Abstract
While most blood lineages are assumed to mature through a single cellular and developmental route downstream of HSCs, dendritic cells (DCs) can be derived from both myeloid and lymphoid progenitors in vivo. To determine how distinct progenitors can generate similar downstream lineages, we examined the transcriptional changes that accompany loss of in vivo myeloid potential as common myeloid progenitors differentiate into common DC progenitors (CDPs), and as lymphoid-primed multipotent progenitors (LMPPs) differentiate into all lymphoid progenitors (ALPs). Microarray studies revealed that IFN regulatory factor 8 (IRF-8) expression increased during each of these transitions. Competitive reconstitutions using Irf8−/− BM demonstrated cell-intrinsic defects in the formation of CDPs and all splenic DC subsets. Irf8−/− common myeloid progenitors and, unexpectedly, Irf8−/− ALPs produced more neutrophils in vivo than their wild-type counterparts at the expense of DCs. Retroviral expression of IRF-8 in multiple progenitors led to reduced neutrophil production and increased numbers of DCs, even in the granulocyte-macrophage progenitor (GMP), which does not normally possess conventional DC potential. These data suggest that IRF-8 represses a neutrophil module of development and promotes convergent DC development from multiple lymphoid and myeloid progenitors autonomously of cellular context.
Introduction
HSC differentiation occurs through a series of intermediate stages, or progenitors, whereby self-renewal capacity is progressively lost and genetic programs limit the development of alternate cell lineages until commitment to a single cell type is reached. The ability to identify distinct progenitor populations via differential surface marker expression has allowed lineage potentials to be determined, and a hierarchical map of lineage restriction to be inferred. The classic model of hematopoietic development suggests that the order in which cells progress though intermediate stages, and thus the order in which developmentally restrictive programs are placed on progenitor cells to limit lineage potential, is fixed. The discovery of the common lymphoid progenitor (CLP), that yields all lymphoid cells, and the common myeloid progenitor, which propagates myeloid, erythroid, and megakaryocytic cells, provided initial evidence for the first division in the hematopoietic developmental hierarchy.1,2
Subsequent studies, however, led to the identification of progenitors that could not easily be developmentally placed in relation to CLPs, common myeloid progenitors, and other progenitors identified in early studies. For example, in contrast to the lineage potential of common myeloid progenitors,1,3 lymphoid-primed multipotent progenitors (LMPPs) generate lymphocytes, macrophages, and granulocytes, yet most of these cells lack the ability to generate erythrocytes and megakaryocytes at the clonal level.4 Similarly, while granulocyte-macrophage progenitors (GMPs) generate granulocytes and macrophages in vivo and in vitro,1 macrophage–dendritic cell (DC) progenitors (MDPs) generate macrophages and DCs, but not neutrophils.5 These findings have generated considerable debate as to whether the existence of one progenitor precludes the existence of another progenitor that has partially overlapping developmental potential, whether multiple developmental paths lead to maturity, and whether the mature lineages that ultimately arise from different developmental routes are functionally distinct.
The lineage potentials for some of the progenitors mentioned above are still in dispute because of differences in purification strategies and between findings from in vitro and in vivo assays. However, other cases in which a simple hierarchical developmental scheme cannot be applied have been unequivocally proven by multiple groups and complementary approaches. For example, while ∼ 90% of steady-state splenic DCs are thought to be derived from myeloid progenitors, transplantation, genetic tracing, and clonal in vitro assays have all demonstrated that CLPs generate ∼ 10% of the same subsets of DCs in both humans and mice,6–13 although the continued identification of new DC subsets necessitates the validation of these findings.14 The DCs derived from common myeloid progenitors and CLPs appear to have similar transcriptional and functional properties.9,12,13,15 Given that the details of other stages of hematopoietic development remain controversial, DC development is perhaps the best example that hematopoietic differentiation need not necessarily follow a single cellular and developmental route in vivo. A refined cellular and molecular understanding of DC lineage commitment and development could therefore provide broad insight on the mechanisms by which hematopoietic differentiation proceeds.
Steady-state DCs are found in murine lymphoid tissues and include the conventional DCs (cDCs) and plasmacytoid DCs (pDCs). Conventional DCs can be subdivided based on the surface expression of CD4 and CD8α, into CD8α+CD4−, CD8α−CD4+, and CD8α−CD4− subsets.16,17 Elegant studies have demonstrated that a DC-restricted progenitor, termed the common DC progenitor (CDP), gives rise to all of these DC subsets, suggesting a clonal origin of all steady-state DCs.18,19 CDPs branch into either a pDC-committed pathway or a conventional DC pathway that follows the development of pre-cDCs located in the BM and spleen.20 The splenic pre-cDCs then diverge into CD8α+ or CD8α− conventional DC subsets through the coordinated action of specific transcriptional regulators. Each of these subsets is relatively short-lived and requires continuous replenishment from the BM-derived precursors described above.21–23 Monocytes and macrophages can also differentiate into specific subsets of DCs, such as Langerhans cells and inflammatory DCs on exposure to GM-CSF, but are not thought to contribute substantially to the steady-state splenic DC subsets.20,24–27
Several transcription factors have been found to regulate the development of specific mature DC subsets. For example, BATF3 and ID2 are required for the development of CD8α+ DCs, IFN regulatory factor 4 (IRF-4) is required for CD4+ DC development, and E2-2 is required for pDC development.28–32 Yet, little is known about the molecular mechanisms that regulate the initial commitment to the DC lineage and whether these mechanisms are shared between myeloid and lymphoid progenitors. Here, we demonstrate that IRF-8, a transcription factor known to be involved in later stages of CD8α+ DC and pDC development,33–35 regulates CDP development and the initial commitment to the DC lineage through the suppression of neutrophil potential in both the myeloid and lymphoid pathways. These findings help explain the existence of hematopoietic progenitors with partially overlapping potentials and the similarities in the mature progeny that arise from these distinct developmental pathways.
Methods
Animals
All animal procedures were carried out according to the Institutional Animal Care and Use Committee at Washington University. C57BL/6 (CD45.2+) and B6.SJL (CD45.1+) mice were obtained from The Jackson Laboratory and subsequently maintained in our animal facility. Irf8−/− mice36 were initially provided by K. Ozato (National Institutes of Health, Bethesda, MD); subsequent colonies were obtained from the European Mutant Mouse Archive and maintained in our animal facility. All animals used were between 4 and 10 weeks of age.
Abs, flow cytometry, and cell sorting
An Ab list can be found in supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Stains were carried out in 1%-2% adult bovine serum (Hyclone)/PBS. For staining CDPs, buffer was supplemented with 1mM EDTA. Propidium iodide (Sigma-Aldrich) was added to cells before acquisition/sorting to gate out dead cells. Cells were acquired/sorted on the FACSAria (BD Biosciences). Data analysis was carried out using FlowJo software (TreeStar).
Progenitor cell isolation
BM was isolated from the femur, tibia, humerus, and pelvis of mice. Bones were crushed using a mortar and pestle, and debris was removed by gradient centrifugation using Histopaque 1119 (Sigma-Aldrich). Common myeloid progenitors were identified as Lineage−c-kithiSca-1−CD11c− CD34+ Flk2+CD16/32−CD115− while CDPs were identified as Lin−c-kitintSca-1−CD34+Flk2+CD16/32−CD115+. LMPPs were identified as Lin-B220−c-kithiSca−1+Flk2+IL7Rα− while ALPs were sorted as Lin-Ly6D−B220−c-kit+Flk2+IL7Rα+ cells. GMPs were identified as Lin−B220−Flk2−IL7Rα−c-kithiSca-1−CD16/32+.
Microarray
For common myeloid progenitor and CDP arrays, RNA was isolated from 10 000-30 000 cells using TRIzol (Invitrogen) and linear acrylamide (Ambion) according to the manufacturer's instructions. RNA was subjected to 2 rounds of amplification using Affymetrix Two-Cycle Amplification and IVT kits. Fragmented cRNA (10 μg) was hybridized to Affymetrix Mouse Genome 430 2.0 microarrays. Background subtraction and quantile-based normalization was performed using the GCRMA algorithm, available from Bioconductor.org. For all other microarrays, RNA was processed using the Qiagen RNeasy Mini kit. Cells (2000-15 000) were sorted into RLT buffer and RNA was processed per the manufacturer's instructions. RNA was amplified using the Nugen pico-amplification kit per manufacturer's instructions antisense RNA (750 ng) was hybridized to Illumina MouseRef-8 Version 2.0 bead chips. Data were analyzed using Arraystar software (DNAstar) and quantile-based normalization. All microarray data are available in the Gene Expression Omnibus (GEO) under accession number GSE34917.
Quantitative real-time PCR
Cells were double sorted using the FACSAria (BD Biosciences) into TRIzol reagent (Invitrogen). cDNA was generated using the SuperScript III First Strand kit (Invitrogen) per the manufacturer's instructions using random hexamers. Real-time PCR was carried out using SybrGreen PCR master mix (Applied Biosystems) per the manufacturer's instructions. Transcript levels were quantified using the ABI 7000 Sequence Detection System (Applied Biosystems). Primer sequences were: IRF8-1: 5′-GACCATGTTCCGTATCCCCTGGAAG-3′ and 5′-GGGACCGGTCAGTCACTTCTTCA-3′; β-actin: 5′-GTCTGAGGCCTCCCTTTTT-3′ and 5′-GGGAGACCAAAGCCTTCATA-3′.
Retrovirus production
293FT cells (Invitrogen) were cultured to ∼ 60% density on 10-cm2 tissue-culture plates and transfected using calcium phosphate coprecipitation with 3.5 μg of pMD2.G (a VSV-G–encoding expression vector), 6.5 μg of an expression plasmid encoding MMLV gag-pol, and 10 μg of MSCV-IRES-GFP or MSCV-IRF-8-IRES-GFP. Media was changed 4-6 hours after transfection, and supernatant was harvested at 48 and 72 hours after transfection and filtered through 0.45-μm syringe filters. Twelve milliliters of viral supernatant was mixed with 3 mL of 1× PBS/25% polyethylene glycol 8000 (Sigma-Aldrich) and left overnight at 4°C. The mixture was spun at 2000g for 10 minutes, supernatant was removed, and the pellet was resuspended in 100 μL of PBS. Aliquots were frozen and stored at −80°C until use.
In vitro culture and retroviral transductions
LMPPs, common myeloid progenitors, and GMPs were double sorted as described above and 1000 cells/well were plated in triplicate and cultured in DME/F12 + 10mM HEPES (SAFC Biosciences) and supplemented with 10% defined FBS (Hyclone), Glutamax (Invitrogen), NEAA (Lonza), 1mM sodium pyruvate (Lonza), penicillin/streptomycin (Sigma-Aldrich), and 50μM 2-Mercaptoethanol (Invitrogen). Cells were cultured in media containing 10 ng/mL each IL-3 (PeproTech), Flt3 ligand (PeproTech), GM-CSF (PeproTech), and stem cell factor (Peprotech). In some experiments, cells were cultured in cytokines for 24 hours then transduced with 5 μL of purified virus, and cultured for an additional 6 days at 37°C before harvesting cells and analyzing DC and neutrophil production by flow cytometry. For GMP cultures, GFP+ cells were sorted, cytospins were prepared and stained with May-Giemsa, and cells were visually scored by microscopy for lineage output.
Cell transplantation assays
Four thousand ALP or common myeloid progenitor from Irf8+/+ or Irf8−/− donors were injected intravenously via the retro-orbital sinus into 800 cGy irradiated wild-type mice. Ten days after transplantation, BM and spleen were harvested. BM was processed as described in “Progenitor cell isolation.” Spleens were digested with 300 U/mL DNAse I (Sigma-Aldrich) and 250 μg/mL collagenase B (Roche) for 20 minutes at 37°C on a stir plate. Viable cells were isolated by gradient centrifugation using Histopaque 1119 (Sigma-Aldrich). DC and neutrophil development was analyzed by flow cytometry. For mixed BM chimeras, 2 × 106 wild-type B6.SJL (CD45.1+CD45.2−) BM cells were injected with Irf8−/− or wild-type (CD45.1−CD45.2+) BM cells that were matched for HSC numbers, into a sublethally (800 rad) irradiated (C57BL/6 × B6.SJL) F1 recipient (CD45.1+CD45.2+). Seven weeks after transplantation, BM and spleens were harvested as described in this section and in “Progenitor cell isolation,” and analyzed for progenitor, DC, or neutrophil development by flow cytometry.
Results
IRF-8 expression increases during the common myeloid progenitor to CDP transition
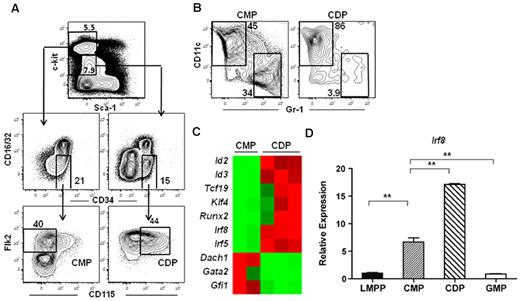
To determine the molecular mechanisms responsible for DC lineage commitment, we analyzed the gene expression profiles of highly purified Flk2+ common myeloid progenitor and CDP subsets isolated from the BM of wild-type mice. Common myeloid progenitors were isolated as Lineage (Lin)−c-kithi Sca-1−CD16/32−CD34+Flk2+CD115− and CDPs were isolated from the Lin−c-kitintermediateSca-1−CD16/32−CD34+Flk2+CD115+ fraction (Figure 1A). Flk2+ common myeloid progenitors, which possess macrophage, neutrophil, and DC potential,6 have been shown to lie developmentally upstream of MDPs and CDPs.19,37 Indeed, common myeloid progenitors were able to produce both DCs and neutrophils, whereas CDPs were largely restricted to producing DCs in an in vitro culture assay (Figure 1B), consistent with previous reports describing these progenitors.6,18,19 Microarray analysis identified several transcription factors that exhibited at least a 5-fold difference in expression between the common myeloid progenitor and CDP subsets (Figure 1C, supplemental Table 1). We chose to focus on IRF-8 because previous studies have demonstrated that IRF-8 expression also increases during differentiation to CLPs.38,39 IRF-8 expression was validated by quantitative RT-PCR in LMPPs, common myeloid progenitors, GMPs, and CDPs. IRF-8 expression in common myeloid progenitors was 10-fold higher than that of LMPPs, and continued to increase during the common myeloid progenitor to CDP transition (Figure 1D). IRF-8 expression in GMPs, on the other hand, was comparable with that of LMPPs (Figure 1D).
IRF-8 expression increases during thecommon myeloid progenitorto CDP transition. Microarray analysis was carried out on double-sorted common myeloid progenitors (CMPs) and CDPs from BM. (A) Common myeloid progenitors were lineage (Lin)−c-kithiSca-1−CD16/32−CD34+ Flk2+CD115− and CDP were were Lin−c-kitintermediateSca-1−CD16/32−CD34+Flk2+CD115+. (B) In vitro differentiation of common myeloid progenitors and CDPs. Cells were cultured for 7 days with IL-3, GM-CSF, Flt3 ligand, and stem cell factor (all 10 ng/mL). DC (CD11c+Gr-1−) and neutrophil (CD11c−Gr-1+) potential was analyzed by flow cytometry. (C) Heat map of microarray data showing selected transcription factors that were differentially regulated at least 5-fold between the common myeloid progenitor and CDP subsets. (D) Quantitative RT-PCR analysis of IRF-8 in double-sorted LMPP, common myeloid progenitor, GMP, and CDP. Data represent 3 or 6 independent sorts for GMP and CDP or LMPP and common myeloid progenitor, respectively, analyzed in 3 separate assays; **P < .05.
IRF-8 expression increases during thecommon myeloid progenitorto CDP transition. Microarray analysis was carried out on double-sorted common myeloid progenitors (CMPs) and CDPs from BM. (A) Common myeloid progenitors were lineage (Lin)−c-kithiSca-1−CD16/32−CD34+ Flk2+CD115− and CDP were were Lin−c-kitintermediateSca-1−CD16/32−CD34+Flk2+CD115+. (B) In vitro differentiation of common myeloid progenitors and CDPs. Cells were cultured for 7 days with IL-3, GM-CSF, Flt3 ligand, and stem cell factor (all 10 ng/mL). DC (CD11c+Gr-1−) and neutrophil (CD11c−Gr-1+) potential was analyzed by flow cytometry. (C) Heat map of microarray data showing selected transcription factors that were differentially regulated at least 5-fold between the common myeloid progenitor and CDP subsets. (D) Quantitative RT-PCR analysis of IRF-8 in double-sorted LMPP, common myeloid progenitor, GMP, and CDP. Data represent 3 or 6 independent sorts for GMP and CDP or LMPP and common myeloid progenitor, respectively, analyzed in 3 separate assays; **P < .05.
CDPs are reduced in Irf8−/− mice compared with wild-type mice
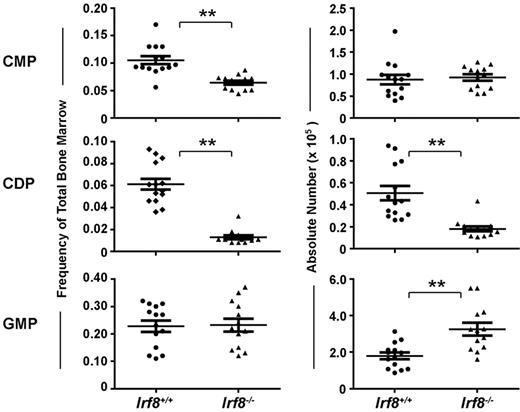
The development of progenitor cell subsets in the BM of Irf8−/− and wild-type mice was analyzed by flow cytometry (supplemental Figure 1). Strikingly, CDPs were significantly reduced both by frequency and by absolute number in Irf8−/− mice compared with controls (Figure 2). In contrast, although common myeloid progenitor frequency was reduced by ∼ 50% in Irf8−/− compared with Irf8+/+ BM, the absolute number of common myeloid progenitor was normal in Irf8−/− mice (Figure 2). This discrepancy was because of increased cellularity in the BM in Irf8−/− animals (supplemental Figure 2A), consistent with previous reports.36 Furthermore, although the frequency of GMPs was similar between Irf8−/− and control mice, the absolute number of GMPs was elevated in Irf8−/− mice (Figure 2). We also analyzed monocyte populations in the BM, blood, and spleen. We detected comparable frequencies of CD11b+CD115+Ly6Chigh and CD11b+CD115+ Ly6Clo monocytes in the BM and spleen in both wild-type and Irf8−/− mice (supplemental Figure 3). However, CD11b+CD115+Ly6Chigh monocytes were selectively reduced the peripheral blood of Irf8−/− mice compared with wild-type mice, indicating possible defects in intravasation or intravascular residence time. Nevertheless, Irf8−/− mice are clearly able to produce monocytes, consistent with previous reports.36 Taken together, these data suggest that IRF-8 regulates initial DC lineage commitment downstream of common myeloid progenitors by promoting CDPs and limiting the GMP fate.
Irf8−/− mice have reduced numbers of CDPs and increased numbers of GMPs. Progenitor cells in the BM of Irf8−/− and wild-type mice were analyzed by flow cytometry. Graphs show the frequency (left) and absolute number (right) of common myeloid progenitor (CMP), GMP, and CDP. Horizontal lines indicate the mean and error bars show SEM. Each data point represents one mouse, with data for 14 wild-type and 13 IRF8−/− shown; **P < .05.
Irf8−/− mice have reduced numbers of CDPs and increased numbers of GMPs. Progenitor cells in the BM of Irf8−/− and wild-type mice were analyzed by flow cytometry. Graphs show the frequency (left) and absolute number (right) of common myeloid progenitor (CMP), GMP, and CDP. Horizontal lines indicate the mean and error bars show SEM. Each data point represents one mouse, with data for 14 wild-type and 13 IRF8−/− shown; **P < .05.
The reduction of CDPs in Irf8−/− mice is because of a cell-intrinsic function of Irf8
To determine whether the requirement for IRF-8 in DC commitment is cell-intrinsic, we analyzed the development of common myeloid progenitors, CDPs, and GMPs in competitive mixed BM chimeras. Chimeric mice were generated by transplanting a mixture of Irf8−/− or wild-type (CD45.2+) BM and BM from wild-type congenic donors (CD45.1+) into irradiated F1 congenic recipients (CD45.1+CD45.2+). The number of BM cells injected from Irf8−/− and wild-type animals was normalized for HSC numbers, as Irf8−/− mice exhibit a reduction in this compartment relative to controls (supplemental Figure 2B). The development of progenitors and mature lineages was analyzed 7 weeks after transplantation.
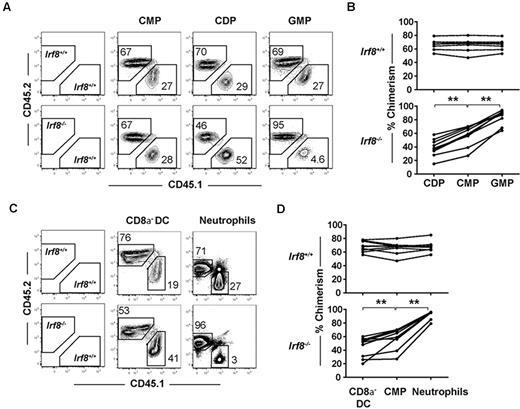
Donor contributions to the CDP and GMP compartments were compared with the common myeloid progenitor donor contribution in each recipient. As expected, within each animal receiving wild-type CD45.2+ BM, CDP and GMP chimerism values were indistinguishable from upstream common myeloid progenitor chimerism levels, indicating normal common myeloid progenitor differentiation to downstream progeny (Figure 3A-B). In contrast, contributions to the CDP compartment from Irf8−/− BM were consistently reduced relative to contributions to the common myeloid progenitor compartment (Figure 3A-B). Reciprocally, Irf8−/− GMP donor chimerism increased compared with common myeloid progenitor chimerism. Together, these findings suggest that IRF-8 regulates DC commitment in a cell-intrinsic manner by regulating the GMP versus CDP decision downstream of common myeloid progenitors.
IRF-8 drives DC commitment in a cell-intrinsic manner. Competitive mixed BM chimeras were carried out by injecting 2 × 106 B6.SJL (CD45.1+) wild-type whole BM cells with Irf8−/− or wild-type (CD45.2+) BM that were matched for HSC numbers into 800 cGy-irradiated (C57BL/6 × B6.SJL) F1 recipients (CD45.1+CD45.2+). Seven weeks after transplantation, BM and spleens were harvested, and progenitor, DC, and neutrophil development was analyzed by flow cytometry. (A,C) Representative plots showing CD45.2 and CD45.1 chimerism in one representative mouse. Graphs show (B) chimerism of common myeloid progenitor (CMPs), CDPs, and GMPs or (D) splenic CD8α− DC and BM neutrophils from wild-type (top) or Irf8−/− (bottom) reconstituted mice. Each point represents 1 mouse, with lines connecting values in a single mouse. Data are pooled from 3 experiments of 1-4 mice per group. Data for all 9 mice in each group are shown; **P < .05 based on paired students 2-tailed t test.
IRF-8 drives DC commitment in a cell-intrinsic manner. Competitive mixed BM chimeras were carried out by injecting 2 × 106 B6.SJL (CD45.1+) wild-type whole BM cells with Irf8−/− or wild-type (CD45.2+) BM that were matched for HSC numbers into 800 cGy-irradiated (C57BL/6 × B6.SJL) F1 recipients (CD45.1+CD45.2+). Seven weeks after transplantation, BM and spleens were harvested, and progenitor, DC, and neutrophil development was analyzed by flow cytometry. (A,C) Representative plots showing CD45.2 and CD45.1 chimerism in one representative mouse. Graphs show (B) chimerism of common myeloid progenitor (CMPs), CDPs, and GMPs or (D) splenic CD8α− DC and BM neutrophils from wild-type (top) or Irf8−/− (bottom) reconstituted mice. Each point represents 1 mouse, with lines connecting values in a single mouse. Data are pooled from 3 experiments of 1-4 mice per group. Data for all 9 mice in each group are shown; **P < .05 based on paired students 2-tailed t test.
Although previous studies have reported normal numbers of CD8α− DCs in Irf8−/− mice,34 the reduction in CDP formation in Irf8−/− mice led us to hypothesize that CD8α− DC development was also affected, and that this defect would be readily observed in a competitive transplantation setting. Indeed, CD8α− DC chimerism derived from Irf8−/− donors was statistically significantly reduced compared with common myeloid progenitor chimerism in the same recipients, suggesting a previously unappreciated defect in the development of CD8α− DCs in Irf8−/− mice (Figure 3C-D). Homeostatic expansion of CD8α− DCs or their committed progenitors thus likely explains the normal numbers of these cells found under steady-state conditions in Irf8−/− mice. Donor contribution of Irf8−/− cells to both pDC and CD8α−DC was negligible, consistent with previous reports (supplemental Figure 4A).33–35 In these same animals, statistically significant increases were also noted in the chimerism of Irf8−/− neutrophils compared with Irf8−/− common myeloid progenitors (Figure 3C-D). The development of lymphoid cells from Irf8−/− BM was comparable with wild-type controls (supplemental Figure 4B).
Common myeloid progenitors from Irf8−/− mice have reduced DC and increased neutrophil developmental potential
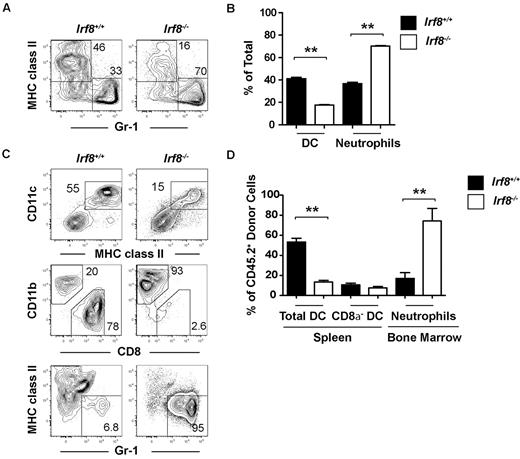
To further dissect the altered lineage potential of Irf8−/− progenitors, we analyzed DC and neutrophil production from common myeloid progenitors both in vitro and in vivo. Highly purified common myeloid progenitors from either Irf8−/− or wild-type animals were cultured for 7 days in the presence of Flt3 ligand, stem cell factor, IL-3, and GM-CSF, before analyzing DC and neutrophil output by flow cytometry. Irf8−/− common myeloid progenitors produced significantly more neutrophils compared with wild-type common myeloid progenitors, while DC production was reduced in Irf8−/− common myeloid progenitors compared with controls (Figure 4A-B).
Common myeloid progenitors from Irf8−/− mice overproduce neutrophils at the expense of DCs in vitro and in vivo. Common myeloid progenitors (CMP) were double-sorted from the BM of Irf8−/− or wild-type controls. (A-B) One thousand progenitors were cultured for 7 days in the presence of Flt3 ligand, IL-3, GM-CSF, and SCF (all 10 ng/mL). DC and neutrophil output was analyzed by flow cytometry. Data show averages from 5 wells and represent 3 independent assays. (C-D) Four thousand common myeloid progenitors from Irf8+/+ or Irf8−/− were injected intravenously into each 800 cGy-irradiated (C57BL/6 × B6.SJL) F1 recipient (CD45.1+CD45.2+). BM and spleens were harvested 10 days after transplantation and the development of DCs and neutrophils was analyzed by flow cytometry. Data shown are for 6 mice in each experimental line pooled from 2 independent experiments with 3 mice per group; **P < .05.
Common myeloid progenitors from Irf8−/− mice overproduce neutrophils at the expense of DCs in vitro and in vivo. Common myeloid progenitors (CMP) were double-sorted from the BM of Irf8−/− or wild-type controls. (A-B) One thousand progenitors were cultured for 7 days in the presence of Flt3 ligand, IL-3, GM-CSF, and SCF (all 10 ng/mL). DC and neutrophil output was analyzed by flow cytometry. Data show averages from 5 wells and represent 3 independent assays. (C-D) Four thousand common myeloid progenitors from Irf8+/+ or Irf8−/− were injected intravenously into each 800 cGy-irradiated (C57BL/6 × B6.SJL) F1 recipient (CD45.1+CD45.2+). BM and spleens were harvested 10 days after transplantation and the development of DCs and neutrophils was analyzed by flow cytometry. Data shown are for 6 mice in each experimental line pooled from 2 independent experiments with 3 mice per group; **P < .05.
To analyze the in vivo consequence of Irf8 deficiency on these progenitors, common myeloid progenitors were sorted from either wild-type or Irf8−/− mice (CD45.2+) and 4000 cells were injected into each sublethally irradiated F1 congenic wild-type recipient (CD45.1+CD45.2+). Ten days after transplantation, DC and neutrophil production was analyzed by flow cytometry in the spleen and BM, respectively. Neutrophil output was increased by Irf8−/− common myeloid progenitors compared with wild-type common myeloid progenitors (Figure 4C-D). Total DC development was reduced after transplantation of Irf8−/− common myeloid progenitors, but this appeared to be because of the selective inability of these cells to produce CD8α+ DCs and pDCs (Figure 4C-D, supplemental Figure 5A). Irf8-deficient CD8α− DCs were formed in normal proportions relative to controls, likely demonstrating the homeostatic expansion of CD8α− DC precursors in this noncompetitive transplantation setting, similar to that observed in unmanipulated Irf8−/− animals. These data suggest that, as a part of DC lineage commitment, IRF-8 limits neutrophil development from common myeloid progenitors.
CLPs from Irf8−/− mice aberrantly produce neutrophils in vivo
Next, we determined whether IRF-8 was also controlling DC lineage commitment in the lymphoid pathway. Compared with LMPPs, IRF-8 expression increased > 10-fold in the Ly6d− all-lymphoid progenitor (ALP) fraction of CLPs, which retains DC potential39,40 (Figure 5A). Although previous studies have suggested that IRF-8 is required for the expression of B-lineage factors such as Ebf1 in CLPs,38 ALPs do not yet express Ebf1,39,40 implying a distinct role for IRF-8 in these cells. To test this prediction, ALPs were double-sorted from wild-type or Irf8−/− mice (CD45.2+) and injected into sublethally irradiated congenic wild-type F1 recipients (CD45.1+CD45.2+). DC and neutrophil potential was analyzed 10 days after transplantation. ALPs from Irf8−/− mice were phenotypically similar to Irf8+/+ ALPs (supplemental Figure 6). Unexpectedly, ALPs from Irf8−/− mice produced a large number of neutrophils (Figure 5B-C). In contrast, wild-type ALPs produce few neutrophils in vivo, as expected based on previous studies.39 Similar to the observations above with Irf8−/− common myeloid progenitors, total DC production from Irf8−/− ALPs was proportionally reduced, while CD8α− DC frequency was normal (Figure 5B-C). Again, the overall reduction in DC potential in Irf8−/− mice was likely because of a significant reduction in pDC and CD8α+ DC (supplemental Figure 7A). While Irf8−/− ALP produced a significant fraction of neutrophils, these cells were still able to produce B cells, demonstrating the retention of lymphoid potential (supplemental Figure 7B). This is also consistent with previous reports that did not note a deficit in lymphoid cells in Irf8−/− mice.36
ALPs from Irf8−/− mice but not wild-type mice produce neutrophils in vivo. (A) LMPPs and ALPs were double-sorted from the BM of Irf8−/− or wild-type controls and quantitative real-time PCR analysis of IRF-8 expression was carried out. Data are representative of 6 independent experiments. (B-C) Four thousand ALPs purified from either Irf8−/− or wild-type mice were injected intravenously into each 800 cGy-irradiated F1 recipient (CD45.1+CD45.2+). BM and spleen were harvested 10 days after transplantation and the development of DCs, neutrophils, and B cells was analyzed by flow cytometry. (B) Representative contour plots. (C) Pooled data from 3 independent experiments. Data represent 5 wild-type and 6 Irf8−/− recipients. Error bars show calculated SEM; **P < .05.
ALPs from Irf8−/− mice but not wild-type mice produce neutrophils in vivo. (A) LMPPs and ALPs were double-sorted from the BM of Irf8−/− or wild-type controls and quantitative real-time PCR analysis of IRF-8 expression was carried out. Data are representative of 6 independent experiments. (B-C) Four thousand ALPs purified from either Irf8−/− or wild-type mice were injected intravenously into each 800 cGy-irradiated F1 recipient (CD45.1+CD45.2+). BM and spleen were harvested 10 days after transplantation and the development of DCs, neutrophils, and B cells was analyzed by flow cytometry. (B) Representative contour plots. (C) Pooled data from 3 independent experiments. Data represent 5 wild-type and 6 Irf8−/− recipients. Error bars show calculated SEM; **P < .05.
Our data imply that the stage-specific expression of IRF-8 in ALPs is required to suppress neutrophil potential. However, as the Irf8−/− mice used in these experiments completely lack IRF-8 in all tissues, it is possible that an earlier developmental phenotype exists in which Irf8 deficiency indirectly affects ALPs. To test this possibility, we retrovirally expressed IRF-8 in both wild-type and Irf8−/− ALPs. Ectopic expression of IRF-8 increased DC development in both Irf8+/+ and Irf8−/− ALP and completely abrogated neutrophil development from Irf8−/− ALPs (supplemental Figure 7C). To exclude the possibility that contamination with other neutrophil producing progenitors was driving the phenotype observed in Irf8−/− ALP, we sorted single Irf8+/+ or Irf8−/− ALPs per well of a 96-well plate containing OP-9 stromal cells, Flt3L, IL-7, and SCF. We found that ∼ 70% of colonies derived from a single Irf8−/− ALP produced neutrophils, whereas neutrophils were not detected in any of the colonies clonally derived from Irf8+/+ ALPs (supplemental Figure 7D). As previously published, ALPs can be forced to produce myeloid cells when cultured in myeloid cytokines.11 To determine whether retroviral expression of IRF-8 was sufficient to block neutrophil development from wild-type ALPs, we added IL-3 to Irf8+/+ cultures to induce neutrophil development, and then infected cells with a control or IRF-8–expressing retrovirus. Wild-type ALPs infected with a control retrovirus produced a substantial fraction of neutrophils in these culture conditions; however, neutrophil development was significantly reduced while DC development was increased in cells that retrovirally expressed IRF-8 (supplemental Figure 7E). Taken together, our data support a critical stage-specific role for IRF-8 in restricting neutrophil potential and promoting DC commitment in ALPs, although further experiments with conditional Irf8−/− mice will be required to conclusively demonstrate this.
IRF-8 blocks neutrophil development and induces DC differentiation at multiple stages of progenitor cell development
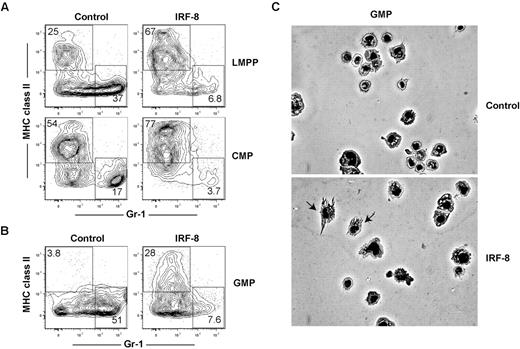
Our in vivo and in vitro CMA and ALP data indicate that IRF-8 limits neutrophil production, consistent with previous studies,36,41 but does not establish whether IRF-8 may also actively promote DC commitment and differentiation. Previous studies have demonstrated that retroviral expression of IRF-8 can promote CD8a+ DC maturation in Irf8−/− BM cells, but did not address whether this affected initial DC lineage commitment.42 To determine whether ectopic expression of IRF-8 could block neutrophil development and promote DC commitment in progenitor cells with known neutrophil potential, double-sorted LMPP, common myeloid progenitor, and GMP were infected with control or IRF-8–expressing retroviruses. Cells were cultured for 7 days in the presence of Flt3 ligand (Flt3L), IL-3, stem cell factor, and GM-CSF, transduced with retroviruses 24 hours after the initiation of culture, and neutrophil and DC production from GFP+ retrovirally transduced cells was assessed by flow cytometry. IRF-8 expression in LMPPs, common myeloid progenitors, and GMPs reduced the production of neutrophils compared with controls (Figure 6A-B). Strikingly, IRF-8 expression also induced the production of MHCII+ Gr-1− DCs from GMPs, a cell type that does not normally produce conventional DCs under these conditions (Figure 6B). IRF8-GMP DC expressed CD11c (supplemental Figure 8A), and induced expression of CD80 after TLR stimulation (supplemental Figure 8B). These cells also produced IL-12 after stimulation with either LPS or CpG-B (supplemental Figure 8C), consistent with the fact that the majority of the DCs derived in these assays were CD24hiCD11bloSirpαlo, or CD8α+ DC equivalents (supplemental Figure 8D).26 These data contrast with previous studies based on cell lines, which found that retroviral IRF-8 expression primarily drives macrophage development.41
Ectopic expression of IRF-8 extinguishes neutrophil potential and inducesDCpotential in LMPPs,common myeloid progenitors, and GMPs. LMPPs, common myeloid progenitors (CMPs), or GMPs were double-sorted from the BM of wild-type mice. One thousand cells per well were cultured in the presence of Flt3 ligand, GM-CSF, IL-3, and stem cell factor (all 10 ng/mL) for 7 days, and transduced by retrovirus as described in “In vitro culture and retroviral transductions.” Cells were harvested and analyzed for neutrophil (MHC class II−GR-1+) and DC (MHC class II+Gr-1−) production in virus-infected (GFP+) cells. Representative FACS plots showing the development of GFP+ MHC class II− Gr-1+ neutrophils and GFP+ MHC class II+ Gr-1− DCs in (A) LMPPs and common myeloid progenitors or (B) GMPs. Data represent 4 independent experiments with progenitors analyzed in triplicate. (C) GFP+ cells were sorted from GMP cultures and morphology of cells was analyzed by Giemsa stain and microscopy.
Ectopic expression of IRF-8 extinguishes neutrophil potential and inducesDCpotential in LMPPs,common myeloid progenitors, and GMPs. LMPPs, common myeloid progenitors (CMPs), or GMPs were double-sorted from the BM of wild-type mice. One thousand cells per well were cultured in the presence of Flt3 ligand, GM-CSF, IL-3, and stem cell factor (all 10 ng/mL) for 7 days, and transduced by retrovirus as described in “In vitro culture and retroviral transductions.” Cells were harvested and analyzed for neutrophil (MHC class II−GR-1+) and DC (MHC class II+Gr-1−) production in virus-infected (GFP+) cells. Representative FACS plots showing the development of GFP+ MHC class II− Gr-1+ neutrophils and GFP+ MHC class II+ Gr-1− DCs in (A) LMPPs and common myeloid progenitors or (B) GMPs. Data represent 4 independent experiments with progenitors analyzed in triplicate. (C) GFP+ cells were sorted from GMP cultures and morphology of cells was analyzed by Giemsa stain and microscopy.
GFP+ cells were sorted and analyzed for morphology. As shown in Figure 6C, GMP that overexpressed IRF-8 generated morphologically identifiable DCs, but control-transduced GMPs did not. We next determined whether the DCs produced from GMP cultures were more similar to conventional, Flt3L-derived, or inflammatory GM-CSF–derived DCs.43 Phenotypically, all IRF8-GMP DCs expressed Mac-3 at similar levels as DCs derived from GM-CSF cultures (supplemental Figure 8E). Functionally, however, these cells produced only low levels of nitric oxide at the highest doses of LPS stimulation (supplemental Figure 8F), consistent with conventional Flt3L-dependent DCs. Morphologically, cells from IRF8-GMP cultures were heterogeneous and contained cells representative of both conventional and monocyte-derived DC subtypes (supplemental Figure 8G). These data suggest that while GM-CSF could be driving the surface expression of Mac-3, IRF-8 regulates the development of both conventional and a limited number of monocyte-derived DC under these culture conditions. Regardless, it is clear that IRF-8 is driving DC development in GMP cultures. These data show that IRF-8 is sufficient to block neutrophil development and induce DC commitment at multiple progenitor cell stages.
IRF-8 regulates distinct transcriptional programs in lymphoid and myeloid progenitors
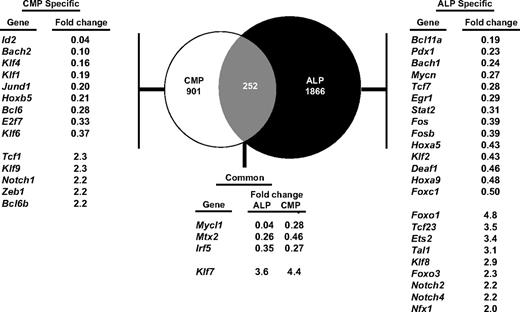
Because DCs can be generated from both lymphoid and myeloid pathways, and because Irf8 deficiency causes similar defects in ALPs and common myeloid progenitors, we hypothesized that IRF-8 controls similar genetic programs in lymphoid and myeloid progenitors. To test this, we performed microarray analysis using Irf8+/+ and Irf8−/− common myeloid progenitors and ALPs. We identified genes that were at least 2-fold differentially expressed between Irf8-deficient ALPs or common myeloid progenitors compared with wild-type cells (supplemental Tables 2-3). We then quantified the extent of overlap of these IRF8–dependent genes between the myeloid and lymphoid pathways. Surprisingly, the majority of IRF-8–dependent genes were unique to lymphoid or myeloid progenitors, as only 252 genes were coordinately dysregulated in both ALP and common myeloid progenitor (Figure 7). Of these genes, only a small number of known and putative transcription factors were identified. None of these genes are known to have effects on neutrophil or DC development. These data suggest that although IRF-8 mediates similar developmental functions in lymphoid and myeloid progenitors, it does so through distinct transcriptional programs.
IRF-8 regulates distinct transcriptional programs in myeloid and lymphoid progenitors. ALPs or common myeloid progenitors (CMPs) were double-sorted from Irf8+/+ or Irf8−/− mice and gene expression profiling was carried out by microarray analysis. Venn diagram shows genes that are specifically modulated in the common myeloid progenitor or ALP pathways, as well as genes that are coordinately dysregulated in both pathways. The genes listed in the tables show known transcription factors. Numbers represent fold changes in Irf8−/− compared with Irf8+/+ cells.
IRF-8 regulates distinct transcriptional programs in myeloid and lymphoid progenitors. ALPs or common myeloid progenitors (CMPs) were double-sorted from Irf8+/+ or Irf8−/− mice and gene expression profiling was carried out by microarray analysis. Venn diagram shows genes that are specifically modulated in the common myeloid progenitor or ALP pathways, as well as genes that are coordinately dysregulated in both pathways. The genes listed in the tables show known transcription factors. Numbers represent fold changes in Irf8−/− compared with Irf8+/+ cells.
To gain insight on the mechanism of IRF-8 function in lymphoid and myeloid progenitors, we identified several transcription factors whose expression was dependent on IRF-8 in either the common myeloid progenitor or the ALP pathway. In common myeloid progenitors, IRF-8 is required for the proper expression of Klf4, which has been shown to limit neutrophil production44 ; of Id2, which is required for the development of CD8α+ DCs29 ; and of Bcl6, which is required for the development of both CD4+ and CD8α+ DCs.45 In ALP, IRF-8 was required for the proper expression of Tcf7 (also known as Tcf1), which was recently shown to inhibit myeloid and B-lineage commitment.46
Discussion
Classically, hematopoietic differentiation is thought to be a fixed and linear progression from one progenitor to the next, until development results in the formation of a single mature subset. This is largely based on the placement of progenitors in a logical order based on the differences and similarities of their developmental restrictions, rather than by direct evidence of one progenitor leading to the finite development of another progenitor. While this provides a useful and tangible model of development, progenitor cells that have partially overlapping developmental potentials disrupt this paradigm, and until recently, have provided grounds for arguing the nonexistence of other progenitors. The first major branch in this pathway is thought to be the CLP and common myeloid progenitor, which yield all lymphoid and myeloid cell, subsets, respectively.1,2 However, the discovery of the LMPPs, which were presumed to lie upstream of the common myeloid progenitor/CLP branchpoint, yet lacked some of the developmental potential imparted by common myeloid progenitor, suggested an inconsistency in a linear developmental model.4,47 As a result of these and other studies, the existence of the common myeloid progenitors was questioned because common myeloid progenitors no longer logically fit into the hierarchical model. However, common myeloid progenitors and CLPs were found to potentiate the development of all cDC and pDC subsets.7,8,12,15 Furthermore, Gata1 and PU.1-based reporter mice were used to demonstrate the simultaneous existence of both lymphoid-myeloid progenitors and myeloid-erythroid progenitors.3 These data provided evidence that progenitor cells with overlapping developmental potential do, in fact, exist. Since this discovery, other progenitor cell subsets have been described that do not easily fit into the classic model of hematopoietic differentiation because of the fact that they have some common developmental potential, but are in nonlinear positions within the developmental hierarchy. One recent example of this is the discovery of MDP.5 Like GMP, MDP yield macrophages, yet they retain DC potential and cannot generate neutrophils. Thus, these subsets cannot be linked in a simple precursor-progeny relationship.
The data described in our study provide a mechanism whereby progenitor cells with overlapping developmental potential can exist. IRF-8 is induced asynchronously in the lymphoid and myeloid pathways, where it restricts neutrophil potential, a process necessary for DC commitment. Furthermore, our data suggest that IRF-8 could induce DC development, by either directly or indirectly activating genes necessary for DC commitment. This is supported by the finding that IRF-8 expression instructed GMPs to produce DCs. The fact that IRF-8 induced DC development from this subset also suggests that there is not necessarily an obligatory order in which fate restrictions must be placed on cells.
The next question that arose from the discovery of progenitor cells with partially overlapping developmental potential was whether the cells developing from distinct developmental pathways represented the same, or functionally distinct, cell subsets. One study analyzing human cDCs and pDCs developing from either common myeloid progenitors or CLPs found that these subsets exhibited similar gene expression profiles, implying functional similarity.13 While other studies have suggested subtle functional differences between CLP-derived and common myeloid progenitor–derived pDCs,48 these cells are clearly similar at the global transcriptional level.13,15 Based on the similarities in mature DCs derived from lymphoid and myeloid progenitors, we initially hypothesized that IRF-8 would induce critical transcriptional events common to both the lymphoid and myeloid pathways. Surprisingly, however, our data instead provide evidence that distinct transcriptional events occur as lymphoid and myeloid progenitors initially commit to the DC lineage, although IRF-8 is induced and similar mature DCs are produced from both pathways. These data are consistent with previous studies demonstrating that the differential order of transcription factor induction can lead to distinct outcomes.49
Previous studies have shown that CD8α− DC development is normal in Irf8−/− mice.34 However, our studies suggest that there is a reduction in the efficiency of CD8α− DC development from Irf8−/− mice, likely resulting from the reduction in the development of the upstream CDPs. Given that this deficiency is only readily observed in competitive settings, we propose that homeostatic expansion of CD8α− DC-committed progenitors occurs in Irf8−/− mice in the absence of competition for extrinsic factors such as Flt3L.23 The stage at which commitment to the CD8α− DC lineage occurs is thought to be at the pre-cDC stage20 ; however, the heterogeneity in functional outcomes of single CDPs suggests that some of these cells may already be committed to a particular DC subset.18,19 Whether a functional subset of the CDP population is preferentially affected by Irf8 deficiency remains to be determined.
IRF-8 has also been shown to be important for some human myeloid immunodeficiencies and leukemias. A recent study showed that patients with complete deficiencies in monocytic and/or DC compartments had 2 distinct point mutations found in the IRF-8 DNA-binding domain, presumably blocking binding to target genes.50 Although these data would seem to conflict with phenotypes observed in mice with Irf8 deficiency, which clearly still retain CD8α− DCs, our data demonstrate that even in mice, IRF-8 does play a role in the generation of all conventional DCs in part by regulating initial lineage commitment. Our data thus suggest that IRF-8 mutations could play a causative role in human immunodeficiencies and leukemia, at least in part, by dysregulating progenitor cell, and subsequently, mature DC and neutrophil development.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank O. Malkova and D. Atibalentja for assistance with flow cytometry, L. Herzenberg for providing the ThB α-Ly6d hybridoma, B. T. Edelson for assistance with the nitric oxide assays, K. Murphy and W. KC for IRF-8–expressing retrovirus, and K. Ozato for initially providing Irf8-deficient mice.
This work was supported by National Institutes of Health grants K01DK078318 (D.B.) and T32CA009547 (A.M.B.). The authors thank the Alvin J. Siteman Cancer Center at Washington University School of Medicine and Barnes-Jewish Hospital (St Louis, MO), for the use of the Molecular and Genomics Analysis Core and Genome Technology Access Center, which provided microarray hybridization service. The Siteman Cancer Center is supported in part by National Cancer Institute (NCI) Cancer Center Support Grant P30 CA91842.
National Institutes of Health
Authorship
Contribution: A.M.B. designed and executed experiments, analyzed data, and wrote the manuscript; D.G.M. designed and executed experiments and analyzed data; A.T.S. executed experiments; R.S. and H.S. initially contributed Irf8−/− BM and spleens and revised the manuscript; and D.B. supervised the project, analyzed data, and revised the manuscript.
Conflict-of-interest disclosure: H.S. is employed by Genentech. The remaining authors declare no competing financial interests.
The current affiliation for D.G.M. is Department of Computer Science and Engineering, Washington University School of Medicine, St Louis, MO. The current affiliation for R.S. is Department of Surgery, University of Chicago, Chicago, IL. The current affiliation for H.S. is Department of Discovery Immunology, Genentech, South San Francisco, CA.
Correspondence: Deepta Bhattacharya, Department of Pathology and Immunology, Washington University School of Medicine, St Louis, MO 63110; e-mail: deeptab@pathology.wustl.edu.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal