Abstract
Routine incorporation of FISH into multiple myeloma (MM) diagnostic testing has led to a better appreciation of the heterogeneity of genetic abnormalities associated with this disease. We studied a group of 484 patients with newly diagnosed symptomatic MM to better understand the prevalence of the various abnormalities and the prognostic significance of the overlapping abnormalities. A translocation involving the IgH locus and 1 of the 5 recurrent partner chromosomes was seen in 161 (33%) patients, and 275 (57%) had trisomy of at least 1 odd-numbered chromosome. High-risk FISH, defined as the presence of t(4;14), t(14;16), t(14;20), or loss of P53, was seen in 115 (24%) patients; the median overall survival for this group was 3.9 years, compared with “not reached” for standard-risk patients (P < .001). Among the patients with high-risk FISH, 49 patients who also had at least 1 trisomy had a median overall survival that was not reached, compared with 3 years for high-risk patients without a concurrent trisomy (P = .01). Based on the current findings, we conclude that the presence of trisomies in patients with t(4;14), t(14;16), t(14;20), or p53 deletion abnormalities in MM ameliorates the usual adverse impact associated with these prognostic markers.
Introduction
Studies over the past decade have revealed numerous overlapping and nonoverlapping genetic abnormalities in the myeloma cell and have elucidated their impact on patient outcome.1–4 Given the low proliferative nature of the malignant plasma cell, conventional metaphase cytogenetics reveal the presence of karyotypic abnormalities in only a small number of multiple myeloma (MM) patients.5,6 With the advent of FISH studies and with the increasing number of probes used for the study of various abnormalities, it has become clear that nearly all patients with MM have one or more abnormalities that can be detected by this methodology.3,7 Currently, MM patients are broadly grouped into a non-hyperdiploid group, in which the majority have a translocation involving the IgH locus on chromosome 14 and 1 of the 5 recurrent translocation partners (on chromosomes 4, 6, 11, 16, or 20), or into a hyperdiploid group,1,2 which is typically characterized by trisomies of 1 or more of the odd-numbered chromosomes 3, 7, 9, 11, 15, or 17. Other abnormalities, such as deletions involving chromosome 1, monosomy/deletion of chromosome 17 (which leads to the loss of the p53 gene), monosomy of chromosome 13 or interstitial deletion (which involves chromosome 13q), and abnormalities involving the myc locus, are often considered to be secondary abnormalities that increase in prevalence with disease evolution. These abnormalities often overlap with each other or with any one of the primary cytogenetic abnormalities.8–12
Prior studies have shown that abnormalities such as t(4;14), t(14;16), t(14;20), and del 17p predict for significantly shortened survival in patients with newly diagnosed MM, whereas hyperdiploidy has been associated with better survival.3,4,10,12–16 However, the prognostic impact of overlapping primary cytogenetic abnormalities is unclear, especially the concurrent presence of trisomies and translocations. To address this issue, we studied a large group of patients with newly diagnosed MM who were seen at our institution and who had complete FISH studies available.
Methods
Patients
We identified 500 patients with MM who were seen at the Mayo Clinic within 90 days of their diagnosis. Only patients who had BM FISH studies performed within 1 year before or 6 months after their diagnosis were included in the study. Among this group, 16 patients did not have sufficient plasma cells observed during the FISH analysis and were excluded from the analysis. The patients received a variety of different treatments depending on the prevailing standard practice at the time of their diagnosis. A regimen containing at least 1 of the novel agents (ie, thalidomide, lenalidomide, or bortezomib) was used for initial therapy in 78% of the patients. The study was approved by the Mayo Clinic Institutional Review Board and was done in accordance with the Declaration of Helsinki.
FISH Studies
Aspirate samples were enriched for mononuclear cells using the Ficoll method and cytospin slides were prepared. FISH analysis was performed as described previously using the following probes: 3cen (D3Z1), 7cen (D7Z1), 9cen (D9Z1), 15cen (D15Z4), 11q13 (CCND1-XT), 14q32 (IGH-XT), 13q14 (RB1), 13q34 (LAMP1), 14q32 (5′IGH,3′IGH), 17p13.1 (p53), and 17cen (D17Z1).6 The specificity of the detection process was improved with immunofluorescent detection of the cytoplasmic Ig light chain in the plasma cells, as described previously. Patients were considered to have high-risk disease if FISH studies demonstrated one of the following abnormalities: t(4;14), t(14;16), t(14;20), or loss of the p53 gene locus (del 17p or monosomy 17; described on www.msmart.org).15,17 Patients with any of the other abnormalities or a normal FISH were considered to have standard-risk MM.
Statistical analysis
The Fisher exact test was used to test differences in nominal variables. Differences in continuous variables between groups were compared using Wilcoxon signed-rank test. Overall survival (OS) was defined as the time from diagnosis to death, with patients alive at the time of last follow-up censored at that date. Survival curves were constructed according to the Kaplan-Meier method and compared using the log-rank test. All analyses were performed using JMP Version 9.0 software (SAS Institute).
Results
The current analysis includes 484 patients diagnosed with MM between January 1, 2004 and December 31, 2009. The median age of the patients at diagnosis was 65 years (range, 22-91) and 60% were male. The median estimated follow-up time for the entire group was 3 years from diagnosis (95% confidence interval [95% CI], 2.8-3.2), with 358 (74%) patients alive at the time of this analysis.
Distribution of abnormalities
No abnormality was found by FISH testing in 15 patients (3%), and the remaining 469 patients had 1 or more abnormalities. The overall frequencies of the common abnormalities are shown in Table 1. One-third of the patients had 1 of the 5 recurrent translocations involving the IgH region, with the common abnormalities being t(11;14), t(4;14), and t(14;16), seen in 18%, 10%, and 5% of the patients, respectively. In addition, 59 (12%) patients had an abnormality involving the IgH locus that might represent a translocation involving a chromosome other than the 5 recurrent translocation partners or a germline abnormality, and are therefore referred to herein as “other IgH abnormality”). Trisomy of at least 1 of the odd-numbered chromosome (3, 7, 9, 11, 13, 15, or 17) was observed in 275 (57%) patients, and 233 (48%) patients had trisomy of at least 2 of the odd-numbered chromosomes, which is conventionally termed as hyperdiploidy.18 The most commonly observed trisomy was that of chromosome 9 (42%), followed by those of chromosomes 15 (37%), 11 (36%), 3 (33%), and 7 (27%). Monosomy or deletion involving chromosome 13 was seen in 228 (47%) patients, and loss of the p53 gene either because of deletion involving chromosome 17p or monosomy of chromosome 17 was seen in 62 (13%) patients. In addition, we observed monosomy of chromosome 14 in 38 (8%) patients and monosomy of chromosome 16 in 14 (3%) patients; 31 and 9 of these patients, respectively, also had a concurrent monosomy of chromosome 13. Finally, there were 3 patients with none of the above abnormalities, but who had a tetraploid clone.
Distribution of various abnormalities of FISH testing and number of patients with overlapping trisomies
| FISH abnormality . | Frequency, n (%) . | Trisomy also present, n (%)* . |
|---|---|---|
| Any translocations | 220 (46) | |
| t(11;14) | 86 (18) | 12 (14) |
| t(4;14) | 47 (10) | 19 (40) |
| t(14;16) | 24 (5) | 5 (21) |
| t(6;14) | 3 (< 1) | 3 (100) |
| t(14;20) | 1 (< 1) | 0 |
| Other IgH locus abnormality | 59 (12) | 35 (60) |
| Any trisomy | 275 (57) | NA |
| 1 chromosome | 42 (9) | NA |
| 2 chromosomes | 52 (11) | NA |
| 3 chromosomes | 55 (11) | NA |
| 4 chromosomes | 66 (14) | NA |
| >‖= 5 chromosomes | 60 (12) | NA |
| Monosomy | 236 (49%) | |
| Monosomy 13/Del 13q | 228 (47) | 98 (43) |
| Monosomy 14 | 38 (8) | 11 (29) |
| Monosomy 16 | 14 (3) | 6 (43) |
| P53 abnormality | 62 (13) | |
| Del 17p | 49 (10) | 26 (53) |
| Monosomy 17 | 13 (3) | 5 (38) |
| Other (all tetraploidy) | 3 (< 1%) | NA |
| Normal | 15 (3%) | NA |
| FISH abnormality . | Frequency, n (%) . | Trisomy also present, n (%)* . |
|---|---|---|
| Any translocations | 220 (46) | |
| t(11;14) | 86 (18) | 12 (14) |
| t(4;14) | 47 (10) | 19 (40) |
| t(14;16) | 24 (5) | 5 (21) |
| t(6;14) | 3 (< 1) | 3 (100) |
| t(14;20) | 1 (< 1) | 0 |
| Other IgH locus abnormality | 59 (12) | 35 (60) |
| Any trisomy | 275 (57) | NA |
| 1 chromosome | 42 (9) | NA |
| 2 chromosomes | 52 (11) | NA |
| 3 chromosomes | 55 (11) | NA |
| 4 chromosomes | 66 (14) | NA |
| >‖= 5 chromosomes | 60 (12) | NA |
| Monosomy | 236 (49%) | |
| Monosomy 13/Del 13q | 228 (47) | 98 (43) |
| Monosomy 14 | 38 (8) | 11 (29) |
| Monosomy 16 | 14 (3) | 6 (43) |
| P53 abnormality | 62 (13) | |
| Del 17p | 49 (10) | 26 (53) |
| Monosomy 17 | 13 (3) | 5 (38) |
| Other (all tetraploidy) | 3 (< 1%) | NA |
| Normal | 15 (3%) | NA |
As a proportion of the primary abnormality.
NA indicates not applicable.
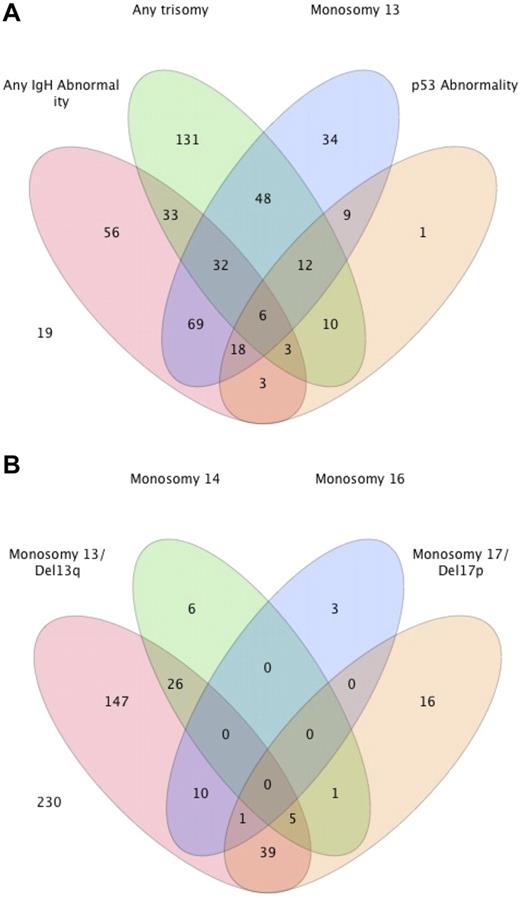
We then examined the overlap between the different abnormalities in individual patients, and the results are shown in Figure 1A. Because there was no overlap between patients with 1 of the 5 common IgH translocations and the presence of another IgH abnormality, we grouped them together. Similarly, patients with trisomy of any of the odd-numbered chromosomes were grouped together. Overall, 74 (15.3%) patients had a concurrent IgH translocation/abnormality and a trisomy. Monosomy 13/Del13q was seen in 57% of patients with an IgH abnormality, compared with 36% of patients with a trisomy, and 43 (9%) patients had monosomy 13/Del13q without either an IgH abnormality or trisomy. Similarly, a p53 abnormality (monosomy 17/Del17p) was seen in 14% and 11% of the groups of patients with a translocation and any trisomy, respectively, with only 1 patient having a p53 abnormality in the absence of an IgH abnormality or trisomy. The overlap between patients with one or more of the monosomies/deletions are shown in Figure 1B. The majority of the patients with a monosomy 14 or monosomy 16 had concurrent monosomy 13/Del 13q. In comparison, no overlap was seen between monosomy 14 and monosomy 16, and minimal overlap existed between monosomy 14 and the presence of any IgH abnormality (data not shown).
Distribution of various genetic abnormalities among patients with MM. (A) Venn diagram demonstrating the overlapping nature between the common abnormalities seen with FISH in patients with newly diagnosed MM. The actual number of patients with different abnormalities is presented from among 484 patients. The remaining 19 patients either had a normal FISH (n = 15) or another abnormality (n = 4). (B) Distribution of various monosomies/deletions and their overlap.
Distribution of various genetic abnormalities among patients with MM. (A) Venn diagram demonstrating the overlapping nature between the common abnormalities seen with FISH in patients with newly diagnosed MM. The actual number of patients with different abnormalities is presented from among 484 patients. The remaining 19 patients either had a normal FISH (n = 15) or another abnormality (n = 4). (B) Distribution of various monosomies/deletions and their overlap.
Prognostic relevance of overlapping abnormalities
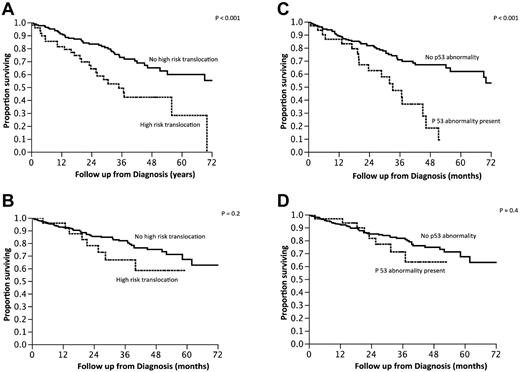
We then examined the outcome of patients based on the commonly used criteria for high-risk MM: the presence of t(4;14), t(14;16), t(14;20), or p53 deletion (monosomy 17 or deletion 17p). High-risk FISH was seen in 115 (24%) patients, and the median OS of this group was 3.9 years (95% CI, 2.9-5.8) compared with median not reached for the standard-risk group of patients (P < .001; Figure 2A). Given the observed overlap between the presence of trisomies and other types of abnormalities and the good prognosis generally associated with hyperdiploidy, we further grouped the patients with high-risk FISH into those with and without a concurrent trisomy of one or more of the odd-numbered chromosomes. The median OS for patients with high-risk FISH without concurrent trisomy (n = 66) was 3 years (95% CI, 2.2-4.3), compared with median not reached for those high-risk patients who also had a trisomy (n = 49; P = .01; Figure 2B). Conversely, when the impact of the current high-risk stratification was examined within the group of patients with a trisomy (n = 275), there was no difference in the OS between the high-risk and standard-risk groups (Figure 2C). The beneficial effect of trisomies was seen irrespective of whether patients were classified as high risk based on the presence of p53 abnormalities or the presence of the high-risk translocations (Figure 3A-D). We also examined whether any one of the odd chromosomes had more impact compared with the others, but did not find any such unique relationship (data not shown).
Kaplan-Meier curves demonstrating OS from diagnosis based on various risk factors. (A) Comparison of OS between patients with standard-risk MM (n = 370) based on FISH testing with those with high-risk MM (n = 114). (B) Comparison of OS between those with standard-risk MM (n = 370), high-risk MM with any trisomy (n = 48), and high-risk MM without any concurrent trisomy (n = 66). (C) Comparison of OS among patients with any trisomy (n = 275) with or without high-risk FISH features.
Kaplan-Meier curves demonstrating OS from diagnosis based on various risk factors. (A) Comparison of OS between patients with standard-risk MM (n = 370) based on FISH testing with those with high-risk MM (n = 114). (B) Comparison of OS between those with standard-risk MM (n = 370), high-risk MM with any trisomy (n = 48), and high-risk MM without any concurrent trisomy (n = 66). (C) Comparison of OS among patients with any trisomy (n = 275) with or without high-risk FISH features.
Kaplan-Meier curves demonstrating OS from diagnosis based on the presence or absence of high-risk IgH translocations or P53 loss. (A-B) Survival of patients with high-risk IgH translocations in the absence (A) or presence (B) of trisomies. (C-D) OS of patients with P53 loss in the absence (A) or presence (B) of trisomies.
Kaplan-Meier curves demonstrating OS from diagnosis based on the presence or absence of high-risk IgH translocations or P53 loss. (A-B) Survival of patients with high-risk IgH translocations in the absence (A) or presence (B) of trisomies. (C-D) OS of patients with P53 loss in the absence (A) or presence (B) of trisomies.
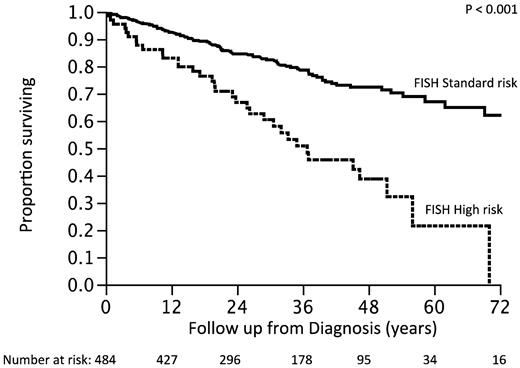
We then reclassified the patients into a new high-risk group, moving those patients previously classified as high-risk but with a trisomy into the standard-risk group. The survival of patients reclassified as high-risk (n = 66; 14%) was 3 years compared with not reached for the standard-risk group (P < .001; Figure 4). The hazard ratio associated with the high-risk status was 2.2 (95% CI, 1.5-3.2) with the older risk stratification and 2.9 (95% CI, 1.9-4.2) with the revised model.
Survival of patients according to the revised classification. Previous standard plus high-risk with trisomies (new FISH standard-risk) versus high-risk with no trisomies (new FISH high-risk)
Survival of patients according to the revised classification. Previous standard plus high-risk with trisomies (new FISH standard-risk) versus high-risk with no trisomies (new FISH high-risk)
Classification of FISH abnormalities
The current FISH classification broadly groups patients into a hyperdiploid group and a non-hyperdiploid group that includes all translocations and other abnormalities. Given the pattern of overlapping abnormalities and the good prognostic effect of trisomies even in the presence of conventional high-risk abnormalities, we reclassified the FISH abnormalities seen in MM into mutually exclusive groups in terms of the common genetic abnormalities (Table 2). Based on the results of this study, we have reclassified patients into those with: (1) trisomies with no concurrent IgH abnormalities; (2) IgH locus abnormalities including translocations with the 5 common recurring partner chromosomes; (3) both an IgH abnormality and trisomy; (4) monosomy 14 without trisomy or IgH abnormalities; (5) other abnormalities; and (6) a normal FISH. The “other abnormalities” group consists primarily of patients with chromosome 13 abnormalities and loss of p53, abnormalities that are thought to have later onset and also can be seen within any of the other 4 categories.
Nonoverlapping classification of primary molecular cytogenetic abnormalities in MM
| FISH abnormality . | Frequency (%) . |
|---|---|
| Trisomy(ies) without IgH abnormality | 201 (42%) |
| IgH abnormality without trisomy(ies) | 146 (30%) |
| t(11;14) | 74 (18) |
| t(4;14) | 28 (10) |
| t(14;16) | 19 (5) |
| t(14;20) | 1 (< 1) |
| Unknown partner/ deletion of IgH region | 24 (5) |
| IgH abnormality with trisomy(ies) | 74 (15%) |
| t(11;14) | 12 (18) |
| t(4;14) | 19 (10) |
| t(14;16) | 5 (5) |
| t(6;14) | 3 (< 1) |
| Unknown partner/deletion of IgH region | 35 |
| Monosomy 14 in absence of IgH translocations or trisomy(ies) | 22 (4.5%) |
| Other cytogenetic abnormalities in absence of IgH translocations or trisomy(ies) or monosomy 14* | 26 (5.5%) |
| Normal | 15 (3%) |
| FISH abnormality . | Frequency (%) . |
|---|---|
| Trisomy(ies) without IgH abnormality | 201 (42%) |
| IgH abnormality without trisomy(ies) | 146 (30%) |
| t(11;14) | 74 (18) |
| t(4;14) | 28 (10) |
| t(14;16) | 19 (5) |
| t(14;20) | 1 (< 1) |
| Unknown partner/ deletion of IgH region | 24 (5) |
| IgH abnormality with trisomy(ies) | 74 (15%) |
| t(11;14) | 12 (18) |
| t(4;14) | 19 (10) |
| t(14;16) | 5 (5) |
| t(6;14) | 3 (< 1) |
| Unknown partner/deletion of IgH region | 35 |
| Monosomy 14 in absence of IgH translocations or trisomy(ies) | 22 (4.5%) |
| Other cytogenetic abnormalities in absence of IgH translocations or trisomy(ies) or monosomy 14* | 26 (5.5%) |
| Normal | 15 (3%) |
These included primarily monosomy 13 and p53 abnormalities.
Discussion
The introduction of FISH testing and its routine incorporation into the evaluation of MM has resulted in a greater understanding of MM biology in the past decade.2,3,12 There is no doubt that genetic abnormalities are the main drivers of the significant heterogeneity seen in this disease in terms of the clinical features, response to therapy, and the eventual survival outcomes. Whereas more sophisticated and advanced technologies, such as gene-expression profiling using high-density oligonucleotide arrays, array comparative genomic hybridization, and, more recently, whole-genome sequencing of the tumor cells, have continued to unravel the mysteries of the MM genome, FISH-based risk classification remains the mainstay of clinical evaluation and is a practical approach to risk assessment.14,19,20
Overall, the frequency of the different abnormalities found in the present study is very similar to that reported by Avet-Lousseu et al in a large number of MM patients included in the Intergroupe Francophone du Myelome studies, reflecting consistency with the prior reported frequencies.3 However, the present study makes several unique observations that have profound clinical implications in terms of risk assessment and decisions regarding therapeutic approaches, as well as for our understanding of the pathogenesis of MM. We show that approximately 10% of patients are in an overlapping category, with the presence of both IgH translocations and trisomies, and that this overlap has a significant impact on outcome. FISH-based risk assessment has been criticized for the significant heterogeneity in outcome among high-risk patients defined by the presence of t(4;14), t(14;16), t(14; 20), and deletion of 17p. Previous studies have suggested that high-risk features such as t(4;14) may be further classified using other prognostic markers such as B2M or hemoglobin levels, thus explaining the heterogeneity (although this was not confirmed in a second study).21,22 Our present results suggest that the heterogeneity in patient outcome may be explained by the presence of the overlap between different genetic abnormalities.
The findings herein may also impact treatment decisions. Several studies have shown that treatment with bortezomib may improve the outcome of patients with the t(4;14) abnormality, but this has not been a uniform finding.23,24 It is possible that the therapeutic benefit of bortezomib may be restricted to one of the subgroups and thus may explain the heterogeneity seen between the different studies. However, the relatively small number of patients in the present study, and specifically the low numbers who received bortezomib-based therapies, precludes this analysis. Clearly, this question merits evaluation in the larger datasets, such as that of the Intergroupe Francophone du Myelome, which showed a benefit for bortezomib.
Hyperdiploidy has typically been considered to result from trisomies of odd-numbered chromosomes and has been demonstrated to be a good prognostic feature.2,9,25 However, when examined using conventional cytogenetics or flow-based ploidy evaluation, trisomies of 1-3 chromosomes can often be “hidden” by the frequent presence of monosomies, especially those involving chromosomes 13 and 14. With FISH testing, patients are termed hyperdiploid only when trisomies of 2 or more chromosomes are observed, and trisomy indices have been developed using specific trisomies, such as those involving 9, 11, and 15, to identify those with hyperdiploid MM.18 The results of the present study suggest that it is not necessarily the presence of “hyperdiploidy” that imparts a favorable prognosis, but rather trisomies of one or more of the odd-numbered chromosomes. This raises a question as to whether we should emphasize hyperdiploidy (ie, numeric excess on karyotype studies) or the presence of trisomies as the primary pathogenetic marker that is found more often but not exclusively in the non-IgH–translocated subtype of MM. The frequency of trisomies of different chromosomes seen in the present study is similar to that reported previously in FISH-based studies.4,26 Interestingly, a good prognostic impact of trisomies was also reported previously by Perez-Simon et al in the context of trisomies involving chromosomes 6 and 9.4
The underlying mechanisms for the development of trisomies in odd-numbered chromosomes in the MM cells and their favorable prognostic impact remain poorly understood. It is possible that the additional copies of these odd-numbered chromosomes may increase the copy number of gene loci critical for tumor suppression or of gene loci mediating drug sensitivity. Increased expression of genes from the trisomic chromosomes has been reported from gene-expression studies, suggesting that the impact of the trisomies may be mediated through a gene dose effect.25 For example, the tumor-suppressor genes p15 and p16 are present on 9p21 and play a role in MM cell proliferation because of their effect on cyclins and cyclin-dependent kinases.27 Detailed gene-expression studies of hyperdiploid patients have demonstrated subgroups within hyperdiploid MM that are characterized by overexpression of certain groups of genes and with differing survival outcomes.25 One cannot discount the possibility that the presence of trisomies is a surrogate marker for some hitherto undiscovered genetic abnormality that hampers tumor growth in some fashion. The presence of overlapping translocations and trisomies suggests that different, but not mutually exclusive, mechanisms or underlying characteristics contribute to their development. Clearly, this remains an area requiring further investigation.
Given the differences in outcome among patients with both trisomies and high-risk translocations compared with those without a trisomy, we should consider classifying patients into nonoverlapping categories, as shown in Table 2. Currently, we classify patients with any translocation as being in the “translocated group” irrespective of the presence of concurrent trisomies.2 However, if we take into consideration the results of the present study, a better approach would be to group patients into those with trisomy(ies), those with IgH abnormalities, those with both abnormalities, and those with monosomy 14 in the absence of other abnormalities. This would still exclude nearly 10% of patients in whom either the single abnormality is monosomy 13/Del13q and/or monosomy 17/Del17p or who have a normal FISH. The current concept is that trisomies and IgH abnormalities are the primary abnormalities present from the earliest stages of development of the monoclonal process, and monosomies and deletions involving chromosomes 13 and 17 occur later, with increasing prevalence during disease progression for chromosome 17 abnormalities. We have shown previously in a temporal analysis of various abnormalities in hyperdiploid myeloma that trisomies are likely the earliest events, followed by translocations and monosomies/deletions.28 The results of the present study placed in the context of this previous work suggests an evolutionary framework in which trisomies develop early on in the plasma cell evolution in more than one-half of the patients, followed by development of IgH translocations in nearly one-half of the patients, including some who have already developed a trisomy. This is likely followed by loss of or deletions involving chromosomes such as 13 and 17, events that continue to occur late into the course of the disease and can occur in all MM cells. Given this scenario, isolated monosomy 13 likely represents patients with neither an IgH abnormality nor trisomy who acquired this abnormality with disease evolution.
In conclusion, we have further refined the category of high-risk MM, identifying a group of patients with poor prognosis and removing some of the heterogeneity of the current stratification. In the present study, we have shown for the first time the beneficial impact of trisomies on patients otherwise considered to have high-risk MM, thus allowing for a more stringent definition of high-risk status among MM patients. Our data suggest a rethinking of the hierarchy of genetic findings in terms of risk stratification, with importance given to trisomies over the translocations, reflecting a shift in the current concepts. We are now able to suggest an alternate FISH-based classification in MM, enabling a better grouping for the future that will allow more accurate comparisons across the datasets. Based on the findings of this study, we recommend that the FISH testing panel should have the probes necessary to identify the presence of any 1 of the 5 recurrent IgH translocations, 17p deletion, trisomy of any of the odd-numbered chromosomes, and chromosome 13 abnormalities. These findings should be confirmed in other datasets that include patients undergoing different types of treatment strategies to better delineate the interaction between treatments and the presence of these abnormalities. The impact of the type of treatment on the findings herein cannot be analyzed with clarity given the different types of treatments used, but a significant number of patients received an immunomodulatory drug–based regimen. It is also unclear how the new “higher” risk patients relate to those identified as high risk using gene-expression profiling. Finally, future studies need to specifically examine whether the beneficial effects of therapies such as bortezomib are restricted to patients in one or the other group.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the Mayo Clinic Hematologic Malignancies Program, the National Institutes of Health (Paul Calabresi K12 Award CA96028 and grant P01 CA62242), the Predolin Foundation, the Mayo Clinic Cancer Center, and the Mayo Foundation.
National Institutes of Health
Authorship
Contribution: S.K. designed the study, collected and analyzed the data, and wrote the manuscript; S.V.R. contributed to writing the manuscript; R.F., A.D., M.Q.L., M.A.G., S.R.H., F.K.B., D.D., S.J.R., S.R.Z., J.A.L., R.A.K., L.B., and S.V.R. contributed patients and contributed to writing the manuscript; A.G. contributed to writing the manuscript; and R.P.K. and R.A.K. performed the FISH testing and contributed to writing the manuscript.
Conflict-of-interest disclosure: R.F. has received a patent for the prognostication of MM based on genetic categorization of the disease; has received consulting fees from Medtronic, Otsuka, Celgene, Genzyme, BMS, and AMGEN; and has sponsored research from Cylene and Onyx. S.K. has research support for clinical trials from Celgene, Millennium, Novartis, and Genzyme and is a consultant for Merck. A.D. and M.Q.L. received clinical trial support from Celgene. The remaining authors declare no competing financial interests.
Correspondence: Shaji Kumar, MD, Associate Professor of Medicine, Division of Hematology, Mayo Clinic, 200 First St SW, Rochester, MN 55906; e-mail: kumar.shaji@mayo.edu.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal