Abstract
HSC function depends on the tight control of proliferation and the balance between self-renewal and differentiation. Here, we report that the trimeric transcription factor NF-Y is critical for the survival of cycling, but not quiescent HSCs. With the use of a conditional knockout mouse model, we demonstrate that NF-Ya deletion creates an accumulation of HSCs in G2/M and prompts apoptosis, causing hematopoietic failure and death of the animal. These defects are accompanied by the dysregulation of multiple genes that influence cell cycle control (cyclin b1 and p21), apoptosis (Bcl-2), and self-renewal (HoxB4, Notch1, Bmi-1) and are independent of p53. Our results identify NF-Y as a pivotal upstream participant in a regu-latory network necessary for the pre-servation of cycling HSCs.
Introduction
HSCs have the unique potential to give rise to all lineages of blood cells. To ensure the proper generation of blood cells throughout the lifetime of an organism, differentiation versus self-renewal decisions of HSCs need to be tightly regulated. Numerous genes and signaling pathways, including HoxB4, Notch1, Bmi-1, and the Wnt signaling pathway, have been implicated in this process.1-11 HSCs cycle at varying rates with 2 kinetic groups identified: a quiescent pool dividing every 149-193 days and a more rapidly dividing pool with an average cycle time of 28-36 days.12 The basis for modulation of cell cycling among HSCs appears to be both determined by microenvironment and cell intrinsics.13,14
NF-Y is a heterotrimeric transcription factor consisting of 3 subunits, NF-Ya, -Yb, and -Yc, all of which are necessary for DNA binding at CCAAT-containing promoters.15 NF-Yb and NF-Yc are constitutively expressed, whereas NF-Ya encodes the regulatory subunit that is differentially expressed. Because the CCAAT NF-Y recognition site is a common promoter motif,16,17 NF-Y activity is thought to be highly influenced by cell state and direct binding partners. For example, upstream stimulating factor-1/-2 (USF-1/-2) and p53 have been shown to form a transcriptional complex with NF-Y before binding to their target promoters.18,19
Numerous in vitro studies implicate NF-Y in the regulation of the cell cycle.20-24 Despite these reports, there is controversy about how NF-Y inactivation affects the cell cycle, perhaps because different cell lines were used. The early lethality of constitutively deleted NF-Ya in mice, causing embryonic death at E8.5,25 has heretofore impeded the ability to address the role of NF-Y in specific cell lineages and cell stages in vivo.
By generating mice overexpressing NF-Ya in HSCs, we previously identified NF-Y to be a potential activator of genes regulating HSC behavior, such as HoxB4, HoxC4, D4, Notch1, and Lef-126 . These mice had an increased pool of HSCs. Similarly, TAT-NF-Ya protein transduction of human CD34+ cells led to increased proliferation in the presence of myeloid cytokines by 4-fold.27 Interestingly, the overexpression of NF-Ya in the hematopoietic system showed no effect on the cell cycle.26 These studies, however, could not address the role, if any, of endogenously regulated NF-Y on normal HSC behavior. To directly assess the constitutive role of NF-Y on normal HSC physiology in vivo, we now report the effects of conditional deletion of NF-Ya within the hematopoietic compartment on the fate of quiescent and proliferating HSCs. The results show that normal expression of NF-Y is essential for HSC proliferation and survival, probably through its role in the coordinated activation of cell cycle regulatory genes as well as genes regulating HSC differentiation and survival.
Methods
Generation of mice with a deletion of NF-Ya in the hematopoietic system
Mice carrying a floxed NF-Ya gene (NF-Yafl/fl)25 were mated with mice carrying the cre-recombinase transgene controlled by the Mx1 promoter.28 When indicated we used NF-Ya control, heterozygous, and mutant BM chimeras generated by BM transplantation as described.29 Six to 8 weeks after the transfer, deletion of NF-Ya was induced by pIpC injection as described previously28 unless stated otherwise.
Animal treatment
Mice were maintained in the animal facilities of the University of Pennsylvania and the Massachusetts General Hospital. The ethic committees of both institutions approved all animal experiments.
Flow cytometry
Single-cell suspensions were prepared and analyzed as described.29 Erythrocytes in blood and BM samples were lysed before analysis.29
The following primary and secondary Abs (clones) were used in flow cytometry: rat anti–B220-FITC (RA3-6B2), rat anti–CD19-PE (1D3), rat anti–IgM-PE (R6-60.2), rat anti–IgD-FITC (11-26c.2a), rat anti–CD4-PE (H129.19), rat anti–CD8-FITC (53-6.2), rat anti–CD3-APC (allophycocyanin; 17A2), rat anti–Gr-1–APC (RB6-8C5), rat anti–Mac-1–PE (M1-70), rat anti–Ter-119–PE (Ter119), mouse anti–Nk1.1-PE (PK136), these lineage Abs biotinylated, streptavidin-APC or streptavidin-PerCP–Cy5.5, rat anti–sca-1-PE–Cy7 (D7), rat anti–c-kit–APC-Alexa 750 (2B8), rat anti–Ly-5.1–APC (A20), rat anti–Ly-5.2–PE (104), rat anti–CD150-PE (9D1), hamster anti–CD48-APC (HM48-1), rat anti–CD34-Alexa Fluor 700 (RAM34; all eBioscience), and anti–Ki-67–FITC (35; BD Biosciences). For HSC cycle analysis we first stained 20 × 106 cells with surface markers. Then cells were resuspended in 200 μL of PBS and fixed by adding 1 mL of ice-cold fixation solution (0.25% Saponin, 2.5% paraformaldehyde, 2% FCS in PBS) and 30 minutes of incubation on ice and in the dark. Cells were washed 2 times in ice-cold Saponin wash buffer (0.25% Saponin, 2% FCS in PBS) and resuspended in 1 mL of Saponin wash buffer. Twenty microliters of Ki-67–FITC Ab and 1 μL of RNase (100 mg/mL) were added and vortexed. After 30 minutes of incubation cells were washed 2 times in Saponin wash buffer and resuspended for flow analysis in 250 μL of Saponin wash buffer. Immediately before flow cytometric analysis 250 μL of DAPI (4,6-diamidino-2-phenylindole; Sigma-Aldrich) solution was added (to a final concentration of 10 μg/mL). Annexin V (BD Biosciences) staining was performed according to the manufacturer's instructions.
Flow cytometric data were collected with a FACSCalibur or a LSR II bench top (BD Biosciences) and evaluated with FlowJo 8.8.6 software (TreeStar).
Colony formation assay
Pre-B, GM, and erythroid CFU (CFUe) colony formation assays were performed as described previously.29
Cell-separation by MACS
Single-cell suspensions were isolated and washed in MACS buffer (0.5% BSA, 2mM EDTA in PBS). Cells were stained with biotin-labeled Abs, washed, and subsequently labeled with Streptavidin beads (Miltenyi Biotec). Magnetic sorting was performed with MACS beads according to the manufacturer's instructions. For HSC analysis, lineage-positive cells were depleted with biotinylated lineage Abs and subsequent magnetic labeling with streptavidin-labeled Dynal magnetic beads followed by separation in a 15-mL tube with the use of a VarioMACS separator magnet.
Western blot analysis
Protein lysates from the indicated cells were separated with 12% SDS-PAGE and subsequently blotted onto a nitrocellulose membrane (Bio-Rad). The membrane was blocked in 5% skim milk (Bio-Rad) in PBS plus 0.2% Tween 20. The membranes were probed with rabbit polyclonal anti mouse NF-Ya (H-209), p53 (FL-393-G), and goat polyclonal anti–mouse actin (I-19) primary Abs and HRP-labeled secondary Abs (all Santa Cruz Biotechnology), which were detected with the enhanced ECL reagent (GE Healthcare) before the resulting signals were detected with a X-OMAT film (Kodak) and developed. The resulting bands were quantified with ImageJ software (National Institutes of Health).
RT-PCR and quantitative RT-PCR analyses
For reverse transcriptions, we used the Superscript III reverse transcription kit (Invitrogen). Quantitative expression analysis was performed with TaqMan Gene expression assays (Applied Biosystems). Assay numbers for the quantified expression of genes are as follows: NF-Ya, Mm00477814_m1; HoxB4, Mm00657964_m1; Notch-1, Mm00435249_m1; Lef-1, Mm00550265_m1; Bmi-1, Mm00776127_gH; Ccnb1, Mm00838401_g1; Cdkn1a, Mm00432448_m1; Bax, Mm00432050_m1; Bcl-2, Mm00477631_m1. GapDH (4352932E) was used as internal control. For the (nonquantitative) NF-Ya RT-PCR reaction we used the primers 3′-TGGAGCCTCTGATTGGGTTTCG-5′ and 3′-GGAATGTGGTCAACTCAGGA GGA-5′.
For the determination of the amount of NF-Ya null allele in DNA samples we used the QuantiFast SYBR green PCR system (QIAGEN). Primers used to detect the NF-Ya null allele were forward, 5′-CTGTCTCAGGCCAGGATTTC-3′; reverse, 5′-GCTCTCATCTCTCCTGGAGC-3′. Internal control primers specific for the cdc42 gene were used forward, 5′-ATGTAGTGTCTGTCCATTGG-3′; reverse, 5′-TCTGCCATCTAC-ACATACAC-3′.
Statistical analysis
Error bars in all figures represent the SD. P values are based on an unpaired Student t test.
Results
NF-Ya is expressed in the hematopoietic system; its expression can be efficiently deleted in the hematopoietic system
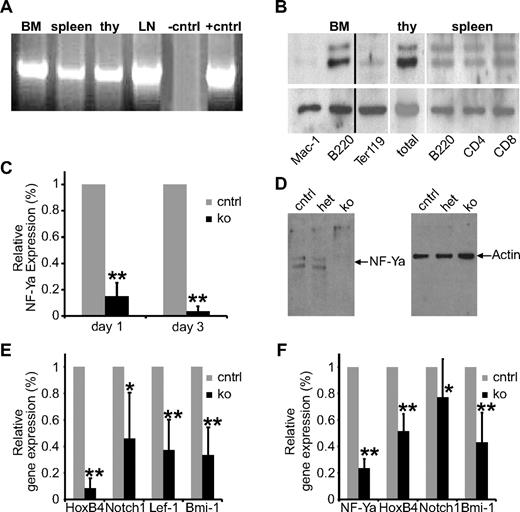
The constitutive deletion of NF-Ya in mice leads to lethality before E8.5 of embryonic development,25 and the first definitive HSCs in mouse cannot be found before E10.5.30 Therefore, we sought to address the role of NF-Y in HSCs by an inducible NF-Ya knockout (ko) within the adult hematopoietic system, including HSCs. First, we demonstrated NF-Ya mRNA expression by RT-PCR in BM, spleen, thymus, and lymph nodes (Figure 1A). NF-Ya protein was detected by Western blot analysis in B cells and T cells at high levels and in erythroblasts and myeloid cells at lower levels, the latter concurrent with previous observations.18 Thus, NF-Ya is expressed in multiple blood cell lineages with protein levels varying, depending on cell type (Figure 1B).
NF-Ya is expressed in all hematopoietic tissues and lineages and can be deleted efficiently. (A) RNA from BM cells, splenocytes, thymocytes (thy), and cells from the lymph nodes (LNs) of wt mice was analyzed for NF-Ya expression by RT-PCR. (B) Total BM cells, splenocytes, and thymocytes were isolated from wt mice and different blood cell lineages purified with the indicated Abs and magnetic beads. Cell lysate from these cells was subjected to Western blot analysis for NF-Ya (top) and actin (as a loading control; bottom). A black vertical line has been inserted to indicate a repositioned gel lane. (C) RNA from total BM of control and ko mice 1 day and 3 days after a single pIpC injection was subjected to quantitative RT-PCR for NF-Ya. GAPDH was used as an internal control; results were normalized to control samples (n = 6); **P < .01. (D) Protein from BM cells of control, het, and ko mice 4 days after the first pIpC injection was subjected to Western blot analysis with the use of Abs against NF-Ya (left) and actin (as loading control; right). (E) RNA from total BM of control and ko mice 1 day after a single pIpC injection was subjected to quantitative RT-PCR for the indicated genes. GAPDH was used as an internal control; the results were normalized to control samples (n = 6); *P < .05 and **P < .01. (F) RNA from HSCs (LSK/SLAM) of control and ko mice 1 day after a single pIpC injection was subjected to quantitative RT-PCR for the indicated genes. GAPDH was used as an internal control; the results were normalized to the control samples (n = 6); **P < .01.
NF-Ya is expressed in all hematopoietic tissues and lineages and can be deleted efficiently. (A) RNA from BM cells, splenocytes, thymocytes (thy), and cells from the lymph nodes (LNs) of wt mice was analyzed for NF-Ya expression by RT-PCR. (B) Total BM cells, splenocytes, and thymocytes were isolated from wt mice and different blood cell lineages purified with the indicated Abs and magnetic beads. Cell lysate from these cells was subjected to Western blot analysis for NF-Ya (top) and actin (as a loading control; bottom). A black vertical line has been inserted to indicate a repositioned gel lane. (C) RNA from total BM of control and ko mice 1 day and 3 days after a single pIpC injection was subjected to quantitative RT-PCR for NF-Ya. GAPDH was used as an internal control; results were normalized to control samples (n = 6); **P < .01. (D) Protein from BM cells of control, het, and ko mice 4 days after the first pIpC injection was subjected to Western blot analysis with the use of Abs against NF-Ya (left) and actin (as loading control; right). (E) RNA from total BM of control and ko mice 1 day after a single pIpC injection was subjected to quantitative RT-PCR for the indicated genes. GAPDH was used as an internal control; the results were normalized to control samples (n = 6); *P < .05 and **P < .01. (F) RNA from HSCs (LSK/SLAM) of control and ko mice 1 day after a single pIpC injection was subjected to quantitative RT-PCR for the indicated genes. GAPDH was used as an internal control; the results were normalized to the control samples (n = 6); **P < .01.
To generate mice with an inducible NF-Ya deletion in the hematopoietic system, we intercrossed mice carrying the floxed (fl) NF-Ya allele25 with mice carrying the Mx-cre transgene.28 We used NF-Ya+/++Mx-cre mice as control and NF-Yafl/fl+Mx-cre as ko mice. NF-Ya deletion was induced by 3 intraperitoneal poly(I):poly(C) (pIpC) injections. Twenty-four hours after ko induction, quantitative RT-PCR for NF-Ya showed the successful abrogation of NF-Ya transcription in the BM (Figure 1C). Western blot analysis confirmed the lack of NF-Ya protein 96 hours after induction of the gene deletion (Figure 1D). We examined heterozygote cells because others had reported a phenotype in vitro among heterozygote cells or cells in which a knockdown strategy was used.19,25 Although we found NF-Ya expression levels reduced to ∼ 50% in the heterozygous (het) compared with wild-type (wt) samples (supplemental Figure 1A, available on the Blood Web site; see the Supplemental Materials link at the top of the online article), NF-Ya protein levels were indistinguishable between wt and het mice (supplemental Figure 2B). Not surprisingly, hematopoiesis in het mice was phenotypically indistinguishable from that of wt mice. We conclude that het BM cells compensate for the reduced expression posttranscriptionally and that 1 functional NF-Ya allele is sufficient for normal hematopoiesis and HSC function.
NF-Ya deletion reduces the expression of HoxB4, Notch1, Lef-1, and Bmi-1
Our prior studies that used NF-Y overexpression26 showed increased expression of HoxB4, as well as Notch1 and Lef-1. To determine whether these genes are indeed targets of basal NF-Ya expression in vivo, we examined mRNA levels after NF-Ya deletion. HoxB4, Notch1, Lef-1 and, unexpectedly, Bmi-1 transcripts were significantly down-regulated 24 hours after a single pIpC injection (Figure 1E). Because each of these genes is known to play an important role in HSC proliferation, we confirmed their altered mRNA expression specifically in (Lin−, Sca1+, c-kit+ [LSK], CD48−, CD150+) sorted HSCs (Figure 1F). This result suggests that NF-Y might play a key role in the support of basal HSC function in vivo.
NF-Ya ko mice die of a rapid decrease in peripheral blood cell counts caused by BM failure
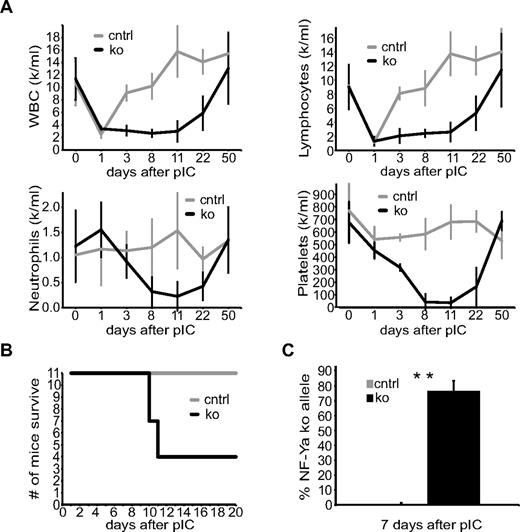
To restrict effects of NF-Y depletion with the Mx1-Cre model to the hematopoietic system, we transplanted control and ko BM into wt host animals. We then induced NF-Ya deletion with 3 injections of pIpC (0.025 mg pIpC/g of mouse weight) in the BM chimeric mice 8 weeks after BM transplantation (BMT) and followed their complete blood counts over time. White blood, lymphocyte, neutrophil, and platelet counts fell sharply to reach the lowest values as soon as 11 days after pIpC treatment (Figure 2A). By that time, 7 of 11 ko mice had died probably because of BM failure (Figure 2B). The blood counts of the surviving mice normalized over time, suggesting recovery because of either residual nonablated host HSCs or undeleted donor HSC cell activity.
NF-Ya–deficient BM chimeric mice die with severely reduced complete blood counts. (A) PB from one cohort of control (gray) and ko (black) BM chimeric mice 1, 3, 8, 11, 22, and 50 days after the first pIpC injection was isolated and counted (n ≥ 4 for every time point). (B) Survival curve of control (gray) and ko BM chimeric mice (black) of 2 cohorts not used in any other experiment. (C) DNA from control and ko BM chimeric mice from panel A was isolated from PB 7 days after the first pIpC injection and tested quantitatively for the presence of the NF-Ya ko allele (NF-Ya floxed allele after cre recombination; n ≥ 4); **P < .01.
NF-Ya–deficient BM chimeric mice die with severely reduced complete blood counts. (A) PB from one cohort of control (gray) and ko (black) BM chimeric mice 1, 3, 8, 11, 22, and 50 days after the first pIpC injection was isolated and counted (n ≥ 4 for every time point). (B) Survival curve of control (gray) and ko BM chimeric mice (black) of 2 cohorts not used in any other experiment. (C) DNA from control and ko BM chimeric mice from panel A was isolated from PB 7 days after the first pIpC injection and tested quantitatively for the presence of the NF-Ya ko allele (NF-Ya floxed allele after cre recombination; n ≥ 4); **P < .01.
Genomic DNA from the peripheral blood (PB) of control and ko BM chimeras was isolated 7 days after the first pIpC injection, and 77% ± 7% NF-Ya deletion was observed (Figure 2C). The presence of NF-Ya null PB cells indicates that mature blood cells of the periphery do not depend on NF-Y activity for survival. To confirm that NF-Y inactivation results in BM failure, we analyzed total BM cellularity. The amount of dead cells, determined by propidium iodide (PI) staining was increased in the BM of ko versus control mice, starting as early as 1 day after NF-Ya deletion (Figure 3B). The percentage of live cells in the BM of ko mice (characterized by forward scatter–side scatter and PI negativity) decreased progressively from 70% ± 16% at 24 hours to 51% ± 15% at 60 hours and to 26% ± 18% at 96 hours after a single pIpC treatment compared with control mice (Figure 3A). With BM cellularity decreasing so rapidly, we postulated that the BM was probably acellular at the time of death. H&E stains of femur bones isolated 10 days after pIpC confirmed the severe decline in ko BM cellularity (Figure 3C). These results show that NF-Ya–deficient BM chimeric mice die because of BM failure and imply that NF-Ya is important for cycling BM cells but dispensable for the survival of noncycling PB cells.
BM cellularity decreases after pIpC-mediated NF-Ya deletion in the BM because of cell death and apoptosis. (A) Four days after pIpC treatment control and ko BM cells were isolated. After erythrocyte lysis cells were resuspended in equal volumes for analysis by FACS for equal amounts of time. Representative dot plots are shown. The percentage of cells within the gate is shown on the upper right corner of each plot, and the total amount of cells counted in equal periods of time is shown in the lower right. (B) BM cells from panel A were stained with PI and analyzed by FACS. One representative histogram overlay is shown (left; control [gray], ko [black]). On the basis of this analysis the scatter plot (right) shows the relative amount of dead cells for control and ko samples at indicated time points after pIpC treatment. Single data points and the averages, including SDs, are shown (n ≥ 3); *P < .05 and **P < .01. (C) Ten days after ko induction femur bones of control and ko BM chimera were fixed in formalin for 48 hours, decalcified in 0.5M EDTA for 5 days, paraffin embedded, sectioned, and H&E stained. The images show representative control and ko femur bones at a magnification of ×200, Nikon Eclipse 80i, Qimage Micropublisher 5.0 camera and software, 20×/0.50 NA objective. (D) Twenty-four hours after pIpC treatment BM cells were isolated from control and ko mice, erythrocytes were lysed, cells were stained with PI and annexin V and analyzed by FACS. Annexin V staining was determined among PI-negative cells (left). The histogram overlay (middle) shows a representative annexin V staining of live cells of control (gray) and ko (black). Results of 4 independent experiments are shown in the scatter plot (right). Single data points and the averages, including SDs, are shown (n = 4); **P < .01. (E) Twenty-four hours after pIpC treatment BM cells were isolated from control and ko mice, stained with DAPI, and analyzed by FACS. The histogram overlay shows a representative control and ko sample of the cell cycle analysis according to DNA content. The bar graph (right) shows the relative amount of cells in the respective phase of the cell cycle 24 hours after pIpC-induced NF-Ya deletion (n = 4); **P < .01.
BM cellularity decreases after pIpC-mediated NF-Ya deletion in the BM because of cell death and apoptosis. (A) Four days after pIpC treatment control and ko BM cells were isolated. After erythrocyte lysis cells were resuspended in equal volumes for analysis by FACS for equal amounts of time. Representative dot plots are shown. The percentage of cells within the gate is shown on the upper right corner of each plot, and the total amount of cells counted in equal periods of time is shown in the lower right. (B) BM cells from panel A were stained with PI and analyzed by FACS. One representative histogram overlay is shown (left; control [gray], ko [black]). On the basis of this analysis the scatter plot (right) shows the relative amount of dead cells for control and ko samples at indicated time points after pIpC treatment. Single data points and the averages, including SDs, are shown (n ≥ 3); *P < .05 and **P < .01. (C) Ten days after ko induction femur bones of control and ko BM chimera were fixed in formalin for 48 hours, decalcified in 0.5M EDTA for 5 days, paraffin embedded, sectioned, and H&E stained. The images show representative control and ko femur bones at a magnification of ×200, Nikon Eclipse 80i, Qimage Micropublisher 5.0 camera and software, 20×/0.50 NA objective. (D) Twenty-four hours after pIpC treatment BM cells were isolated from control and ko mice, erythrocytes were lysed, cells were stained with PI and annexin V and analyzed by FACS. Annexin V staining was determined among PI-negative cells (left). The histogram overlay (middle) shows a representative annexin V staining of live cells of control (gray) and ko (black). Results of 4 independent experiments are shown in the scatter plot (right). Single data points and the averages, including SDs, are shown (n = 4); **P < .01. (E) Twenty-four hours after pIpC treatment BM cells were isolated from control and ko mice, stained with DAPI, and analyzed by FACS. The histogram overlay shows a representative control and ko sample of the cell cycle analysis according to DNA content. The bar graph (right) shows the relative amount of cells in the respective phase of the cell cycle 24 hours after pIpC-induced NF-Ya deletion (n = 4); **P < .01.
NF-Ya–deficient BM cells undergo apoptosis and accumulate in the G2/M phase of the cell cycle
Loss of BM cellularity after NF-Ya deletion could theoretically be caused by induction of apoptosis or deceleration in cell cycling. Evaluating cells by annexin V staining, we noted that NF-Ya deficiency increased the fraction of annexin V+ cells ∼ 1.5-fold 24 hours after the induction of NF-Ya deletion (Figure 3D). It has been suggested from in vitro studies that NF-Y is involved in the progression through the cell cycle; however, one study reported that cells lacking functional NF-Y accumulate in the G2/M phase of the cell cycle,20 whereas another reported the opposite.19 In the only in vivo study, NF-Ya overexpression did not result in aberrations of the cell cycle.26 To distinguish whether the BM failure is caused solely by apoptotic cell death or additionally by reduced proliferation of progenitor cells in the BM, and to test whether NF-Y is involved in the progression of the cell cycle in our system, we used flow cytometry to measure the cell cycle of BM cells 24 hours after NF-Ya deletion. In these experiments, we found a significant increase of ko cells in the G2/M phase compared with control cells (Figure 3E) as well as a slight increase in the percentage of cells found in the S phase. We conclude that NF-Ya–deficient BM cells have a block at the G2/M stage of the cell cycle. Thus, NF-Y inactivation leads to a rapid loss in BM cellularity by a combination of apoptotic cell death and cell cycle arrest in G2/M phase.
HSCs deficient for NF-Ya fail to compete with wt HSCs to replenish hematopoietic cells in the BM
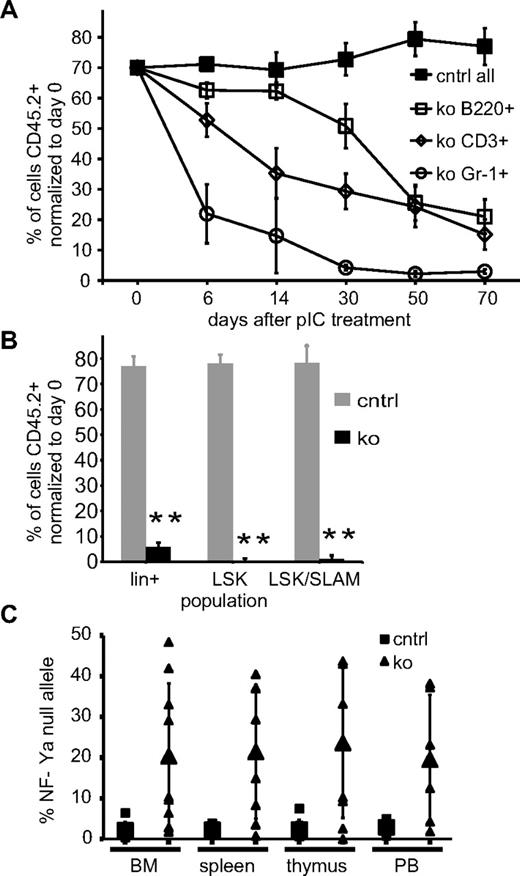
We directly tested the function of NF-Y–deficient HSCs in the context of competitive BMT by transplanting 70% NF-Ya mt BM (CD45.2+) cells together with 30% NF-Ya wt competitor cells (CD45.1+) into wt recipient mice. Eight weeks after BMT, we induced NF-Ya deletion by 3 intraperitoneal pIpC injections. Six, 14, 30, 50, and 70 days later, PB was isolated from these mice, and the ratio of donor to competitor cells was determined among B220+, CD3+, and Gr-1+ subpopulations by flow cytometry (Figure 4A). Recipients of NF-Ya+/++Mx-cre BM served as controls. In these mice, we found the CD45.1-to-CD45.2 ratio remained unchanged over the 70-day period. In BM chimera receiving ∼ 70% NF-Yafl/fl+Mx-cre BM with ∼ 30% wt BM, the percentage of lymphoid and myeloid cells derived from the NF-Ya mt compartment, labeled with the markers Gr-1, B220, and CD3, respectively, and CD45.2, declined over time with the fastest decline in Gr-1+ cells. For both myeloid and lymphoid cells, the wt compartment had taken over the blood system entirely 70 days after NF-Ya deletion. At the same time point, we observed that ko BM cells within lin+, LSK, and LSK/SLAM (signaling lymphocyte activation molecule) populations, were devoid of NF-Ya ko (CD45.2+) cells (Figure 4B). The lack of long-term sustenance of NF-Ya–deficient hematopoiesis in all lineages combined with the complete absence of any ko HSCs indicates a significant defect in HSC function in the absence of NF-Ya in a competitive setting.
HSCs cannot support hematopoiesis in the absence of NF-Ya and various genes implicated in HSCs behavior are dysregulated in HSCs. (A) CD45.1+ recipients received a BM transplant from NF-Ya+/++Mx-cre (control) and NF-Yafl/fl+Mx-cre (ko, all CD45.2+) mice together with 30% CD45.1+ NF-Ya wt competitor cells. The mice were treated with pIpC at day 0. The CD45.1-to-CD45.2 ratio of Gr-1+, B220+, and CD3+ PB-cell populations was determined and normalized to day 0 (n = 5 for all data points except for ko samples day 30, 50, and 70; n = 4). (B) Mice from panel A were killed, and the indicated BM subpopulations were analyzed for CD45.1 versus CD45.2 expression and are shown in a bar graph, control (gray) and ko (black). (C) DNA from BM, spleen, thymus, and PB of NF-Ya+/++Mx cre (control) and NF-Yafl/fl+Mx cre (ko) mutant BM chimeras 10 weeks after pIpC treatment was subjected to quantitative PCR analysis that measured the amount of NF-Ya–null allele. Single data points and averages, including SDs, are shown.
HSCs cannot support hematopoiesis in the absence of NF-Ya and various genes implicated in HSCs behavior are dysregulated in HSCs. (A) CD45.1+ recipients received a BM transplant from NF-Ya+/++Mx-cre (control) and NF-Yafl/fl+Mx-cre (ko, all CD45.2+) mice together with 30% CD45.1+ NF-Ya wt competitor cells. The mice were treated with pIpC at day 0. The CD45.1-to-CD45.2 ratio of Gr-1+, B220+, and CD3+ PB-cell populations was determined and normalized to day 0 (n = 5 for all data points except for ko samples day 30, 50, and 70; n = 4). (B) Mice from panel A were killed, and the indicated BM subpopulations were analyzed for CD45.1 versus CD45.2 expression and are shown in a bar graph, control (gray) and ko (black). (C) DNA from BM, spleen, thymus, and PB of NF-Ya+/++Mx cre (control) and NF-Yafl/fl+Mx cre (ko) mutant BM chimeras 10 weeks after pIpC treatment was subjected to quantitative PCR analysis that measured the amount of NF-Ya–null allele. Single data points and averages, including SDs, are shown.
NF-Ya–deficient HSCs cannot sustain hematopoiesis
To test whether NF-Ya–deficient HSCs might be functional but have reduced proliferative capacity versus normal HSCs, we performed noncompetitive BMT with NF-Ya−/− BM as a sole source of rescue. Under these circumstances, similar to Figure 2, we predicted that either all mice would die or that some fraction would survive, solely on the basis of recovery of residual undeleted donor BM cells after NF-Ya deletion.
Eight weeks after BMT, we found that > 98% of blood cells were donor derived (CD45.2+). Deletion was induced by pIpC, and 70 days later we analyzed ≥ 3 mice per genotype (control, ko) of 3 different transplantations. The initial analysis indicated that the number and lineage composition of cells in the BM, spleen, thymus, and PB were indistinguishable between ko and control BM chimera (supplemental Figure 2). However, quantification of NF-Ya-null allele by quantitative PCR for each sample, as shown in the scatter plot (Figure 4C), showed that NF-Ya was deleted in ≤ 50% of the NF-Ya–floxed alleles. This suggests that the BM cells in the surviving mice might have proliferated from those rare HSCs in which NF-Ya was not fully deleted after pIpC. Indeed, NF-Ya deletion in het (NF-Yafl/++Mx-cre) BM chimeric mice after pIpC was highly efficient (supplemental Figure 3). Taken together, these results suggest that NF-Ya–deficient HSC cannot support hematopoiesis and that only cells with ≥ 1 intact NF-Ya allele provide hematopoietic support.
To definitely exclude the possibility of any functional NF-Ya–null HSC/HPCs cells being present after pIpC, we subjected total BM cells from control and ko mice 70 days after pIpC treatment to methylcellulose assays that support colony formation from pre-B cells, myeloid, and erythroid progenitors. The resulting colonies were picked and genotyped by PCR (Table 1). All colonies obtained from wt BM carried 2 NF-Ya wt alleles, as expected. Eight of 235 colonies from ko BM chimeric mice were found to have 2 NF-Ya wt alleles, indicating that these colonies were derived from BM cells that survived irradiation. Strikingly, in the ko samples, not a single colony of any lineage could be found carrying 2 NF-Ya null alleles, despite the high recombination efficiency of the NF-Ya–floxed allele in this experimental system shown in het mice (supplemental Table 1). Together, these results indicate that NF-Y activity is necessary for HSC/HPC function.
Genotyping of pre-B, GM, and CFUe colonies by PCR shows that no NF-Ya−/− colonies were obtained
| . | +/+ +Mx cre . | fl/fl +Mx cre . | |||||
|---|---|---|---|---|---|---|---|
| +/+ . | fl/+ . | −/− . | +/+ . | fl/fl . | fl/− . | −/− . | |
| Pre-B | 65 | 0 | 0 | 2 | 36 | 29 | 0 |
| GM | 87 | 0 | 0 | 5 | 52 | 47 | 0 |
| CFUe | 57 | 0 | 0 | 1 | 42 | 21 | 0 |
| . | +/+ +Mx cre . | fl/fl +Mx cre . | |||||
|---|---|---|---|---|---|---|---|
| +/+ . | fl/+ . | −/− . | +/+ . | fl/fl . | fl/− . | −/− . | |
| Pre-B | 65 | 0 | 0 | 2 | 36 | 29 | 0 |
| GM | 87 | 0 | 0 | 5 | 52 | 47 | 0 |
| CFUe | 57 | 0 | 0 | 1 | 42 | 21 | 0 |
B cell, myeloid, and erythroid precursors of BM cells of NF-Ya+/++Mx cre (control) and NF-Yafl/fl+Mx cre (ko) BM chimera were grown in methylcellulose. The resulting colonies were randomly picked and genotyped.
Quiescence protects HSCs from defects because of loss of NF-Y
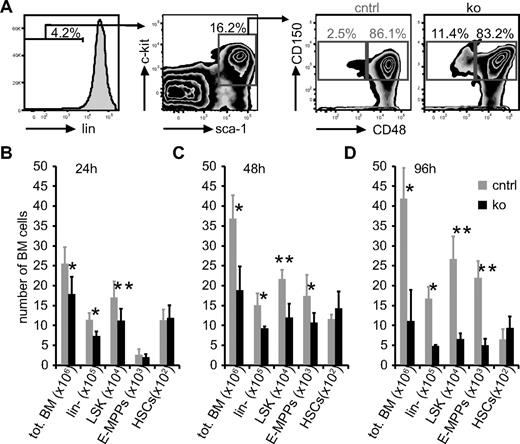
To assess the acute effects of loss of NF-Ya, we quantified BM subpopulations 24, 60, and 96 hours after a single pIpC treatment. Although we observed a frequency of HSCs similar to previously published reports (Figure 5A),31 we found that HSCs recovered after pIpC treatment were less quiescent. This change in cell cycle status was accompanied by up-regulation of Sca-1, confirming the results of recent publications studying the effects of IFNs on HSCs.32,33 In addition, we tested whether NF-Y regulates the expression of CD34 on HSCs, because human CD34 was shown to be induced by NF-Y in vitro.34 We found that 66% ± 10% of control HSCs (LSK, CD48−, CD150+) were positive for CD34. However, within 24 hours after NF-Ya ko induction, only 47% ± 6% of the HSCs were CD34+ (supplemental Figure 4). These data show that murine CD34 expression is regulated by NF-Y in vivo. Thus, we did not use CD34 to characterize HSCs in subsequent experiments.
After pIpC-mediated NF-Ya deletion total BM cells of ko samples decrease in numbers, and HSC numbers remain unchanged. (A) BM cells were collected 96 hours after pIpC treatment; stained with lineage Abs, sca-1, c-kit, and CD48, CD150 (SLAM markers); and analyzed by FACS. The gating strategy to enumerate LSK, CD48+, CD150+ cells (E-MPPs) and LSK, CD48−, CD150+ (HSCs) is shown. Note, ko samples contain relatively more HSCs. Twenty-four (B), 60 (C), and 96 (D) hours after pIpC treatment BM cells were isolated from control and ko mice, and absolute cell numbers of different populations were enumerated as shown in panel A. Absolute cell numbers as indicated are shown (n = 5 in B and C; n = 3 in D); *P < .05 and **P < .01).
After pIpC-mediated NF-Ya deletion total BM cells of ko samples decrease in numbers, and HSC numbers remain unchanged. (A) BM cells were collected 96 hours after pIpC treatment; stained with lineage Abs, sca-1, c-kit, and CD48, CD150 (SLAM markers); and analyzed by FACS. The gating strategy to enumerate LSK, CD48+, CD150+ cells (E-MPPs) and LSK, CD48−, CD150+ (HSCs) is shown. Note, ko samples contain relatively more HSCs. Twenty-four (B), 60 (C), and 96 (D) hours after pIpC treatment BM cells were isolated from control and ko mice, and absolute cell numbers of different populations were enumerated as shown in panel A. Absolute cell numbers as indicated are shown (n = 5 in B and C; n = 3 in D); *P < .05 and **P < .01).
The quantification of BM cell subsets showed that in ko mice, lin− and LSK cells were decreased in number compared with controls, similar to the total BM cellularity 24 (Figure 5B) and 60 (Figure 5C) hours after ko induction. Interestingly, early multipotent progenitors (E-MPPs; LSK, CD48+, CD150+) and HSCs (LSK, CD48−, CD150+) numbers appeared to be unaffected by the deletion of NF-Ya at the 24-hour time point. Sixty and 96 hours after induction of NF-Ya deletion (Figure 5C-D) we additionally detected fewer E-MPPs in ko compared with control mice, whereas the number of HSCs still remained unaffected by NF-Ya ablation.
These results imply that quiescent HSCs do not depend on NF-Y activity for their acute survival, whereas more mature BM progenitors and precursors do require NF-Y activity. Because BM cell numbers decline because of a combination of apoptosis and a cell cycle defect, our results suggest that the quiescent state protects HSCs from NF-Y–mediated defects. Alternatively, it is also conceivable that apoptosing HSCs are replaced by symmetric HSC division, masking the loss of HSCs.
To clarify whether NF-Ya–deficient HSCs can progress through the cell cycle and whether NF-Ya deletion leads to apoptosis of HSCs, we exploited the effect of pIpC to induce cycling in quiescent HSCs.32,33 With the use of intracellular Ki67 staining in combination with a DAPI staining of DNA, we found that 24 hours after a single pIpC treatment only 24% ± 5% of control HSCs remained in G0, in agreement with recent reports.32 In NF-Ya ko HSCs, 19% ± 1.4% remained in G0, indicating that NF-Ya–deficient HSCs are activated at least as efficiently as control HSCs (P = .09; Figure 6A). Twenty-four hours after pIpC treatment, at the same time that we observed accumulation of total ko BM cells in G2/M, ko HSC cycle distribution was unaltered (Figure 6A). The same analysis 60 and 96 hours after a single pIpC treatment, however, indicated that NF-Ya–deficient HSCs also accumulate in G2/M. At those later time points, more control HSCs started to return to G0 compared with ko HSCs, possibly indicating that ko HSCs respond to the loss of overall BM cellularity by remaining in cycle.
HSCs accumulate 60 and 96 hours but not 24 hours after pIpC-mediated NF-Ya deletion in the G2/M phase of the cell cycle and present with increased apoptosis. (A left) A representative Ki67 versus DAPI staining of control and ko LSK/SLAM HSCs is shown 60 hours after pIpC treatment. (Right) Quantitative evaluation of the cell cycle analysis is based on DAPI and Ki67 stainings of viable LSK/SLAM HSCs 24, 60, and 96 hours after pIpC treatment (control [gray], n = 5; ko [black], n = 4; *P < .05; G0 24 hours, P = .09; G0 60 hours, P = .06; G0 96 hours, P = .06). (B left) A representative FACS plot of LSK/SLAM HSCs stained for annexin V and 7AAD is shown for control and ko. (Right) This bar graph shows the percentage of apoptotic (annexin V positive, 7AAD negative) and dead HSCs (annexin V, 7AAD double positive) for control (gray) and ko (black) samples 96 hours after pIpC treatment (n = 4); *P < .05.
HSCs accumulate 60 and 96 hours but not 24 hours after pIpC-mediated NF-Ya deletion in the G2/M phase of the cell cycle and present with increased apoptosis. (A left) A representative Ki67 versus DAPI staining of control and ko LSK/SLAM HSCs is shown 60 hours after pIpC treatment. (Right) Quantitative evaluation of the cell cycle analysis is based on DAPI and Ki67 stainings of viable LSK/SLAM HSCs 24, 60, and 96 hours after pIpC treatment (control [gray], n = 5; ko [black], n = 4; *P < .05; G0 24 hours, P = .09; G0 60 hours, P = .06; G0 96 hours, P = .06). (B left) A representative FACS plot of LSK/SLAM HSCs stained for annexin V and 7AAD is shown for control and ko. (Right) This bar graph shows the percentage of apoptotic (annexin V positive, 7AAD negative) and dead HSCs (annexin V, 7AAD double positive) for control (gray) and ko (black) samples 96 hours after pIpC treatment (n = 4); *P < .05.
In addition, we tested whether HSCs undergo apoptosis as do total BM cells. We found a significant increase of annexin V+, 7-amino-actinomycin D (7AAD)–negative HSCs from 9.5% ± 3.6% in control compared with 15.5% ± 3.5% in ko samples 96 hours after NF-Ya deletion. This was accompanied by increased numbers of dead HSCs (Figure 6B). Taken together, we conclude that NF-Y activity is dispensable for the survival of quiescent HSCs, but on activation, NF-Ya–deficient HSCs are arrested in the G2/M phase of the cell cycle and undergo apoptosis.
Finally, we controlled possible IFN-mediated side effects caused by pIpC treatment by deleting NF-Ya in BM cells in vitro. BM cells present with a cell cycle arrest and apoptotic cell death phenotype 24 and 48 hours after NF-Ya deletion by retroviral cre delivery (supplemental Figure 5). These results indicate unequivocally that the described phenotypes are effects of the NF-Ya deletion under physiologic conditions.
HSC depletion is p53 independent
NF-Ya target CCAAT boxes are present in 30% of eukaryotic gene promoters.16,17 To explore potentially relevant mechanistic pathways of NF-Y function, we used data from previous reports to focus on probable candidate molecular pathways underlying the observed defects. Cyclin b1 plays a central role in the G2/M transition of the cell cycle and has also been implicated in apoptosis.33 In addition, the cyclin b1 promoter has 2 CCAAT boxes and has been reported to be regulated by NF-Y.20,23 We used quantitative RT-PCR analysis from RNA isolated from total BM cells 1 and 3 days after pIpC treatment and from sorted HSCs to measure gene expression. We observed a reduction to 48% ± 6.4% of cyclin b1 expression in BM cells from NF-Ya ko mice compared with controls, corresponding well with the observed cell cycle arrest in the G2/M phase (Table 2). Likewise, the cyclin-dependent kinase inhibitor, Cdkn1α (p21), is involved in cell cycle arrest and can inhibit apoptosis.35 We found expression levels reduced 1 day after ko induction in HSCs and reduced to ∼ 30% of control levels 3 days after ko induction in ko BM cells, indicating that this gene may be directly regulated by NF-Y.
Expression levels of cell cycle– and apoptosis-related genes are dysregulated in NF-Ya ko BM cells
| . | Day 1 HSCs . | Day 1 total BM . | Day 3 total BM . | |||
|---|---|---|---|---|---|---|
| Average ± SD . | P . | Average ± SD . | P . | Average ± SD . | P . | |
| Ccnb1 | 92.6 ± 28.4 | .29 | 92.3 ± 10.5 | .08 | 48.3 ± 6.4 | < .01 |
| Cdkn1α | 63.5 ± 17.4 | < .01 | 40.0 ± 34.0 | < .01 | 29.3 ± 20.0 | < .01 |
| Bax | 55.3 ± 14.2 | < .01 | 41.6 ± 29.1 | < .01 | 66.3 ± 17.1 | .04 |
| Bcl-2 | 62.8 ± 25.9 | .02 | 54.7 ± 28.6 | .05 | 22.5 ± 5.9 | < .01 |
| Bax/Bcl-2 | 0.9 ± 0.6 | 0.8 ± 0.8 | 3.0 ± 1.5 | |||
| . | Day 1 HSCs . | Day 1 total BM . | Day 3 total BM . | |||
|---|---|---|---|---|---|---|
| Average ± SD . | P . | Average ± SD . | P . | Average ± SD . | P . | |
| Ccnb1 | 92.6 ± 28.4 | .29 | 92.3 ± 10.5 | .08 | 48.3 ± 6.4 | < .01 |
| Cdkn1α | 63.5 ± 17.4 | < .01 | 40.0 ± 34.0 | < .01 | 29.3 ± 20.0 | < .01 |
| Bax | 55.3 ± 14.2 | < .01 | 41.6 ± 29.1 | < .01 | 66.3 ± 17.1 | .04 |
| Bcl-2 | 62.8 ± 25.9 | .02 | 54.7 ± 28.6 | .05 | 22.5 ± 5.9 | < .01 |
| Bax/Bcl-2 | 0.9 ± 0.6 | 0.8 ± 0.8 | 3.0 ± 1.5 | |||
RNA was isolated from BM cells 1 or 3 days after pIpC treatment. Equal amounts of RNA were reverse transcribed and subsequently subjected to quantitative PCR. The table shows average ± SD expression levels of the indicated genes in ko samples in percentage of control samples (n ≥ 4).
Because we previously showed that NF-Y interacts biochemically with inverted CCAAT motifs (Y-box), and, through its protein-protein interactions with USF-1/2, CACGCG motifs (E-box),18 we also looked for relevant genes harboring these sequences and found a Y-box and multiple E-boxes in the promoter region of the proapoptotic gene Bax. Expression levels were reduced significantly in BM cells and in HSCs, making it a candidate for a direct target of NF-Y. Thus, we aimed to determine the ratio of Bax and Bcl-2 expression as parameters stimulating/inhibiting apoptosis, similar to Benatti et al.19 With Bcl-2 expression modestly decreased after 1 day and reduced to 22.5% ± 5.0% 3 days after the NF-Ya deletion, the ratio of expression of these proapoptotic/antiapoptotic genes increased on day 3 by ∼ 3-fold (Table 2).
It is well established that p53 plays a key role in the transcriptional regulation of apoptosis [in part through Bax and Cdkn1α (p21)]. In addition, it has been shown in human colon carcinoma cells that the balance of NF-Y and p53 is critical to maintain transcription of antiapoptotic genes and that p53 inactivation rescued apoptosis in NF-Yb knockdown cells.19 Furthermore, experiments that used gain-of-function p53 mutants have shown that p53/NF-Y complexes play an important role in transcriptional cell cycle regulation.36 Therefore, we tested whether the apoptotic response in NF-Ya–deficient hematopoietic cells depends on p53 by generating inducible NF-Ya, p53 double ko (dko) mice. We bred mice carrying the conditional p53 ko allele37 with our ko mice to obtain NF-Yafl/fl, p53fl/fl+Mx-cre (dko) mice and confirmed p53 deletion in p53 mutant mice after pIpC treatment with the use of Western blot analysis (Figure 7A). Two days after a single pIpC injection, we analyzed BM cells and found 3.6% ± 1% in control versus 12.2% ± 3.8% and 11.5% ± 4.5% apoptotic BM cells in ko and dko mice, respectively, with the use of a TUNEL assay (Figure 7B-C). In addition, in competitive BM chimeric mice transplanted with 30% wt BM (CD45.1+) and 70% NF-Yafl/fl, p53fl/fl+Mx-cre (CD45.2+) the mutant compartment declined after pIpC treatment over time in the PB similar to the NF-Ya (single) ko transplants (Figure 7D). The fact that after 70 days the lin+, LSK, and LSK/SLAM populations in the BM were comprised exclusively of CD45.1+ (wt) cells indicates that NF-Ya, p53 dko hematopoietic cells, including HSCs, failed to sustain long-term hematopoiesis (Figure 7E). These data indicate that the induction of apoptosis and the failure of NF-Ya–deficient hematopoietic cells to maintain hematopoiesis long term are independent of p53. Rather, the defects observed in NF-Ya–deficient BM cells may be related to the observed down-regulation of Cyclin b1, p21, and Bcl-2. The increased ratio of Bax/Bcl-2 gene expression 3 days after NF-Ya deletion particularly represents a shift in the genetic program promoting apoptosis in NF-Ya–deficient BM cells.
Deletion of p53 in addition to NF-Ya does not rescue the defects of NF-Ya–deficient BM cells and HSCs. (A) Protein from wt and p53 conditional ko BM 7 days after pIpC treatment was subjected to Western blot analysis and probed with a p53 (top) and actin (bottom) Ab. (B) Representative flow cytometric staining of control (left) and NF-Ya ko (right) for TUNEL and PI. Note, NF-Ya sko and NF-Ya, p53 dko (not shown) stainings are indistinguishable. (C) Results from panel B are summarized in this bar graph (n = 5); **P < .01. (D) CD45.1+ recipients received a BM transplant from NF-Ya+/++Mx-cre (control), NF-Yafl/fl+Mx-cre (ko), and NF-Yafl/fl, p53fl/fl+Mx-cre (dko), (all CD45.2+) mice together with 30% CD45.1+ NF-Ya wt competitor cells. The mice were treated with pIpC at day 0. The CD45.1-to-CD45.2 ratio of PB cells was determined and normalized to day 0 (n = 8 [control], n = 7 [sko], n = 6 [(dko]). (E) Mice from panel D were killed, and the indicated BM subpopulations were analyzed for CD45.1 versus CD45.2 expression and are shown in a bar graph; control (gray) and ko (black).
Deletion of p53 in addition to NF-Ya does not rescue the defects of NF-Ya–deficient BM cells and HSCs. (A) Protein from wt and p53 conditional ko BM 7 days after pIpC treatment was subjected to Western blot analysis and probed with a p53 (top) and actin (bottom) Ab. (B) Representative flow cytometric staining of control (left) and NF-Ya ko (right) for TUNEL and PI. Note, NF-Ya sko and NF-Ya, p53 dko (not shown) stainings are indistinguishable. (C) Results from panel B are summarized in this bar graph (n = 5); **P < .01. (D) CD45.1+ recipients received a BM transplant from NF-Ya+/++Mx-cre (control), NF-Yafl/fl+Mx-cre (ko), and NF-Yafl/fl, p53fl/fl+Mx-cre (dko), (all CD45.2+) mice together with 30% CD45.1+ NF-Ya wt competitor cells. The mice were treated with pIpC at day 0. The CD45.1-to-CD45.2 ratio of PB cells was determined and normalized to day 0 (n = 8 [control], n = 7 [sko], n = 6 [(dko]). (E) Mice from panel D were killed, and the indicated BM subpopulations were analyzed for CD45.1 versus CD45.2 expression and are shown in a bar graph; control (gray) and ko (black).
Discussion
We demonstrate here that NF-Y is dispensable for noncycling quiescent HSCs and mature blood cells. However, once HSCs are activated into the cell cycle, they depend on NF-Y activity similar to other cycling cells in the BM.
Our data indicate that HoxB4, Lef-1, Notch1, and Bmi-1 are regulated by basal NF-Ya expression in vivo. The fact that these genes are all involved in HSC biology with deletion or overexpression models1-8,38 that result in much milder phenotypes highlights the key role of NF-Y as an upstream regulator of HSC behavior.
Although the changes in expression of these genes, together with previous studies in other settings, might suggest that NF-Ya deletion would promote differentiation over self-renewal, we did not observe that phenotype. Rather, we observed that NF-Ya deletion results in initially stable HSC numbers and a sharp decline of more differentiated cells. A possible explanation for the HSC phenotype is that HSCs differentiate after NF-Ya deletion, and those cells then commit apoptosis, hiding an increase in HPC numbers. In this scenario, the differentiating HSCs would need to be replaced by symmetric HSC division for the HSC number to remain unaltered. We tested this in great detail and found that HSCs, once activated, cannot complete the cell cycle, excluding this possibility. Alternatively, it cannot be ruled out that NF-Y inactivation drives HSCs toward differentiation, which would thus lead to the exhaustion of the HSC pool over a longer term. The apoptosis and cell cycle phenotype of BM cells as well as of activated HSC makes it difficult for us to address this point in more detail. It is possible to focus on quiescent HSCs and screen their gene expression profile for transcriptional signatures promoting differentiation over self-renewal. However, in this ko system, the detrimental apoptosis and cell cycle phenotypes obstruct any functional readout, which is critical for the distinction of HSC versus HPC populations. Therefore, we speculate that there may be a threshold of NF-Ya expression that allows survival and cycling of HSCs while exposing differences in self-renewal versus differentiation decisions. This could be investigated with a NF-Ya knockdown approach. Alternatively, it is possible that leukemic cells differ from normal BM cells in their requirement for NF-Y activity and would present a suitable model to investigate this question.
Of note, our findings of an accumulation of cells in the G2/M phase of the cell cycle, although in agreement with some previous data,20 differ from those of Benatti et al19 who reported a decrease of cells in G2/M and a role for p53 inactivation in HCT116 human colon carcinoma cells. These differences may indicate that NF-Y function may be cell type specific.
Our data argue against, but do not entirely exclude, a role for p53. For example, the competitive BMT experiment in Figure 7D showed a lesser effect on PB cells for the dko compared with NF-Ya single ko BM chimeras 7 days after pIpC treatment, suggesting a possible mitigating effect of p53. However, this difference was below the level of statistical significance and was not sustained; thus, any effect of p53 appears to be modest and insufficient to rescue the crisis induced by loss of NF-Ya.
In summary, we have shown that NF-Ya deletion in vivo causes the aberrant expression of multiple genes known to be important in HSC regulation and results in lethal disruption of cell cycle progression in a manner that is not dependent on p53. NF-Y is thus a key regulatory transcription factor whose activity, mediated through its multiple downstream developmentally regulated and cell cycle–specific targets, plays a critical role in the proliferation of normal HSCs. The role of NF-Y therefore extends beyond its potential as an HSC activator via overexpression to a fundamental integrator of molecular pathways that link cell cycle progression, differentiation, and survival.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Sankar Maity and Anuradha Bhattarcharya for the NF-Ya conditional knockout mice and Martin Carroll for his generous support.
This work was supported by the National Institute of Health (grant 5R01CA090833, S.G.E.) and grants from the Deutsche Forschungs-Gemeinschaft and the Leukemia & Lymphoma Society (G.B.).
National Institutes of Health
Authorship
Contribution: G.B. designed, conducted, and evaluated most of the experiments and wrote the manuscript; H.L. performed and evaluated numerous qRT-PCR experiments; S.G.E. designed the study, supervised experiments, and edited the manuscript; and D.T.S. supervised experiments and edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Stephen G. Emerson, Haverford College, 370 Lancaster Ave, Haverford, PA 19041; e-mail: semerson@haverford.edu; or David T. Scadden, Center for Regenerative Medicine, MA General Hospital and Harvard Medical School CPZN, Boston, MA 02114; e-mail: scadden.david@MGH.Harvard.edu.

![Figure 3. BM cellularity decreases after pIpC-mediated NF-Ya deletion in the BM because of cell death and apoptosis. (A) Four days after pIpC treatment control and ko BM cells were isolated. After erythrocyte lysis cells were resuspended in equal volumes for analysis by FACS for equal amounts of time. Representative dot plots are shown. The percentage of cells within the gate is shown on the upper right corner of each plot, and the total amount of cells counted in equal periods of time is shown in the lower right. (B) BM cells from panel A were stained with PI and analyzed by FACS. One representative histogram overlay is shown (left; control [gray], ko [black]). On the basis of this analysis the scatter plot (right) shows the relative amount of dead cells for control and ko samples at indicated time points after pIpC treatment. Single data points and the averages, including SDs, are shown (n ≥ 3); *P < .05 and **P < .01. (C) Ten days after ko induction femur bones of control and ko BM chimera were fixed in formalin for 48 hours, decalcified in 0.5M EDTA for 5 days, paraffin embedded, sectioned, and H&E stained. The images show representative control and ko femur bones at a magnification of ×200, Nikon Eclipse 80i, Qimage Micropublisher 5.0 camera and software, 20×/0.50 NA objective. (D) Twenty-four hours after pIpC treatment BM cells were isolated from control and ko mice, erythrocytes were lysed, cells were stained with PI and annexin V and analyzed by FACS. Annexin V staining was determined among PI-negative cells (left). The histogram overlay (middle) shows a representative annexin V staining of live cells of control (gray) and ko (black). Results of 4 independent experiments are shown in the scatter plot (right). Single data points and the averages, including SDs, are shown (n = 4); **P < .01. (E) Twenty-four hours after pIpC treatment BM cells were isolated from control and ko mice, stained with DAPI, and analyzed by FACS. The histogram overlay shows a representative control and ko sample of the cell cycle analysis according to DNA content. The bar graph (right) shows the relative amount of cells in the respective phase of the cell cycle 24 hours after pIpC-induced NF-Ya deletion (n = 4); **P < .01.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/119/6/10.1182_blood-2011-06-359406/5/m_zh89991184080003.jpeg?Expires=1767718990&Signature=c7H-bIEE5XhIB6wvlr1OjsCelEYAfRcVFJuatA33VccxgGjUmyO0MMoniw5tLtYQup9VGSi6geXmKqPSYqTPV43z7gzKw9vG12Lb6xAYLa05725h1STN1pugpVC597JDqw3QHCfDPcf4QzlllqMjGBzKaqfajX-sJohMlBStuDHJlak-GgjyhUaXmSSIGdu8lwbNSyFbaKIdRtarJuS-hiNeXhqkR6ezhGDZOCmQcTv~S9R51pBuxhvaUHBZNnbfaxRBGUgamLVMF8oDwfrKVDG~hQg0TmTjQi8E8Wj-s~OBdsN08i1OChQVAGfoYMIGe8nGgvgxAYA5NvnvL5q0eg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 6. HSCs accumulate 60 and 96 hours but not 24 hours after pIpC-mediated NF-Ya deletion in the G2/M phase of the cell cycle and present with increased apoptosis. (A left) A representative Ki67 versus DAPI staining of control and ko LSK/SLAM HSCs is shown 60 hours after pIpC treatment. (Right) Quantitative evaluation of the cell cycle analysis is based on DAPI and Ki67 stainings of viable LSK/SLAM HSCs 24, 60, and 96 hours after pIpC treatment (control [gray], n = 5; ko [black], n = 4; *P < .05; G0 24 hours, P = .09; G0 60 hours, P = .06; G0 96 hours, P = .06). (B left) A representative FACS plot of LSK/SLAM HSCs stained for annexin V and 7AAD is shown for control and ko. (Right) This bar graph shows the percentage of apoptotic (annexin V positive, 7AAD negative) and dead HSCs (annexin V, 7AAD double positive) for control (gray) and ko (black) samples 96 hours after pIpC treatment (n = 4); *P < .05.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/119/6/10.1182_blood-2011-06-359406/5/m_zh89991184080006.jpeg?Expires=1767718990&Signature=xDFjhJdID0gTQWly8Gk70-KYG9EnKPD9rSCcIsR0gbrgf5VVZKmJ3NsMLQl~YdDh67ha8gvnt-Ta8cg9QuvOc0ICNTeOLX7rTjOp8y3uTy5d6OFPqqNgEnN8s2da1a6Ap8q08nCbf5Qyxp2JWDPUfG9om22ugVsYjrT4YeK8APAq-dxjY6AK2Y9xi2ejwnPe6kiuzJBQCq97lirk0EZJgVZTGnWkOmZfp7pKZL8C9nHJpnGS0lRSE6NYIgv5BhhWaKXCI6EowztoWfNY8SXuKAFXl99Vz9OL0IjN1REvSEOM8K1OvWW5DBqWeNSD9NcQhW-CV-9d1VVzj8sRA03o7g__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 7. Deletion of p53 in addition to NF-Ya does not rescue the defects of NF-Ya–deficient BM cells and HSCs. (A) Protein from wt and p53 conditional ko BM 7 days after pIpC treatment was subjected to Western blot analysis and probed with a p53 (top) and actin (bottom) Ab. (B) Representative flow cytometric staining of control (left) and NF-Ya ko (right) for TUNEL and PI. Note, NF-Ya sko and NF-Ya, p53 dko (not shown) stainings are indistinguishable. (C) Results from panel B are summarized in this bar graph (n = 5); **P < .01. (D) CD45.1+ recipients received a BM transplant from NF-Ya+/++Mx-cre (control), NF-Yafl/fl+Mx-cre (ko), and NF-Yafl/fl, p53fl/fl+Mx-cre (dko), (all CD45.2+) mice together with 30% CD45.1+ NF-Ya wt competitor cells. The mice were treated with pIpC at day 0. The CD45.1-to-CD45.2 ratio of PB cells was determined and normalized to day 0 (n = 8 [control], n = 7 [sko], n = 6 [(dko]). (E) Mice from panel D were killed, and the indicated BM subpopulations were analyzed for CD45.1 versus CD45.2 expression and are shown in a bar graph; control (gray) and ko (black).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/119/6/10.1182_blood-2011-06-359406/5/m_zh89991184080007.jpeg?Expires=1767718990&Signature=azsc2DRkz-16LqE-h1Ac9uceHCGuArtRDF34FjuvGYBzsaIAe7AXNZga3zpPqeO-0uktHNaqwtbulEbxyXmH8UYPblTFXGV0hFp6Q~XlzPs~S478dIBm-ne05Iwre4Hgo8p2Il48Nr~sxndziN4VQA18VpSlYBaA5MyUC5NGGriUwLjghjDHeIHRcl3T2hqXFoYXQupEEAsbQKu3gAA2nqarj-HidbI3TDrdH0Klcndno8gOK7c7~EyUY3thgxXgvUl9AlSQJVDZETEXEZU06sBkQDEQayxPENlrPMx-jzxTARI2afmlJ~R5fZ6S1ssAcKq~eUDXczxwZmwiETqYag__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal