exosomes surface labeled with 5 nm of gold particles (IAd+, arrowheads) with the plasma membrane of an acceptor BMDC (B6, IAd−). The arrow shows the tight contact between the surface membrane of the BMDCs and the membrane of the labeled exosome (×100 000). Bar = 100nm. (D) Internalized 5 nm of gold-labeled (arrowheads) exosomes (arrows) inside a BMDC phagosome (×100 000). Bar = 100nm. The area within the dotted line is shown at higher detail on the right. (E) Luciferase reporter construct with 3 tandem copies of the target sequence to miR-451 or miR-148a located in the 3′-untranslated region and the control vector with the inverted sequences. (F-G) Normalized expression of luciferase in 106 DC2.4 cells transfected with pCMV-Luc/3 × PT-miR-451 or control pCMV-Luc/3 × PT-inverted-miR-451 (F), or with pCMV-Luc/3 × PT-miR-148a or control pCMV-Luc/3 × PT-inverted-miR-148a (G), after 18 hours incubation alone, or with increasing concentrations of exosomes in complete medium. Shown results are representative of 4 (A-B), 12 (C-D), and 3 (F-G) independent experiments.
Release of luminal content of exosomes and acquisition of exosome-shuttle miRNAs by DCs. (A) Content-mixing assay between 106 BMDCs transduced with RAd-LUC (Luciferase BMDCs or with RAd-Empty, control BMDCs) and 10 μg exosomes loaded with luciferin (or not, Control-Exosomes) added to the DCs 3 minutes later. The assays were done in a final volume of 2 mL of 2 g/L glucose Ca/Mg HBSS. Luciferase BMDCs incubated alone were included as negative controls. (B) Content-mixing assay between 106 control BMDCs and 10 μg of exosomes loaded with luciferin (or not, Control-Exosomes) added to the DCs 3 minutes later. (C) Ultrastructural analysis of the interaction of exogenously added (BALB/c) BMDC exosomes surface labeled with 5 nm of gold particles (IAd+, arrowheads) with the plasma membrane of an acceptor BMDC (B6, IAd−). The arrow shows the tight contact between the surface membrane of the BMDCs and the membrane of the labeled exosome (×100 000). Bar = 100nm. (D) Internalized 5 nm of gold-labeled (arrowheads) exosomes (arrows) inside a BMDC phagosome (×100 000). Bar = 100nm. The area within the dotted line is shown at higher detail on the right. (E) Luciferase reporter construct with 3 tandem copies of the target sequence to miR-451 or miR-148a located in the 3′-untranslated region and the control vector with the inverted sequences. (F-G) Normalized expression of luciferase in 106 DC2.4 cells transfected with pCMV-Luc/3 × PT-miR-451 or control pCMV-Luc/3 × PT-inverted-miR-451 (F), or with pCMV-Luc/3 × PT-miR-148a or control pCMV-Luc/3 × PT-inverted-miR-148a (G), after 18 hours incubation alone, or with increasing concentrations of exosomes in complete medium. Shown results are representative of 4 (A-B), 12 (C-D), and 3 (F-G) independent experiments.
Fusion/hemifusion of BALB/c BMDC exosomes (labeled with 5 nm of gold anti-IAd antibody) with acceptor B6 (IAd−) BMDCs was investigated by electron microscopy. After 15 or 30 minutes of incubation (37°C), numerous gold-labeled exosomes were detected attached to the BMDC surface (Figure 6C). Labeled exosomes were also found in the lumen or attached to the membrane of phagocytic vesicles (Figure 6D). We did not detect conclusive images of labeled exosomes fusing with the cell surface or phagosome membranes (12 independent experiments), suggesting that complete fusion of exosomes must be an extremely rapid and/or localized phenomenon.
Acquisition of functional exosome-shuttle miRNAs by DCs
We tested whether DCs acquire functional exosome-shuttle miRNAs to evaluate the physiologic relevance of this phenomenon in DC-to-DC communication. To address this, it was critical to establish that the miRNA-mediated regulation was caused by a given exosome-shuttle miRNA in the absence of the homologous cellular miRNA. After testing different exosome-shuttle miRNAs, we selected for the miRNA reporter assay mature miR-451 and miR148a and the DC2.4 cell line (as acceptor cells) because of the following reasons: (1) miR-451 and miR148a were present in BMDC exosomes and absent/expressed at very low levels in DC2.4 cells (supplemental Figure 9A); (2) BMDC-derived exosomes are internalized and fused to DC2.4 cells to a similar extent as BMDCs (supplemental Figure 9B-C); and (3) DNA transfection efficiency in much higher in DC2.4 cells than in BMDCs. For the miRNA reporter assay, we added to a vector encoding firefly luciferase (pCMV-Luc, pMIR-REPORT System; Ambion) 3 tandem copies of the perfectly complementary target (3 × PT) sequence for miR-451 or miR-148a between the 3′-untranslated region and the polyA (pCMV-Luc/3 × PT-miR-451; Figure 6E). DC2.4 cells were cotransfected with the β-galactosidase reporter pMIR-REPORT β-gal (Ambion) to normalize variability because of differences in cell viability and transfection efficiency or with an eGFP reporter plasmid to assess transfection efficiency (≥ 71% of DC2.4 cells). Incubation of pCMV-Luc/3 × PT-miR-451–tranfected DC2.4 cells with BMDC exosomes caused a significant and dose-dependent decrease in normalized expression of luciferase, measured by luminometry (Figure 6F). By contrast, DC2.4 cells transfected with control pCMV-Luc encoding 3 × PT of the inverted sequence for miR-451 did not reduce normalized expression of luciferase after incubation with BMDC exosomes (Figure 6F). Similar results were obtained when pCMV-Luc/3 × PT-miR-148a–tranfected DC2.4 cells were incubated with BMDC exosomes (Figure 6G). Together, these findings indicate that regulation of the reporter gene was specific and dependent on functional miRNAs delivered by the exosomes.
Discussion
Most studies on exosomes have been done with purified exosomes.17-22,31 Therefore, whether transfer of exosomes released endogenously constitutes an efficient mechanism of communication between DCs (and cells in general) is unknown. Moreover, it is presently unclear the mechanism by which the intravesicular content of exosomes is transferred between cells.
Our findings indicate that BMDCs efficiently transfer endogenous exosomes to other DCs and that they are internalized, hemifused, and/or fused with the target DC. Previous reports showed that exosomes labeled with fluorescent dyes are internalized by different cell types.18,22,31 However, because the size of the exosomes is below the limit resolution of conventional microscopy, they probably detected aggregates of exosomes inside phagosomes and were unable to follow the interaction of individual exosomes with the cells. Besides, detection of exosome hemifusion/fusion is expected to be difficult because it is probably a localized event that occurs in milliseconds. By combining spectrofluorimetry, fluorescence time-lapse, and immuno-electron microscopy, we demonstrated that exosomes dock, bind, and fuse with acceptor BMDCs.
The results of the content-mixing assays with the use of luciferin-loaded exosomes showed that exosomes “inject” their intraluminal content into the cytosol of the target DCs. Thus, similarly, exosome-shuttle miRNAs (and other RNAs) are delivered into the cells protected from degradation by extracellular RNAses. The pattern of adsorption and hemifusion/fusion of exosomes with target DCs, and their cholesterol requirement on the DC membrane, resembles those of enveloped viruses such as HIV.34 This suggests that retroviruses not only share the exosome biogenesis pathway for the assembly of infectious particles, as suggested by the “Trojan exosome hypothesis,”35 but also the exosome delivery mechanism for their cell-to-cell transmission. Whether the fusion of extracellular exosomes with membranes of acceptor cells depends on fusogenic lipids or proteins remains to be explored. The exosome membrane contains lipid rafts (enriched in cholesterol, sphyngomyelin, and ganglioside GM3),36 and externalized phosphatidylserine.18 Several enveloped viruses fuse with target cells through lipid rafts,34 and externalized phosphatidylserine facilitates membrane fusion in the presence of Ca++ and cholesterol.37,38 The tetraspanins CD9 and CD81, both constitutive proteins of the exosome membrane, were also shown to participate in membrane fusion events.39
We and others have demonstrated that exosomes are internalized by DCs and other phagocytes.18,22,31,40 However, whether extracellular exosomes release their luminal content into acceptor DCs, and cells in general, and the cellular localization where this event occurs remain unknown. The finding that exosomes labeled with R18 in the outer leaflet of their membranes transferred R18 to DCs does not necessarily indicate complete fusion of the exosomes with the plasma membrane, because passage of R18 also occurs at the hemifusion stage, when only the outer leaflets of both membranes coalesce without formation of the fusion pore. The 8.0 ± 0.6 minutes of delay detected between the R18 assay and content-mixing experiments are sufficiently long to permit endocytosis. This suggests that, at least a percentage of the exosomes, could release their luminal content via a “2-step event” consisting of, first, exosome hemifusion with the cell membrane, and, second, endocytosis and complete fusion of the exosomes with the limiting membrane of the phagosome. Alternatively, exosomes could be internalized as free/attached vesicles (without hemifusion/fusion) and then fuse with the phagosome membrane. Interestingly, HIV-1 infects target cells with the use of similar fusion mechanisms.41
The reason why miRNAs are present in the exosome lumen is unknown. Components of the RNA-induced silencing complex such as the proteins GW182 and Argonaute2, mRNAs, and mature miRNAs were found associated with MVBs and exosomes,42 suggesting that MVBs are sites for assembly of the miRNA-silencing machinery and/or sorting of miRNAs into exosomes. Whether exosome-shuttle miRNAs are sorted differentially into exosomes based on their sequence, or rather represent a snap-shot of the miRNA content of the cell, remains to be solved.
Our findings indicate that the pattern of exosome-shuttle miRNAs varies according to the maturation of the parental DC. Because a determined miRNA can target different mRNAs, we performed a combinatorial analysis of their potential mRNA targets. For simplicity, we only considered those mRNAs recognized by ≥ 4 or 5 miRNAs from immature or mature exosomes, respectively. Our results show that some of the exosome-shuttle miRNAs clusters regulate key DC functions. Interestingly, both types of BMDC exosomes carry miR-34a and miR-21 that direct differentiation of hematopoietic precursors into myeloid DCs (instead of monocytes), and miR-221 and miR-222 that prevent differentiation into plasmacytoid DCs.43,44 Compared with BMDC immature exosomes, mature exosomes contain more miR-125b-5p, miR-146a, and miR-148, all negative regulators of pro-inflammatory transcripts in myeloid cells and DCs (Figure 2B).45-47 miR-155 is known to be strongly up-regulated during DC maturation in humans.48 Accordingly, we consistently found more miR-155 in mature than in immature exosomes, although the difference did not reach the level of statistical significance in all the 4 independent samples examined in our semiquantitative analysis.
Our finding that BMDC-derived exosomes fuse with target cells and deliver their content into the cytoplasm of acceptor DCs (and probably other target cells) opens the possibility that exosomes could function as a device for horizontal propagation of proteins, lipids, and RNAs between cells. However, whether those miRNAs detected in the BMDC exosomes are specifically targeted to the vesicles, or are there because they are present in the parental cells, remains to be elucidated. The microarray and PCR analysis of miRNAs from BMDC exosomes allowed us to select exosome miRNAs that were absent (or expressed minimally) by the target cells and to demonstrate the acquisition of functional exosomal-shuttle miRNAs by DCs. This mechanism could participate in the fine-tuning of the APCs and the immune response, as recently suggested for viral miRNAs, and after postsynaptic interaction between T and B cells.49,50
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgment
The authors thank Dr J. Lamb for assistance with the miRNA arrays.
This work was supported by the National Institutes of Health (grants R01 HL075512 and HL077545, A.E.M; grant R01 AI077511, A.T.L.; grant F30 DK082131, S.J.D.), and by grants from the T. E. Starzl Transplantation Institute.
National Institutes of Health
Authorship
Contribution: A.M., A.T.L., and A.E.M. designed and performed experiments, analyzed data, and made the figures; W.J.S., D.B.S., M.L.G.S., J.K., K.B., G.A.G., G.E., Z.W., J.M., O.A.T., S.J.D., and S.C.W. performed experiments; R.J. and J.L.-W. analyzed data; and A.E.M. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Adrian E. Morelli, E1546 Biomedical Science Tower, 200 Lothrop St, Pittsburgh, PA 15213-2582; e-mail: morelli@imap.pitt.edu.
References
Author notes
A.M. and A.T.L. contributed equally to this study.

![Figure 5. Fusion/hemifusion of exosomes with BMDCs. (A) Structure of R18-labeled exosomes purified by gel filtration (×100 000). Bar = 100 nm. (B) Spectrofluorimetric analysis after addition of (unlabeled) BMDCs to R18-exosomes. When R18-exosomes were incubated alone (control) fluorescence increased only after their disruption with Triton X-100 (T X100). (C-F) Fusion assays of R18-exosomes with BMDCs at different conditions. (B-F) Results are expressed as percentages of maximal fluorescence de-quenching [FD (%)]. Spectrofluorimetric assays were done exposing 106 BMDCs to 10 μg of R18-exosomes in a final volume of 2 mL of 2 g/L glucose Ca/Mg HBSS. (G) Differential interference contrast (top) and fluorescence (bottom) images during fusion/hemifusion of R18-exosomes with BMDCs. Arrow indicates the fusion/hemifusion of one R18-exosome(s) with the BMDC on the right, detected when the area of diffusion of de-quenched R18 on the DC surface reached the limit of resolution of the objective. Images were pseudo-colored to indicate the intensity of light. Arrowhead points to another fusion of an R18-exosome(s) with a neighboring BMDC. The chart shows the quantitative analysis of the cells from the images (×400). For fluorescence time-lapse microscopy, 150 000 BMDCs were incubated with 2.5 μg of R18-exosomes in a final volume of 500 μL. Results (B-G) represent ≥ 3 experiments per condition.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/119/3/10.1182_blood-2011-02-338004/5/m_zh89991183180005.jpeg?Expires=1765942093&Signature=iDFz1dJP7BZYjBJ1GmiJU7ijgwr4ItwdlxClfVq9t2odkyyYFWogIasIOqzS37Tw41f07lW1OECY56cvOz-JBf95-iV4X96~qCEDHH6JnodzOunRRg4YAqRchgWRtio5y5N~wyESteOJZQ-1enukKDJJwVbMav-lo0pPF2Lc3GIUFpJdMV06uaMY-Ebs-QSfI949xGKw6J7oHPiCKpmpMMdWHrWcTaSX6pKBQk1lR1X~-btiUR9zHWrRyx2TGwVq5-ge9Cw25tIizBNfhZ5p7oi08pI5ZBD0lDkwdob0fxCj6p7C-kaHio9jeFugek2fGjYnAhkO2lMQE12RGe6nQA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal