Abstract
The interactions between the bone marrow (BM) microenvironment and acute myeloid leukemia (AML) is known to promote survival of AML cells. In this study, we used reverse phase-protein array (RPPA) technology to measure changes in multiple proteins induced by stroma in leukemic cells. We then investigated the potential of an mTOR kinase inhibitor, PP242, to disrupt leukemia/stroma interactions, and examined the effects of PP242 in vivo using a mouse model. Using RPPA, we confirmed that multiple survival signaling pathways, including the phosphatidylinositol 3-kinase (PI3K)/protein kinase B (AKT)/mammalian target of rapamycin (mTOR), were up-regulated in primary AML cells cocultured with stroma. PP242 effectively induced apoptosis in primary samples cultured with or without stroma. Mechanistically, PP242 attenuated the activities of mTORC1 and mTORC2, sequentially inhibited phosphorylated AKT, S6K, and 4EBP1, and concurrently suppressed chemokine receptor CXCR4 expression in primary leukemic cells and in stromal cells cultured alone or cocultured with leukemic cells. In the in vivo leukemia mouse model, PP242 inhibited mTOR signaling in leukemic cells and demonstrated a greater antileukemia effect than rapamycin. Our findings indicate that disrupting mTOR/AKT signaling with a selective mTOR kinase inhibitor can effectively target leukemic cells within the BM microenvironment.
Introduction
Functional interplay between acute myeloid leukemia (AML) cells and the bone marrow (BM) microenvironment is a distinct feature of this hematologic malignancy. Several studies have provided evidence suggesting that proliferation, survival, and drug resistance of AML can be modulated by mesenchymal stem cells (MSCs) within the BM microenvironment.1-4 Direct contact between AML cells and BM-derived MSCs triggers a pleiotropic spectrum of proliferative and/or antiapoptotic signaling pathways, including the phosphatidylinositol 3-kinase (PI3K)/protein kinase B (AKT)/mammalian target of rapamycin (mTOR)5,6 pathway (PI3K/AKT/mTOR), which attenuates the response of AML to conventional chemotherapy. Thus, in addition to therapies that directly target AML, interruption of leukemia cell-MSC interactions should be considered when designing anti-AML therapeutic strategies.
mTOR is a critical component of PI3K/AKT signaling, forming 2 complexes—mTORC1 and mTORC2—that are defined by their molecular composition and substrate specificity. mTORC1 includes mTOR and raptor,7 whose downstream targets are the eukaryotic translation initiation factor 4E-binding proteins (4EBPs) and S6 kinases (S6K1 and S6K2). 4EBP1 phosphorylation by mTORC1 releases 4EBP1 from eukaryotic translation initiation factor 4E (eIF4E), allowing eIF4E to form the eIF4F complex that promotes cap-dependent mRNA translation. Phosphorylation of S6Ks by mTORC1 is an activating event that potentiates S6K-dependent phosphorylation of ribosomal S6 protein and other substrates that coordinate aspects of protein and lipid biosynthesis while opposing autophagy.8 In comparison, mTORC2 contains mTOR and rictor.9 It phosphorylates AKT at Ser473 and members of the AGC protein kinase family at hydrophobic motifs. These include protein kinase C isoforms and members of the glucocorticoid-induced kinase family.10
Rapamycin and its derivatives (RAD001 and CCI-779) are first-generation mTOR inhibitors which showed only modest efficacy in antitumor clinical trials.11 These compounds affect mTORC1 more than mTORC2, especially in the initial phase of treatment, resulting in increased AKT phosphorylation through blocking negative feedback loops that limit upstream signaling by PI3K.11,12 In addition, these agents do not completely inhibit mTORC1 activity and have little effect on phosphorylation of 4EBP1 at key threonine residues (Thr37/46), resulting in weak attenuation of cap-dependent translation and little effect on overall protein synthesis.13
PP242 is a new small-molecule protein kinase inhibitor that targets the adenosine triphosphate (ATP)–binding site of mTOR, resulting in greater inhibition of mTORC1 and mTORC2 activity than that produced by the mTOR inhibitors discussed above.14 Compared with the other PI3K/mTOR inhibitors, such as PI-103, PP242 is more selective for leukemic cells, as evidenced by its ability to suppress PI3K/AKT/mTOR signaling in Ph+ B-cell acute lymphoblastic leukemia15,16 and T-cell lymphoma cells,17 and prolongation the survival of mice harboring these leukemias.
Microenvironment-mediated chemoresistance of AML prompted us to investigate signaling pathways activated in leukemic cells on contact with stromal cells, and to study the antileukemia potency of mTOR kinase inhibitors under conditions mimicking the BM microenvironment. In this study, we report that stroma activates multiple antiapoptotic signaling through several protein-protein interactions that correlate with stroma-mediated survival in AML cells. We further show that the PP242 effectively inhibits the activity of mTORC1 and mTORC2 and their downstream targets in primary AML cells, inducing apoptosis in both primary AML blasts and CD34+ progenitor cells. Importantly, PP242 disrupts the stroma-leukemia interaction, antagonizing stroma-mediated survival by suppressing expression of CXC chemokine receptor type 4 (CXCR4) and down-regulating mTOR signaling, both in primary AML cells and in stromal cells. Furthermore, PP242 suppresses leukemia progression in a murine leukemia model driven by mutated FLT3 with constitutive activation of mTOR. Taken together, our findings indicate that the PP242 or its analogues are potentially useful therapeutic agents in AML, affecting leukemic cells within the protective BM microenvironment.
Methods
Cells and culture conditions
Peripheral blood and BM samples were obtained from patients with AML or acute lymphoblastic leukemia (ALL) after informed consent was obtained in accordance with The University of Texas MD Anderson Cancer Center Institutional Review Board regulations. Mononuclear cells were separated using Ficoll-Hypaque density gradient centrifugation (Sigma-Aldrich). Primary cells were cultured in RPMI 1640 containing 10% fetal bovine serum (FBS), 1% l-glutamine, and 1% penicillin-streptomycin. Mesenchymal stem cells (MSCs) obtained from healthy BM donors or from patients with leukemia were cultured at a density of 5000 to 6000 cells/cm2 in minimum essential medium (MEM)α supplemented with 20% FBS, 1% l-glutamine, and 1% penicillin-streptomycin as described elsewhere.18 The isolated, cultured MSCs at passage 3 comprised a single phenotypic population, as determined by flow cytometric analysis, positive for SH2 and SH3 and negative for markers of hematopoietic lineage, as described elsewhere.19
Passage 3 or 4 MSCs were used for coculture experiments. The murine stromal cell line MS-5 was provided by Dr Itoh20 (Niigata University, Japan) and maintained in the 10% FBS-containing MEMα.
Cell viability
Cell viability was measured using flow cytometry with CountBright absolute counting beads (Invitrogen). The percentage of apoptosis was estimated by measuring phosphatidylserine externalization in the cells using annexin V flow cytometry (Roche). Apoptosis of bulk leukemic and leukemic progenitor cells was measured using annexin V+ after electronic gating on CD45+ leukemic blasts (CD45-APC; BD Pharmingen) or CD34+ (CD34-APC; BD Pharmingen) AML progenitor cells. The extent of apoptosis was quantified as percentage of annexin V+ cells, and the extent of drug-specific apoptosis was assessed by the formula: % specific apoptosis = (test−control)/(100 − control) × 100. “Control” represent the percentage of annexin V positivity in the group of cells without treatment (spontaneous apoptosis), and “test” represents the percentage of annexin V positivity in the group of cells treated with PP242.
Flow cytometry for detection of the expression level of proteins and intracellular phospho-proteins
Murine whole blood (100 μL), harvested primary AML cells, and harvested AML MSCs were fixed with formaldehyde at a final concentration of 4% for 10 minutes at room temperature, permeabilized with 0.1% Triton X-100 at 37°C for 15 minutes, and then centrifuged at 1000g (Beckman Coulter) in cold 50% methanol for 3 minutes. The pellets were washed twice in ice-cold phosphate-buffered saline (PBS). Cells were then stained for cell surface markers and intracellular signaling markers at room temperature for 30 minutes and washed once with PBS before analysis using a Gallios flow cytometer (Beckman Coulter). The flow cytometric profiles were analyzed using the Kaluza (Beckman Coulter) or FlowJo (Version 7.6.1) software programs. The antibodies against cell surface markers CD34-Cy5 and CD90-APCAlexa750 were kindly provided by Beckman Coulter. The antibodies against CXCR4-phycoerythrin (PE; 12G5), CXCR4-APC (12G5), CXC4-PE (1D9), CD34-PE Cy7, CD38-PE Cy5, VLA4(CD49d)–APC, CD34-APC, and CD44-APC were purchased from BD biosciences. The antibodies against intracellular signaling markers phosphorylated 4EBP1 (p4EPB1; Thr37/43)–Alexa488, phosphorylated AKT (pAKT; Ser473)–Alexa684, and phosphorylated S6K (pS6K; Ser235/236)–Alexa684 were purchased from Cell Signaling Technology. Rapamycin derivative CCI-779 (temsirolimus) was purchased from LC Laboratories.
The mean fluorescent intensities (MFI) of protein markers were measured by flow cytometry. The MFI ratio of a protein marker in a specific sample is calculated as MFI of protein marker/ MFI of unstained cells or isotype stained cells. Statistical analysis of changes in MFI ratios of proteins in response to different treatment or culture settings were assessed using Pearson's correlation coefficient (ρ). The significance of the correlation was determined using Fisher z-transform with a false discovery rate (FDR) cutoff of 0.05.
Leukemia mouse model
All animal experiments were performed in accordance with protocols approved by the MD Anderson Institutional Animal Care and Use Committee. To evaluate the inhibitory effect of PP242 on mTOR in vivo, a mouse model of leukemia was generated using severe combined immunodeficient (SCID) mice. The mice were irradiated at 6 Gy and 6 hours later injected intravenously with Ba/F3-ITD/Luc/GFP cells (kindly provided by Dr D. Small, Department of Oncology, Johns Hopkins University School of Medicine, Baltimore MD)21,22 at a concentration of 0.2 × 106 cells/mouse. To compare the effects of PP242 and rapamycin (Sigma-Aldrich), 1 group of mice (9 mice per group) was injected with rapamycin intraperitoneally at a dose of 0.5 mg/kg every other day. Mice in the other group were given PP242 (60 mg/kg, via gavage, every other day) was initiated on day 3 after the tumor cells injection. Tumor-cell engraftment and progression were then monitored using noninvasive imaging as previously described,23 and flow cytometry to detect the GFP+ leukemia cells in murine peripheral blood samples. To assess the inhibitory effect of PP242 on mTOR signaling in GFP+ cells, murine peripheral blood was collected before and 2 hours after PP242 administration. Effects on mTOR signaling components pAKT (Ser473), and pS6K (Ser235/236) were analyzed using flow cytometry. Antitumor effects were compared using bioluminescent imaging.
Western blot analysis
For Western blot analysis of the inhibitory effects of PP242 on mTOR, primary AML samples were lysed in a phosphoprotein lysis buffer (150mM NaCl, 1mM MgCl2, 1mM CaCl2, 10mM NaF, 5mM sodium pyrophosphate, 10mM β-glycerophosphate, 1% Triton X-100, 10mM iodoacetamide, 1mM Na3VO4, 0.1% NaN3, 3mM phenylmethylsulfonyl fluoride). The buffer was supplemented with a protease inhibitor cocktail (Roche). Lysates were then separated on a 10% or 12% polyacrylamide gel, transferred to Hybond-P membranes (Amersham Pharmacia Biotech), probed with the appropriate antibodies, and visualized using an ECL plus kit (Amersham Pharmacia Biotech). Western blots were analyzed using a Storm 860 phosphorimager (Molecular Dynamics) and quantified by ImageJ (National Institutes of Health). Antibodies against human pAKT(Ser473), pAKT(Thr308), phosphorylated FoxO1a(Thr24)/FoxO3a(Thr32; pFoxO1a[Thr24]/FoxO3a[Thr32]), FoxO3a, pS6K(Ser235/235), phosphorylated PRAS40 (pPRAS40[Ser246]), p4EBP1(Thr37/46), AKT, S6K, and 4EBP1 were purchased from Cell Signaling Technology.
Reverse-phase protein array
Two AML cell lines (OCI-AML3 and U937) and 20 blood samples obtained from patients with primary AML(19) or ALL(1) with more than 50% blasts were cultured alone or cocultured with mouse stromal MS-5 cells for 24 hours. Cells were lysed in a protein lysis buffer (2 ×; 0.5M Tris-HCl, pH 6.8, 2% sodium dodecyl sulfate, 10% glycerol, 4% β-mercaptoethanol), then heated at 95°C for 5 minutes and stored at −80°C. To verify lack of significant contamination in collected leukemic cells, anti–human CD45-APC or CD34-APC were measured by flow cytometry to discriminate between human leukemic cells and mouse stroma cells (MS-5).
Proteomic profiling of the samples was performed using reverse-phase protein arrays (RPPA). The resulting comprehensive profiles included measures of 51 proteins (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article) representing the level of activation and expression of proteins in signal transduction pathways and apoptosis networks in response to different culture conditions. The methods for using and validating RPPAs were fully described previously.24 Briefly, protein lysates obtained from primary samples were printed onto slides, together with normalization and expression controls, in 5 serial dilutions. The slides were probed with strictly validated primary antibodies against total or phosphorylated protein (as described in the supplemental Table 1) and a secondary antibody was used to amplify the signal. The stained slides were scanned using a scanner and the images were analyzed using the MicroVigene Version 3.4 software program (VigeneTech) to produce quantified data.
The “supercurve” algorithm25 was used to estimate the sample-specific protein expression levels. The RPPA data were normalized by median centering the results for each sample across all antibodies. The expression levels of proteins were examined under 2 different culture settings (with or without MS-5), and the effect of culture setting changes in each protein was estimated using analysis of variance (ANOVA). To account for multiple testing, we fit the distribution of P values (raw P values) for the contrast tests with a β-uniform mixture (BUM) model26 and checked whether any proteins showed significant changes at an FDR of 0.05. For each protein, we also compared the number of samples showing detectable up-regulation with the number of samples showing detectable down-regulation using binomial tests (P < = .05), and then calculated significance using an FDR with a cutoff of 0.05. FDR was defined in “Flow cytometry.”
Quantitative real-time PCR
Total RNA was extracted using Trizol (Invitrogen) as directed by the manufacturer; cDNA was then prepared using SuperScript III primed by random hexamers as described by Andreeff et al.27 The level of expression of CXCR4 was assessed by real time polymerase chain reaction (PCR) using the following TaqMan gene expression assays in triplicate as directed by the manufacturer (Applied Biosystems): CXCR4 (Hs00607978_s1) and ABL1 (Hs00245445_m1). The abundance of CXCR4 transcript relative to that of ABL1 was determined by the ΔΔCt method with RQ Manager Version 1.2 software (Applied Biosystems).
Leukemia/MSC/MS-5 coculture
Primary leukemic cells were cocultured with either primary MSCs or MS-5 cells at a ratio of 100:1. In the direct cell-to-cell contact coculture setting, MS-5 cells or primary MSCs were plated in either 6-well plates or 25-cm2 culture flasks in 10% FBS-containing MEMα for 4 hours. Primary leukemic cells were then added on top of the stromal cells for 4 hours, after which cells were treated with PP242 for the indicated time interval. Nonattached leukemic cells were then harvested by collecting the floating cells and some of the detached cells after washing twice with cold 1× PBS. The viability of the collected cells was estimated, and the cells were lysed for Western blot analysis. In the transwell-based coculture setting, a transwell membrane with 0.4-μM pore diameter (Costar; Corning) was used to prevent suspended cells from direct cellular contact with MSCs. Primary AML MSCs were first seeded at a concentration of 3.5 × 104/0.5 mL on the backside of the transwell membrane for 4 hours. After attachment, the transwells were inverted, and the primary AML cells were seeded on top of the transwells. These transwells harboring the cells on both sides of the insert membrane were placed in 1 mL of 10% FBS-containing MEMα containing 10% FBS for 4 hours before initiating treatment. At the indicated time points, AML cells were harvested and lysed for Western blot analysis.
Results
Stroma up-regulates multiple survival signaling in primary leukemic cells
To investigate stroma-mediated signaling pathways in AML cells, we performed RPPA profiling of 51 proteins along with their phosphorylated forms in 19 primary AML, 1 ALL and 2 AML cell lines cultured alone or cocultured with murine stromal MS-5 cells commonly used for support of human hematopoietic stem cells.6 Coculture with MS-5 stromal cells significantly diminished spontaneous apoptosis of leukemic cells in 10 of 12 patient samples (AML cells alone, 33.7% ± 3.8% annexin V+ cells; AML+MS-5, 19.6% ± 3.1% annexin V+ cells; P = .027; supplemental Figure 1), consistent with our published data.5,6,23,24 We statistically compared the expression levels (average density) for each protein in 20 primary samples and 2 AML cell lines under 2 culture conditions (samples cultured alone versus samples cocultured with MS-5 cells); and the frequency of changes in protein expression in response to the coculture with stroma. Stroma up-regulated p4EBP1 (Thr37/46), p4EBP1(Thr70), pmTOR(Ser2448), pS6K(Ser235/236), and pS6K (Ser240/244) in majority of the samples, and significantly modified the phosphorylation of 4 proteins in leukemic cells in the coculture setting (Table 1). Specifically, it up-regulated pAKT (Thr308), phosphorylated extracellular signal-regulated pERK (Thr202/Tyr204), and pSTAT3 (Thr727), and down-regulated pβ-catenin (Ser33/37/Thr41) expression.
Comparison of the average protein-density changes and the frequency of protein-expression changes in leukemic samples cultured alone or cocultured with MS-5 cells
| Protein* . | Average density change† . | P‡ . | ↑Expression (n)§ . | ↓Expression (n)‖ . | P¶ . |
|---|---|---|---|---|---|
| pAKT (Thr308) | +0.880 | .001 | 18 | 2 | < .001 |
| pβ-catenin (Ser33/37/Thr41) | −0.830 | .002 | 3 | 15 | .035 |
| pERK (Thr202/Tyr204) | +0.660 | .002 | 14 | 3 | .039 |
| mTOR | NS | NS | 17 | 0 | < .001 |
| PRAS40 | NS | NS | 0 | 13 | < .001 |
| pSTAT3 (Ser727) | +0.298 | .013 | 12 | 2 | .039 |
| Protein* . | Average density change† . | P‡ . | ↑Expression (n)§ . | ↓Expression (n)‖ . | P¶ . |
|---|---|---|---|---|---|
| pAKT (Thr308) | +0.880 | .001 | 18 | 2 | < .001 |
| pβ-catenin (Ser33/37/Thr41) | −0.830 | .002 | 3 | 15 | .035 |
| pERK (Thr202/Tyr204) | +0.660 | .002 | 14 | 3 | .039 |
| mTOR | NS | NS | 17 | 0 | < .001 |
| PRAS40 | NS | NS | 0 | 13 | < .001 |
| pSTAT3 (Ser727) | +0.298 | .013 | 12 | 2 | .039 |
NS indicates not significant.
Proteins with significant density changes and/or frequent alteration in response to coculture.
Changes in the average protein density in all samples (see “Reverse-phase protein array” for details). The positive sign represents an increase, whereas the negative sign represents a decrease in the average protein density.
Adjusted P values (see “Reverse-phase protein array”). The cutoff P value for significance was ≤ .05.
Number of primary samples with up-regulation of expression of the indicated protein (please see “Reverse-phase protein array” for statistical analysis).
Number of primary samples with down-regulation of expression of the indicated protein.
Comparison of the numbers of samples with detectable up-regulation and down-regulation of protein expression using conditional binomial tests (P value) to confirm that alteration was heavily skewed (P ≤ .05).
Furthermore, 6 proteins were altered in the majority of the samples when cocultured with stroma. Coculture with stroma up-regulated pAKT (Thr308) in 18 of the 22 samples, mTOR in 17 samples, pERK (Thr202/Tyr204) in 14 samples, and pSTAT3 (Ser727) in 12 samples and decreased the expression of pβ-catenin (Ser33/37/Thr41) in 15 samples, and of mTORC1 substrate PRAS40,28 in 13 samples (Table 1). These findings indicate that stroma triggers activation of multiple parallel prosurvival signaling pathways in leukemic cells.
PP242 overcomes protection by stroma and induces apoptosis in primary AML cells
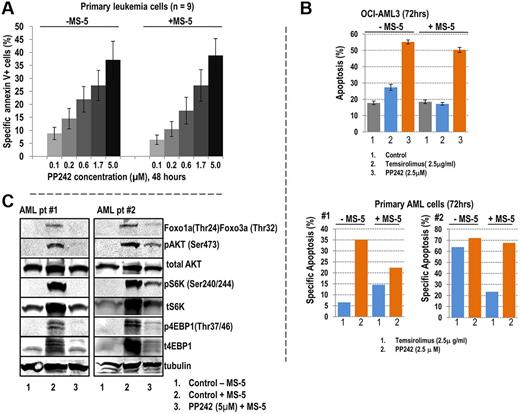
Because activation of pAKT (Thr308) and up-regulation of mTOR were seen in primary AML cells cocultured with stroma, we wanted to assess the antileukemic efficacy of PP242 under stromal coculture conditions. To this end, we cultured cells from 9 AML samples (supplemental Table 2) alone or cocultured with MS-5 cells, in the presence or absence of the indicated concentrations of PP242 for 48 hours. PP242 induced apoptosis in a dose-dependent manner in primary AML cells cultured alone or cocultured with stroma, as determined by annexin V flow cytometry (Figure 1A). In addition, PP242 induced apoptosis in CD34+ AML progenitor cells cultured under these conditions (supplemental Figure 2), although stroma exerted some protective effect in sample no. 2. We previously reported that rapamycin derivatives on prolonged exposure suppress both mTORC1 and mTORC2 signaling in AML.29 In direct comparison, PP242 has more potent inducer of apoptosis than rapamycin derivative temsirolimus in OCI-AML3 cells and in primary AML samples tested, in particular, under conditions of stromal coculture (Figure 1B). Taken together, these results indicate that blockade of mTOR activity with PP242 induces apoptosis in leukemic cells, and overcomes protective signals of stromal cells. Importantly, the significant apoptosis was observed even at lower concentration of PP242 in the range from 0.1μM to 0.6μM, suggesting that PP242 is highly selective for mTOR signaling in the cells.
PP242-induced apoptosis in primary AML cells. (A) Nine samples obtained from patients diagnosed AML with high blast count were treated with PP242 at indicated concentrations in the presence or absence of MS-5 stromal cells for 48 hours. Apoptotic cells were detected using annexin V+ flow cytometry. The percentage of specific annexin V+ apoptosis was calculated as described in “Cell viability.” Data represent average ± SEM of specific apoptosis in 9 AML samples. (B) Comparison of proapoptotic effects of PP242 and temsirolimus. OCI-AML3 cells (top panel) and 2 primary AML samples (bottom panel) were treated with PP242 and temsirolimus with or without MS-5 coculture for 72 hours, and the percentage of apoptotic cells (annexin V+/DAPI+/CD45+) was detected by flow cytometry. (C) AML cells from samples of patient no. 1 and no. 2 cocultured with MS-5 were treated with 2.5μM PP242 for 24 hours. Cells were harvested, and lysates subjected to immunoblotting with indicated antibodies to probe mTOR signaling.
PP242-induced apoptosis in primary AML cells. (A) Nine samples obtained from patients diagnosed AML with high blast count were treated with PP242 at indicated concentrations in the presence or absence of MS-5 stromal cells for 48 hours. Apoptotic cells were detected using annexin V+ flow cytometry. The percentage of specific annexin V+ apoptosis was calculated as described in “Cell viability.” Data represent average ± SEM of specific apoptosis in 9 AML samples. (B) Comparison of proapoptotic effects of PP242 and temsirolimus. OCI-AML3 cells (top panel) and 2 primary AML samples (bottom panel) were treated with PP242 and temsirolimus with or without MS-5 coculture for 72 hours, and the percentage of apoptotic cells (annexin V+/DAPI+/CD45+) was detected by flow cytometry. (C) AML cells from samples of patient no. 1 and no. 2 cocultured with MS-5 were treated with 2.5μM PP242 for 24 hours. Cells were harvested, and lysates subjected to immunoblotting with indicated antibodies to probe mTOR signaling.
We next examined effects of PP242 on intracellular signaling of AML cells cultured under conditions mimicking the BM microenvironment, that is in MS-5 coculture. Consistent with our RPPA findings, stroma stimulated PI3K/mTOR/AKT signaling by significantly up-regulating phosphorylation of AKT (Ser473), S6K (Ser240/244), and 4EBP1 (Thr37/46) in AML cells. PP242 exerted an inhibitory effect on mTORC1 and mTORC2 and partially abrogated stroma-induced activation of their direct downstream substrates (Figure 1C).
PP242 impairs interactions between primary AML cells and AML MSCs through inhibition of mTOR signaling in both cell types
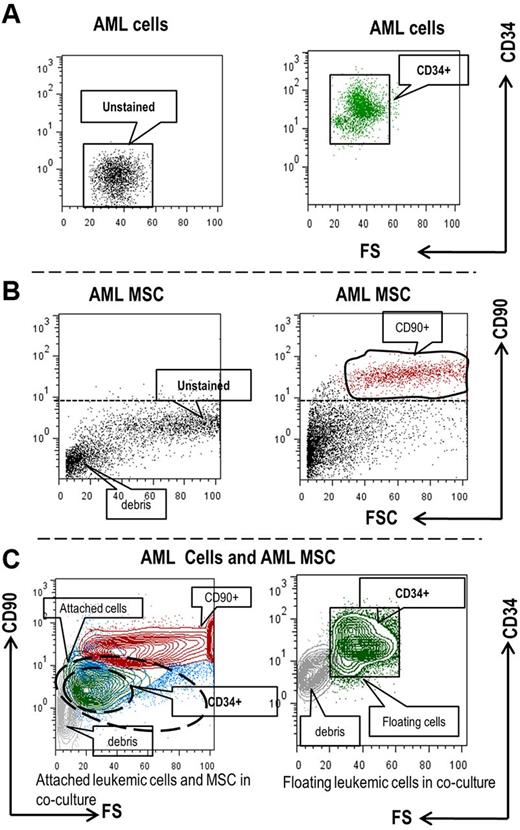
Several studies have demonstrated that leukemia cells reside in close contact with stromal cells in BM microenvironments and that direct cell-cell contact plays an important role in the activation of intracellular signaling. One of the challenges in studying leukemia-stroma interactions is identifying and separating the different cell types in coculture. To avoid physical cell separation while assessing the rapid signaling events on direct leukemia-stroma cell-cell contact, we used phospho-specific multiparameter flow cytometry. To this end, we isolated primary BM MSCs from 11 AML samples and 1 normal healthy donor. Using a coculture system of primary AML (n = 16, supplemental Table 2) with allogenic leukemia or normal BM-derived MSC, we collected (1) floating AML cells that did not attach to the stromal cells; and (2) AML cells that were tightly attached to the stromal cells (by trypsinization). We then fixed and permeabilized cells before staining with the cell surface markers CD90, CD34, and intracellular signaling markers pAKT (Ser473), pS6K (Ser235/236), and p4EBP1 (Thr37/46). We discriminated between leukemic cells and MSCs by electronic gating on CD90+ cells for leukemia-derived MSCs and on CD34+ cells for AML progenitors (Figure 2A-C).
Flow cytometry-based discrimination of primary AML cells and MSCs cultured alone or cocultured. Primary AML cells (A) or AML MSCs (B) cultured alone and used as controls; or harvested from AML/MSC coculture (C) were fixed, permeabilized, and stained with the indicated cell surface markers. (C) Contour blot depicting different population of attached AML cells and MSCs (left) and floating AML cells (right) detected by flow cytometry in the coculture setting. Blue, CD90(−) AML cells; green CD34+ AML progenitor cells; red, CD90+ AML MSCs.
Flow cytometry-based discrimination of primary AML cells and MSCs cultured alone or cocultured. Primary AML cells (A) or AML MSCs (B) cultured alone and used as controls; or harvested from AML/MSC coculture (C) were fixed, permeabilized, and stained with the indicated cell surface markers. (C) Contour blot depicting different population of attached AML cells and MSCs (left) and floating AML cells (right) detected by flow cytometry in the coculture setting. Blue, CD90(−) AML cells; green CD34+ AML progenitor cells; red, CD90+ AML MSCs.
Using this technology, we investigated the effects of PP242 on intracellular signaling in both primary AML cells and MSCs, cultured alone or cocultured (Figure 3A-D, supplemental Figure 3). PP242 significantly inhibited p4EBP1 (Thr37/46) expression in both bulk leukemic cells and CD34+ AML progenitor cells in all 16 samples cultured in medium (P = .01, P = .01, respectively; Figure 3A “alone”). It also down-regulated pAKT (Ser473) expression in 12 of 16 samples (P = .01), consistent with inhibition of mTORC2 complex (Figure 3C “alone”). Coculture with stroma significantly increased the phosphorylation of 4EBP1 (Thr 37/46) in all 16 floating AML cells (P = .03; n = 16; Figure 3A dashed line), and activated pS6K (Ser235/236) in the majority of AML (P = .03; n = 15, dashed line) and in 14 of 16 CD34+ progenitor cells attached to stroma (P = .04; n = 14; Figure 3B dashed line). Treatment with PP242 effectively suppressed the phosphorylation of 4EBP1 (Thr37/46; P < .001; n = 16), AKT (Ser473; P = .02; n = 12), and S6K (Ser235/236; P = .05; n = 15) in floating leukemia cells, and down-regulated the phosphorylation of AKT (Ser473) in the majority of floating CD34+ AML progenitor cells (P = .05, n = 12). Importantly, PP242 exerted its inhibitory effect on p4EBP1 (Thr37/46) and pAKT (Ser473) in both attached AML cells (P = .03, n = 16; P = .01, n = 12, respectively) and most of attached CD34+ AML progenitor cells (P = .02; n = 16; P = .02; n = 12, respectively) and abrogated stroma-mediated up-regulation of pS6K (Ser235/236) in 14 of 16 attached CD34+ AML progenitor cells (P = .05, n = 14). In BM-derived MSCs, PP242 significantly down-regulated p4EBP1 (Thr37/46; P = .002; n = 15) and pS6K (Ser235/236; P = .002; n = 15) in most of MSCs cultured alone and potently inactivated both proteins in most of attached MSCs in the coculture setting (P = .004; n = 15 [p4EBP1 (Thr37/46)]; P = .007; n = 15 [pS6K (Ser235/236)]; Figure 3D). Unlike in primary AML cells, PP242 modestly increased mTORC2-mediated phosphorylation of AKT in the majority of MSCs cultured alone or cocultured (P = .18, P = .88, data not shown), indicating that PP242 affects mTORC2 sensitivity in a cell context-dependent manner.
PP242-mediated intracellular mTOR signaling in primary AML cells and MSCs cultured alone or cocultured. Measurement of the MFI of intracellular p4EBP1 (Thr37/46; A), pAKT (Ser473; C), and pS6K (Ser235/236; B) in primary AML cells, AML CD34+ progenitor cells, and primary AML MSCs (D) cultured alone and in cocultures using flow cytometry, The gray bars represent the MFI of the phosphorylated proteins before, and the black bars after treatment with PP242 for 72 hours. Cells were cultured in medium only (“alone”), or cocultured with MSCs (“floating” or “attached” cells, respectively). Data represent average ± SEM of MFI of the phospho-proteins measured in the indicated number of primary samples. Paired 2-sample t test was used to determine reported P values. (E) Primary AML cells and MSCs in a transwell setting (described in “Leukemia/MSC/MS-5 coculture”) were treated with PP242 for 24 hours. Cells were then lysed, and mTOR signaling targets in the AML cells were detected by immunoblotting.
PP242-mediated intracellular mTOR signaling in primary AML cells and MSCs cultured alone or cocultured. Measurement of the MFI of intracellular p4EBP1 (Thr37/46; A), pAKT (Ser473; C), and pS6K (Ser235/236; B) in primary AML cells, AML CD34+ progenitor cells, and primary AML MSCs (D) cultured alone and in cocultures using flow cytometry, The gray bars represent the MFI of the phosphorylated proteins before, and the black bars after treatment with PP242 for 72 hours. Cells were cultured in medium only (“alone”), or cocultured with MSCs (“floating” or “attached” cells, respectively). Data represent average ± SEM of MFI of the phospho-proteins measured in the indicated number of primary samples. Paired 2-sample t test was used to determine reported P values. (E) Primary AML cells and MSCs in a transwell setting (described in “Leukemia/MSC/MS-5 coculture”) were treated with PP242 for 24 hours. Cells were then lysed, and mTOR signaling targets in the AML cells were detected by immunoblotting.
To validate the findings obtained by flow cytometry, we analyzed expression and phosphorylation of proteins by immunoblotting. To eliminate cross-contamination of the primary AML cells and MSCs in the coculture setting, we cocultured cells from primary AML cells and leukemia-derived MSCs on each side of the transwell membrane, as described in “Leukemia/MSC/MS-5 coculture.” The transwell membrane pores prevent migration of leukemia cells through the membrane, and yet allow direct contact of AML/MSC cell-surface receptors. In agreement with flow cytometry data, Western blot analysis showed that PP242 completely suppressed mTORC1 direct targets p4EBP1 (Th37/Thr46) and pS6K (Ser240/244) and reduced mTORC2/AKT activity as evidenced by decreased phosphorylation of FoxO1a(Thr24)FoxO3a(Thr32) in AML cells cultured alone (Figure 3E). Coculture of primary AML cells and MSCs increased the activity of mTORC1 and mTORC2 in primary AML cells, and this was partially abrogated by PP242. Taken together, these results indicate that treatment with PP242 effectively interferes with the function of both mTORC1 and mTORC2 in primary AML cells, AML progenitor cells, and primary AML MSCs; and partially antagonized MSC-mediated activation of PI3K/mTOR/AKT signaling in primary AML cells and AML progenitor cells in the coculture setting.
PP242 interrupts leukemia-stroma interactions by suppressing surface expression of CXCR4 and simultaneously inhibiting intracellular mTOR signaling in primary AML cells and MSCs
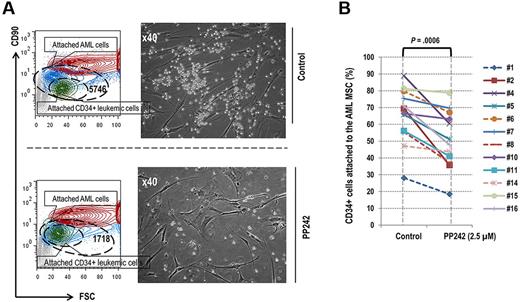
Our findings of PP242-mediated inhibition of mTOR signaling in AML cocultured with stroma prompted us to examine the direct effects of PP242 on adhesion of AML cells to MSC. To this end, we counted primary AML cells directly attached to MSC. As shown in a representative example (Figure 4A left), in the control coculture of primary AML cells and MSCs, 5746 of 51 966 CD34+ AML cells (11%) were attached to the AML MSCs, whereas only 1718 of 31 643 (5.4%) were attached in the sample treated with 2.5μM PP242 for 72 hours. We further confirmed this finding using light microscopy (Figure 4A right). Overall, treatment with PP242 reduced the proportion of CD34+ AML progenitor cells attached to AML MSCs in 12 of 16 patients (Student t test P = .0006; Figure 4B), and in 8 of these the percentage decrease in the fraction of attached cells exceeded the percentage decrease in the total cell number (not shown). These results demonstrated that PP242 effectively disrupted the interactions between leukemia and stroma by reducing the number of leukemic blasts attached to the MSCs.
PP242 reduced the proportion of the leukemic cells attached to primary MSCs in coculture condition. (A). Contour blot depicting different population of attached cells detected by flow cytometry in the coculture setting. Leukemic cells (blue), CD34+ AML cells (green), and MSCs (red). Right, microscopic images display these cocultured AML cells and MSCs before and after PP242 treatment. (B) Percentage of AML CD34+ progenitor cells attached to the primary AML MSCs in the coculture setting before and after PP242 treatment for 72 hours. In samples from 8 patients depicted by solid lines, the percentage decrease in the attached cell number exceeded the percentage decrease in the total number of cells after the PP242 treatment.
PP242 reduced the proportion of the leukemic cells attached to primary MSCs in coculture condition. (A). Contour blot depicting different population of attached cells detected by flow cytometry in the coculture setting. Leukemic cells (blue), CD34+ AML cells (green), and MSCs (red). Right, microscopic images display these cocultured AML cells and MSCs before and after PP242 treatment. (B) Percentage of AML CD34+ progenitor cells attached to the primary AML MSCs in the coculture setting before and after PP242 treatment for 72 hours. In samples from 8 patients depicted by solid lines, the percentage decrease in the attached cell number exceeded the percentage decrease in the total number of cells after the PP242 treatment.
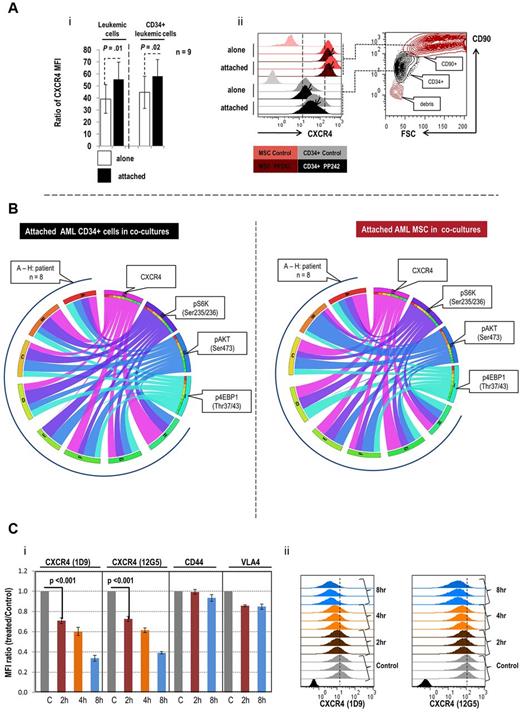
CXCR4, a G-protein–coupled chemokine receptor, plays an important role in leukemia-stroma interactions through regulation of migration and adhesion.5,23,30 We therefore investigated whether the observed decrease in the proportion of attached cells is mediated by inhibition of CXCR4. Notably, expression of CXCR4 increased significantly in both primary AML cells and CD34+ progenitor cells that attached to stroma in samples from 8 of 9 patients examined (P = .01 and P = .02, respectively; Figure 5Ai). In floating cells, the increase was observed only in 4 of 9 samples (P = .46, data not shown). In MSCs, CXCR4 levels were overall higher than in AML cells (Figure 5Aii) and were induced by cocultures only in 3 samples (not shown). In attached cells, the increased expression of CXCR4 was significantly associated with the increased activity of mTORC1, mTORC2 or both as indicated in Table 2. Treatment with PP242 reduced the expression of CXCR4 in the majority of AMLs, AML CD34+ cells and MSCs, cultured alone or cocultured (Figure 5Aii, supplemental Figure 4). Down-regulation of CXCR4 by PP242 significantly correlated with suppression of the intracellular proteins, p4EBP1 (Thr37/46) in attached primary AML cells, AML CD34+ cells and primary MSCs; and inhibition of pS6K (Ser235/236) and pAKT (Ser473) in attached primary MSCs (supplemental Figures 3, 5A-C). The circos diagram (Figure 5B) further illustrates the correlations and the degree of inhibition in cocultured AML CD34+ cells and in their associated MSCs. We observed similar correlations of altered expression of CXCR4 and changes in intracellular levels of p4EPB1 (Thr37/43) and pAKT(Ser473) in AML cells cultured alone (supplemental Figure 5D); and of p4EBP1 (Thr37/43), pS6K (Ser235/236), and pAKT(Ser473) in AML MSC cultured alone (Figure 5E). These data suggest that PP242 disrupts leukemia-stromal interactions by reducing expression of CXCR4 in both cell types, cultured alone or cocultured. To assure that the observed changes in CXCR4 are not secondary to PP242-induced cell death, we evaluated CXCR4 expression in attached live CD34+ primary AML cells, and confirmed decrease of CXCR4 levels on 48 hours PP242 exposure (supplemental Figure 6A-B). We further examined the surface expression of CXCR4 and other adhesion molecules, CD44 and VLA4 in OCI-AML3 cells at short time intervals of PP242 treatment, preceding alterations in cell viability. CXCR4 cell surface expression detected by antibodies against 2 different isotopes (12G5 and 1D9) diminished after only 2 hours of PP242 treatment, with further decrease after 4 hours and 8 hours exposure (Figure 5C). On the contrary, PP242 minimally affected VLA4 and CD44 expression, suggesting specificity of changes in CXCR4 levels. However, PP242 at 30 and 60 minutes had no effect on CXCR4 expression, whereas it inhibited both mTORC1/C2 functions via inactivating the phosphorylation of S6K by 70%, and of AKT (Ser 473) by 50% (supplemental Figure 6C). To evaluate the possibility of PP242 affecting internalization of surface CXCR4, we measured CXCR4 levels using 12G5 and 1D9 antibodies in live (surface) versus fixed/permeabilized (intracellular) cells and found comparable decrease, arguing for changes in total protein levels rather than redistribution phenomenon (supplemental Figure 7A-B). In turn, PP242 down-regulated CXCR4 mRNA levels measured by quantitative reverse transcription (RT)–PCR in 3 AML cell lines as early as 6 hours, with persistent down-regulation at 24 hours, and in primary AML samples at 6 hours (sample no. 2) or 24 hours (samples no. 1-3; supplemental Figure 8A-B). Twenty-four hour exposure to temsirolimus did not affect cell viability and caused modest decrease of cell surface and intracellular CXCR4 levels (supplemental Figure 7A-B). Taken together, these results indicate that PP242 selectively decreases CXCR4 expression after blockade of the intracellular mTOR signaling. Importantly, this effect is seen in primary AML cells and MSCs, cultured alone or cocultured.
PP242-altered expression of CXCR4 and intracellular signaling in primary AML cells and MSCs cultured alone and cocultured. (Ai) Comparison of CXCR4 expression in primary AML and AML CD34+ progenitor cells cultured alone and cocultured with stroma. Results represent the ratio of MFI calculated as described in “Flow cytometry.” P value was calculated using paired 2-sample t test. (Aii) Contour dot plot and histograms demonstrating effects of PP242 on CXCR4 expression in a representative sample. (B) Circos diagram displays the inhibitory effect of PP242 on CXCR4 expression and intracellular signaling in attached AML CD34+ cells and corresponding MSC samples in cocultures. The fold inhibition was calculated by MFI of untreated cells divided by MFI of treated cells. I-IV: surface marker and intracellular markers measured by flow cytometry. I: 4EBP1 (cyan), II: pAKT (blue); III: pS6K (purple); IV: CXCR4 (magenta). (A-H) Eight AML CD34+ samples and 8 MSCs in cocultures. Left panel shows the attached AML CD34+ cells; right panel, their associated MSCs in cocultures. Each ribbon connects a marker and an individual patient. The width of a ribbon represents the degree of inhibition: the wider the ribbon, the greater the degree of inhibition. The color of the ribbon represents an individual marker. (C) The inhibitory effects of short-term PP242 treatment on CXCR4 expression and on mTOR signaling in OCI-AML3 cells. Surface expression of CXCR4, CD44, VLA4 was examined by flow cytometry in OCI-AML3 cells treated with 2.5μM PP242 for the indicated time period. Data represent the MFI ratios (Ci). Histograms of the triplicate experiments measuring changes in CXCR4 levels with 12G5 antibody recognizing the extracellular loop 2 (EC2) domain of CXCR4, and 1D9 antibody recognizing the N-terminal of CXCR4, are shown in panel Cii.
PP242-altered expression of CXCR4 and intracellular signaling in primary AML cells and MSCs cultured alone and cocultured. (Ai) Comparison of CXCR4 expression in primary AML and AML CD34+ progenitor cells cultured alone and cocultured with stroma. Results represent the ratio of MFI calculated as described in “Flow cytometry.” P value was calculated using paired 2-sample t test. (Aii) Contour dot plot and histograms demonstrating effects of PP242 on CXCR4 expression in a representative sample. (B) Circos diagram displays the inhibitory effect of PP242 on CXCR4 expression and intracellular signaling in attached AML CD34+ cells and corresponding MSC samples in cocultures. The fold inhibition was calculated by MFI of untreated cells divided by MFI of treated cells. I-IV: surface marker and intracellular markers measured by flow cytometry. I: 4EBP1 (cyan), II: pAKT (blue); III: pS6K (purple); IV: CXCR4 (magenta). (A-H) Eight AML CD34+ samples and 8 MSCs in cocultures. Left panel shows the attached AML CD34+ cells; right panel, their associated MSCs in cocultures. Each ribbon connects a marker and an individual patient. The width of a ribbon represents the degree of inhibition: the wider the ribbon, the greater the degree of inhibition. The color of the ribbon represents an individual marker. (C) The inhibitory effects of short-term PP242 treatment on CXCR4 expression and on mTOR signaling in OCI-AML3 cells. Surface expression of CXCR4, CD44, VLA4 was examined by flow cytometry in OCI-AML3 cells treated with 2.5μM PP242 for the indicated time period. Data represent the MFI ratios (Ci). Histograms of the triplicate experiments measuring changes in CXCR4 levels with 12G5 antibody recognizing the extracellular loop 2 (EC2) domain of CXCR4, and 1D9 antibody recognizing the N-terminal of CXCR4, are shown in panel Cii.
Pearson correlation coefficient (ρ)–defined association of stroma-altered expression of CXCR4 with changes in intracellular mTOR signaling in 9 pairs of cocultured primary AML cells and MSCs
| Intracellular signaling (N = 9) . | CXCR4 in attached MSCs . | CXCR4 in attached CD34+ cells . | CXCR4 in attached leukemic cells . | |||
|---|---|---|---|---|---|---|
| ρ . | P . | ρ . | P . | ρ . | P . | |
| p4EPB1 (TORC1) | 0.999 | < .001 | 0.724 | .0275 | 0.768 | .0157 |
| pS6K (TORC1) | 0.999 | < .001 | 0.123 | > .05 | 0.232 | > .05 |
| pAKT (TORC2) | 0.995 | < .001 | 0.687 | .0407 | 0.656 | > .05 |
| Intracellular signaling (N = 9) . | CXCR4 in attached MSCs . | CXCR4 in attached CD34+ cells . | CXCR4 in attached leukemic cells . | |||
|---|---|---|---|---|---|---|
| ρ . | P . | ρ . | P . | ρ . | P . | |
| p4EPB1 (TORC1) | 0.999 | < .001 | 0.724 | .0275 | 0.768 | .0157 |
| pS6K (TORC1) | 0.999 | < .001 | 0.123 | > .05 | 0.232 | > .05 |
| pAKT (TORC2) | 0.995 | < .001 | 0.687 | .0407 | 0.656 | > .05 |
Raw and adjusted P values were calculated as described in “Flow cytometry.”
PP242 reduces leukemia burden and extends survival in a mouse model of leukemia
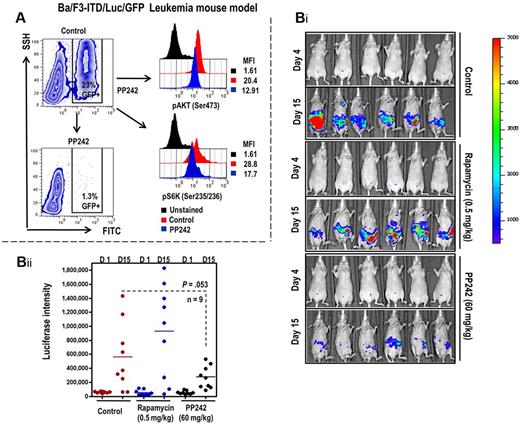
We further evaluated the antileukemic potential of PP242 in vivo using a mouse model of leukemia generated by Ba/F3-ITD/luc/GFP in nonobese diabetic/SCID mice.21,22,31 We monitored leukemia burden in murine peripheral blood using flow cytometry gated on GFP+ circulating cells. As shown in Figure 6A, 2 hours after PP242 administration, PP242 inactivated the mTORC1 downstream target and pS6K (Ser235/236) and mTORC2 direct target pAKT (Ser473) in circulating Ba/F3-ITD/luc/GFP leukemic cells. PP242 at 60 mg/kg reduced leukemia burden as measured by bioluminescence imaging (Figure 6Bi-ii). The antileukemia effect of PP242 was greater than that of rapamycin used at 0.5 mg/kg, the tolerable dose that was previously shown to inhibit mTOR signaling.15
PP242 suppression of AML cell expansion in Ba/F3-ITD/luc/GFP mouse model of leukemia. (A) Peripheral blood obtained from retro-orbital plexus before and 2 hours after PP242 administration. Whole blood was fixed and permeabilized as described in “Flow cytometry.” The cells were then stained with antibodies against pAKT (Ser473), and pS6K (Ser235/236), and the MFI of each intracellular marker in GFP+ leukemic cells was assessed using flow cytometry after gating on GFP+ cells. (Bi-ii) The leukemia burden was monitored using bioluminescent imaging on days 5 and 14 after leukemia-cell inoculation (i). The bioluminescent intensities were averaged the peak light-emitting exposure in each group of mice and displayed as photons/second in the bar graphs (ii).
PP242 suppression of AML cell expansion in Ba/F3-ITD/luc/GFP mouse model of leukemia. (A) Peripheral blood obtained from retro-orbital plexus before and 2 hours after PP242 administration. Whole blood was fixed and permeabilized as described in “Flow cytometry.” The cells were then stained with antibodies against pAKT (Ser473), and pS6K (Ser235/236), and the MFI of each intracellular marker in GFP+ leukemic cells was assessed using flow cytometry after gating on GFP+ cells. (Bi-ii) The leukemia burden was monitored using bioluminescent imaging on days 5 and 14 after leukemia-cell inoculation (i). The bioluminescent intensities were averaged the peak light-emitting exposure in each group of mice and displayed as photons/second in the bar graphs (ii).
Discussion
The interaction of AML cells with stroma within the BM microenvironment attenuates the response of AML cells to conventional chemotherapeutic agents and promotes chemoresistance. Stromal cells can trigger activation of multiple signaling pathways in leukemic cells through secreted growth factors/cytokines and by direct cell-cell interactions, such as ligation of integrins and other cellular receptors.1,5,6,23,32,33 In this study, using RPPA we showed the complex, multidimensional stroma-mediated survival signals conveyed in leukemia, including signals affecting the PI3K/mTOR/AKT pathway. Our results support the notion that AML-stroma interaction is an important factor that must be considered when designing investigational anti-AML therapeutics. Furthermore, our results emphasize the utility of the selective mTOR kinase inhibitors as potential therapeutic adjuncts in AML therapy, particularly under conditions mimicking the BM microenvironment.
PP242, an ATP-competitive inhibitor of mTOR kinase, is one of the new generation of mTOR inhibitors. Compared with rapamycin, PP242 more efficiently inhibits mTORC1 as evidenced by diminished 4EBP1 phophorylation and cap-dependent protein translation.34 Furthermore, PP242 is more effective in preventing the occurrence of negative feedback loops by enhancing its inhibitory effect on mTORC2.11,12 Consequently, PP242 is a more potent inhibitor of protein synthesis, metabolism, and proliferation of cancer cells. In this study, PP242 exhibited significant antileukemic effects in vitro by inducing dose-dependent apoptosis of both, bulk primary AML cells and AML progenitor cells. Most importantly, PP242, but not rapamycin derivative temsirolimus, induced cell death in stromal cocultures, which otherwise reliably support viability of leukemic blasts (supplemental Figure 1) and reduce efficacy of traditional anti-AML chemotherapy agents.35 These data support the recently reported evidence of the superior antileukemic potency of mTOR kinase inhibitors36,37 or dual P13K/mTORK inhibitors38 through suppression of rapamycin-resistant mTORC1 and mTORC2 complexes compared with allosteric mTOR inhibitors. These findings indicate that mTOR signaling plays an essential role in the stroma-leukemia cell interaction, and its blockade is sufficient to abrogate the stroma-mediated survival advantage of leukemic cells.
Consistent with findings in other cancer cell types,15-17 PP242 potently inhibited phosphorylation of 4EBP1 (Thr37/46) and S6K (Ser235/236), 2 direct targets of mTORC1, in AML cells. Importantly, this inhibition is preserved in AML/stromal cells in direct physical contact. These data demonstrate for the first time that PP242 can disrupt AML-stroma interactions by directly targeting not only the intrinsically activated mTOR signaling in AML cells but also additional stroma-induced activation of this pathway. We further observed suppression of the mTORC2 target pAKT (Ser473) in bulk AML cells, AML progenitor cells, and floating and attached AML cells in the coculture setting. Inhibition of mTORC2 was further supported by inactivation of FoxO1a/FoxO3a, direct targets of AKT, in cocultured AML cells after treatment with PP242. Importantly, PP242 also suppressed mTOR activity in stromal cells of supporting microenvironment (MSC). However, PP242 modestly activated pAKT (Ser473) in some AML MSCs under both culture-alone and coculture conditions. Recent findings by Rosen et al demonstrated that mTOR kinase inhibitors may cause activation of PI3K and rephosphorylation of AKT Thr308 through unleashed receptor tyrosine kinase signaling, at least in breast cancer cells with constitutive activation of HER2/EGFR.39 Although this mechanism is unlikely operational in untransformed BM stromal cells, these observations argue for potential advantage of combined PI3K/mTOR blockade to fully inhibit leukemia/stroma interactions. Further studies are required to compare mTOR kinase inhibitors and dual PI3K/mTOR inhibitors in the coculture setting.
In this study, mTOR blockade with PP242 decreased the number of AML progenitor cells attached to MSCs in the coculture setting. This prompted us to examine the effect of PP242 on chemokine receptor CXCR4, which plays a key role in chemotaxis and adhesion of hematopoietic cells to cells of their microenvironment. We previously reported that CXCR4 inhibition interferes with stromal/leukemia cell interactions and in part overcomes stroma-mediated resistance to chemotherapy and FLT3 inhibitors.23,30 Although stroma coculture promoted CXCR4 expression in AML cells, PP242 decreased CXCR4 levels in the majority of primary AML samples and in MSCs, cultured alone and cocultured. PP242-altered surface expression of CXCR4 occurred within hours subsequent to the blockade of intracellular mTOR signaling. Overall, we identified a novel function of PP242 in that it abrogates, at least partially, AML cell-MSC interactions by targeting mechanisms by which AML cells communicate with cells in their microenvironment. Of note, recent ribosome profiling efforts have identified proinvasion mRNA networks translationally controlled by mTOR that were effectively blocked by mTOR ATP site inhibitors.40 Although further studies are warranted to dissect the mechanisms responsible for CXCR4 down-regulation in our system, our preliminary data indicate the effect of PP242 on mRNA transcription (supplemental Figure 8). Notably, prolonged exposure to allosteric mTOR inhibitor temsirolimus also resulted in down-regulation of CXCR4 levels; albeit at a lesser extent compared with PP242. These results would suggest that ATP kinase mTOR inhibitors may have far superior antileukemic effects by virture of their ability to fully inhibit rapamycin-insensitive TORC1/TORC2 complexes and target leukemia/stroma interactions, similar to recent findings with INK128 in metastatic prostate cancer models.40 It is plausible to suspect that PP242 may also target other chemokines, cytokines, and growth factors that mediate the interaction between AML and stroma, and identification of these mediators require further study. We further examined antileukemia effects of PP242 in a FLT3-driven mouse model of leukemia. This in vivo model was selected based on our published data indicating the prominent role of BM microenvironment in survival of FLT3-mutated AML cells, mediated in part through CXCR4 signaling.23 In this model, PP242 suppressed mTOR signaling in mouse leukemia cells and reduced leukemia burden without evidence of toxicity, confirming therapeutic efficacy of this agent.
In summary, we identified several signaling pathways activated by stroma in AML cells that promote leukemia cell survival within its protective microenvironment. Our studies support the key importance of the PI3K/AKT/mTOR pathway in microenvironment-mediated resistance, justifying the use of the selective inhibitors of this pathway as adjunct agents that target leukemia-stroma interactions. We further showed that PP242 has antileukemia activity in vivo, providing a compelling rationale for testing this inhibitor in clinical trials.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This research was supported by Intellikine; an MD Anderson Institutional Research Grant (to M.K.); a Discovery Grant from the University of California Industry, University Cooperative Research Program (to D.A.F.), AML-P01 (CA0055164 to M.A.), and in part by the National Institutes of Health through MD Anderson Cancer Center Support Grant CA016672.
National Institutes of Health
Authorship
Contribution: Z.Z. designed and performed the experiments and wrote the paper; Y.X.S. performed the in vivo study; T.T. and Y.Q. performed RPPA study; S.M.K. supervised the RPPA study and wrote the paper; K.A.B. supervised and performed statistical analysis and wrote the paper; W.L. performed statistical analysis; K.J. provided basic information on the compound; Y.L. provided basic information on the compound, designed the experiments, and wrote the paper; H.K. supervised data analysis and wrote the paper; C.R. provided basic information on the compound, designed the experiments, and wrote the paper; D.A.F. designed the experiments, supervised data analysis, and wrote the paper; M.A. supervised data analysis and wrote the paper; and M.K. designed the experiments, supervised data analysis, and wrote the paper.
Conflict-of-interest disclosure: K.J., Y.L., and C.R. are employed by Intellikine. The remaining authors declare no competing financial interests.
Correspondence: Marina Konopleva, Department of Leukemia, MD Anderson Cancer Center, 1515 Holcombe Blvd, Unit 428, Houston, TX 77030; e-mail: mkonople@mdanderson.org.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal