Abstract
Induced pluripotent stem (iPS) cell technology holds vast promises for a cure to the hemoglobinopathies. Constructs and methods to safely insert therapeutic genes to correct the genetic defect need to be developed. Site-specific insertion is a very attractive method for gene therapy because the risks of insertional mutagenesis are eliminated provided that a “safe harbor” is identified, and because a single set of validated constructs can be used to correct a large variety of mutations simplifying eventual clinical use. We report here the correction of α-thalassemia major hydrops fetalis in transgene-free iPS cells using zinc finger–mediated insertion of a globin transgene in the AAVS1 site on human chromosome 19. Homozygous insertion of the best of the 4 constructs tested led to complete correction of globin chain imbalance in erythroid cells differentiated from the corrected iPS cells.
Introduction
Induced pluripotent stem cell (iPSC) technology has the potential to provide an unlimited source of cells for regenerative medicine.1-3 As shown in an early report in mice, one promising application is the production of gene-corrected autologous transplantable hematopoietic stem cells for patients carrying genetic diseases.4 A recent report has demonstrated the insertion and expression of lentiviruses carrying a therapeutic β-globin transgene in iPSCs from β-thalassemic patients.5 In other reports, the mutation causing sickle cell anemia was corrected in iPSCs using homologous recombination.6-8 In these studies, the gene corrections were demonstrated at the genetic level in undifferentiated iPSCs. In addition, evidence was provided that the correction was functional at the protein or mRNA levels by differentiating the iPSCs into erythroid cells. This revealed that the corrective transgene or the corrected gene were expressed but levels of expression were very low because current iPSC differentiation protocols yield erythroid cells that express mostly embryonic (ζ2ϵ2) and fetal hemoglobins (α2γ2) and only trace amounts of β-globin, the gene that is mutated in both sickle cell disease and β-thalassemia.9,10
α-thalassemias, which are caused by mutations in the α-globin gene cluster,11 are therefore good models to test globin gene correction methods in iPSCs because the α-globin gene is expressed at very high levels in the erythroid cells that can currently be produced from iPSCs. α-thalassemias are divided into 2 major clinical categories: Hb H disease (a relatively severe hemolytic anemia caused by loss or decreased expression of 3 of the 4 α-globin genes) and homozygous α-thalassemia hydrops fetalis (caused by deletion of the 4 genes). This latter form of the disease is generally lethal at the time of birth, although rare individuals have been treated by transfusion or transplantation and survived into adulthood.12
Most of the genetic mutations in the α-globin cluster leading to hydrops fetalis are caused by large deletions that encompass both the α1 and α2 globin genes. Correction of large deletions is difficult to accomplish by homologous recombination. Lentivirus-mediated transgene insertion can be used but is associated with a risk of insertional mutagenesis. Papapetrou et al have shown that this risk can be mitigated by identifying clones in which the virus has integrated into a safe harbor but the procedure is complex and might be difficult to implement clinically.5 To correct these large deletions, we therefore decided to use zinc-finger nucleases (ZFNs) to integrate therapeutic globin transgenes at AAVS1, a landing pad located on chromosome 19.
We chose AAVS1, the preferential integration site of the adeno-associated virus, because several studies have shown that integration into the PPP1R12C gene at AAVS1 leads to high levels of expression,13 because effective ZFN specific for this site have been developed,14 and because the PPP1R12C gene has no known function in hematopoietic cells.
ZFN-mediated site-specific insertion has very attractive characteristics for gene therapy in iPSCs. First, the risk of insertional mutagenesis is eliminated provided that a safe harbor is identified.15 Second, the use of this technology should eventually simplify clinical implementation because a single set of validated constructs could be used to correct the large variety of mutations that cause the hemoglobinopathies. This presents considerable practical and economic advantages over having to design and validate custom constructs for each particular mutation.
We report here that we have successfully tested this strategy and completely corrected homozygous α-thalassemia.
Methods
Cell culture
α-thalassemia fibroblasts were purchased from the Coriell Institute. H1 human embryonic stem cells (hESCs) and α0-thal-iPSCs were maintained by coculture on matrigel (BD Biosciences) in DMEM/F12 media (Invitrogen) with N2 and B27 supplement, 0.5% bovine serum albumin (Sigma-Aldrich), 1mM l-glutamine, 1% penicillin-streptomycin (P/S), 100 ng of bFGF.16 Fourteen- to 16-week-old fetal livers were obtained from the Einstein Fetal Tissue Repository under an approved institutional review board protocol. Adult control and α0-thalassemic circulating peripheral blood samples were also obtained under an approved institutional review board protocol. This study was conducted in accordance with the Declaration of Helsinki.
Reprogramming of α-thalassemia fibroblast into iPSCs
Cells (2 × 106) were used for the nucleofection with 3 μg of plasmid pEP4-EOS2-ET2K, 3 μg of pEP4-EO2S-EN2K and pCEP4-M2L17 (program A-23 on nucleofector II; Lonza). Fibroblasts were transferred directly onto matrigel-coated 6-well plates containing hESC medium. hESC-like colonies could be found ∼ 3 weeks after nucleofection. iPSC colonies were passaged manually by collagenase IV (1 mg/mL) or with laser-enabled analysis and processing (LEAP; Cyntellect). Episomal vector analysis was performed by PCR using primers described in supplemental Table 2 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
ZFN-mediated homologous recombination
The HBA expression construct containing the mini-LCR (locus control region), the β-globin promoter, and the HBA genes was obtained from plasmid pLM18-TNS9.3-HBA1 generously provided by Dr M. Sadelain,18 and the whole expression construct was cloned into pZDonor-AAVS1 Puro vector (Sigma-Aldrich; NotI and SalI sites). For the promoter studies, the β-globin promoter (500 bp upstream of the cap site) was replaced by the α-globin promoter (1000-bp fragment upstream of the cap site). AAVS1 ZFN sequences were as described by Hockemeyer et al14 and were cloned into plasmids pCMV-AAVS1-A and -B.
hESCs and iPSCs were cultured in 10μM ROCK inhibitor (Cayman Chemical)19 1 hour before nucleofection to increase cloning efficiency. Cells were harvested by using accutase (Invitrogen) for 10 minutes at room temperature. Cells (2 × 106) were used for nucleofection14 with 20 μg of donor plasmids and 2 μg of each ZFN-encoding plasmids. Antibiotic selection started 3 days after nucleofection with 0.5 μg/mL puromycin.
Differentiation of hESCs and iPSCs to erythroid cells
Undifferentiated hESCs and iPSCs on feeder plates (1-2 plates per assays) were dissociated with collagenase type IV (1 mg/mL; Invitrogen), transferred onto irradiated FH-B-hTERT feeder layers with RPMI 1640 supplemented with 15% FBS (Invitrogen), 2mM l-glutamine, 1% MEM-nonessential amino acids, 50 μg/mL acscorbic acid, 100μM monothioglycerol (MTG), 100 units/mL penicillin, and 100 μg/mL streptomycin. The medium was changed every 2 to 3 days. On day 14 of coculture, differentiated H1/iPSCs on FH-B-hTERT were dissociated using collagenase IV followed by treatment with trypsin/EDTA (Invitrogen) supplemented with 5% chick serum. CD34 cell separations were performed using the CD34 microbead kit according to the manufacturer's instructions (Milteny Biotec). CD34+ cells obtained either from hESCs or from iPSCs were then seeded in liquid culture as shown in supplemental Figure 3.
Step 1 (amplification of progenitors).
Sorted CD34+ cells (50 000 cells/mL) were placed on a 12-well plate with serum-free basal medium StemSpan (StemCell Technologies) supplemented with hydrocortisone (10−6M), IL3 (13 ng/mL), BMP4 (13 ng/mL), Flt3L (33 ng/mL), SCF (100 ng/mL), and EPO (2.7 U/mL) for 7 days.
Step 2 (differentiation).
Cells were transferred to StemSpan medium supplemented with hydrocortisone (10−6M), IL3 (13 ng/mL), BMP4 (13 ng/mL), SCF (40 ng/mL), EPO (3.3 U/mL), and IGF-1 (40 ng/mL) for 7 days. Cell density was kept below 1 million cells/mL by adding fresh medium every 2 or 3 days as needed.
Step 3 (maturation).
Cells were resuspended in StemSpan containing EPO (3.3 U/mL) for 3 days onto plates containing MS-5 cells.
Step 4.
Medium was switched to StemSpan only for 7 days for final maturation. Yields of erythroid cells were in the range of 5 to 50 million cells per starting plates of iPSCs. Corrected iPSCs used in the differentiation assays were at passages 12 to 16 after the initial transfection of the 4 factors and at passages 4 to 8 after the transfection to introduce the transgene into AAVS1. All corrected clones tested could be differentiated along the erythroid lineage.
Hemoglobin analysis
HPLC analysis was performed to measure globin chain expression. Erythroid cells derived from hESCs or iPSCs were washed twice with PBS, and lysed in water by 3 rapid freeze-thaw cycles. Debris were eliminated by centrifugation at 16 000g. HPLC analysis was performed as described.20
Hemoglobin concentrations were determined using the tetramethylbenzidine (TMB) method.21 Hemolysates (2.5 μL) were mixed with TMB (Sigma-Aldrich) and the reactions were triggered by adding 150 μL of 0.3% H2O2. After 5 minutes, the samples were read at 600 nm, at room temperature. Concentrations of hemolysate of each sample were calculated by comparing with calibrator (Catachem Inc).
Isoelectric focusing (IEF) was performed to identify the types of hemoglobin in the hemolysates. Gels were prepared by following the instructions of the manufacturer (PerkinElmer). Hemolysates (5-7 μL) from erythroid cells derived from iPSCs were dispensed into appropriate wells. The gel was run at constant power (20 W, 1500 V) for 50-60 minutes. Immediately after IEF, the gel was submerged in 10% trichloroacetic acid (TCA) for 10 minutes on a rocking platform. Afterwards, the gel was washed with distilled water 3-5 times and stained using the JB-2 stain system (PerkinElmer).
Flow cytometry
Cells (1 × 105/mL) were stained for 30 minutes on ice with saturating amounts of FITC-conjugated or phycoerythrin (PE)–conjugated monoclonal antibodies against CD34, SSEA-4 (BD Pharmingen), and TRA-1-60, TRA-1-81, SSEA-1, SSEA-4 (eBioscience). Antibodies are described in detail in supplemental Table 2.
Primers for RT PCR and for detection of integration at AAVS1
Primers are described in supplemental Tables 3 and 4.
Results
Reprogramming of homozygote α-thalassemia fibroblasts and erythroblast differentiation
Homozygous α-thalassemia fibroblasts were obtained from the Coriell Institute and the mutations present in the α0-thal fibroblasts were characterized using a PCR assay22 (supplemental Figure 1). This verified that the cells were indeed homozygous for α-thalassemia and revealed the presence of the South-East-Asian (SEA) deletion on one chromosome and of the Filipino deletion on the other one.11 We therefore concluded that the cells contain no α-globin gene and a single ζ-globin gene because the Filipino deletion encompasses both the ζ- and α-globin genes.
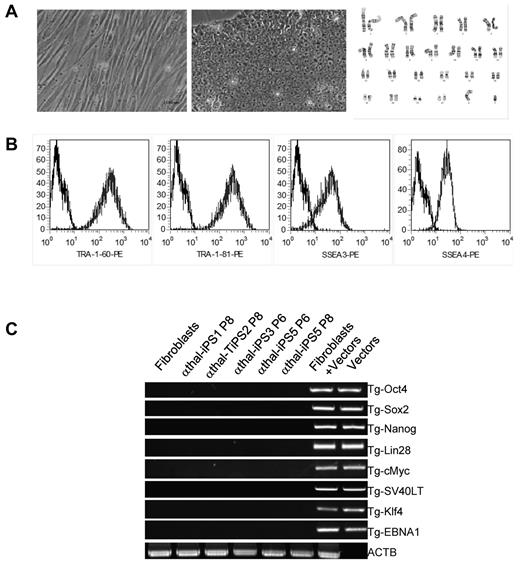
Once we had genotyped the cells, we generated transgene-free iPSCs from these fibroblasts using the episomal vector method developed by the Thomson laboratory.17 Five clones of iPSCs were selected because they had the typical morphology of hESCs and were characterized. After 8 to 10 passages to ascertain that the cells had lost the episomes, the cells were characterized using multiple assays: All lines tested had normal karyotype (Figure 1A), expressed high levels of the cell-surface markers TRA-1-60, TRA-1-81, SSEA-3, and SSEA-4 (Figure 1B), and of the transcription factors Oct4, Nanog, and Sox2 (supplemental Figure 2A). Quantitative RT-PCR demonstrated that mRNA levels of Nanog and Oct4 were comparable with H1 levels (supplemental Figure 2B).
Reprogramming of α-thalassemia fibroblasts and characterization of iPSCs. (A) Phase contrast micrographs illustrating the morphology of fibroblasts (left panel) and iPSCs (middle panel). iPSCs had normal karyotype in culture (right panel). (B) FACS analysis showing that iPSCs express TRA-1-60, TRA-1-80, SSEA-4, and SSEA-3, 4 typical hESCs and iPSCs surface markers. (C) PCR analysis showing loss of the episomal vectors used to reprogram patient-specific fibroblasts after 8 passages in culture. ACTB are control primers that detect a small genomic DNA fragment of the β-actin gene.
Reprogramming of α-thalassemia fibroblasts and characterization of iPSCs. (A) Phase contrast micrographs illustrating the morphology of fibroblasts (left panel) and iPSCs (middle panel). iPSCs had normal karyotype in culture (right panel). (B) FACS analysis showing that iPSCs express TRA-1-60, TRA-1-80, SSEA-4, and SSEA-3, 4 typical hESCs and iPSCs surface markers. (C) PCR analysis showing loss of the episomal vectors used to reprogram patient-specific fibroblasts after 8 passages in culture. ACTB are control primers that detect a small genomic DNA fragment of the β-actin gene.
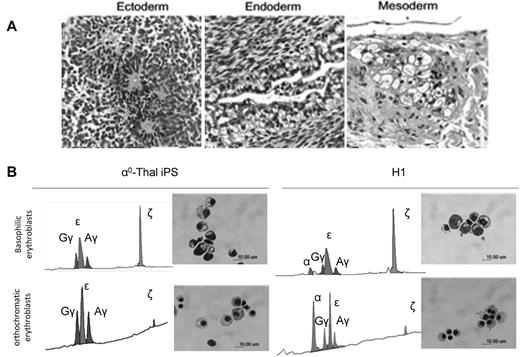
PCR analysis using transgene-specific primers revealed complete loss of the episomes after passage 8 in all 5 α0-thal-iPSC lines (Figure 1C). Pluripotency of the transgene-free α0-thal iPSCs was demonstrated in vivo using teratoma formation and in vitro by embryoid body formation. All the iPSC clones tested could differentiate into cells from the 3 germ layers in both assays (Figure 2A, supplemental Figure 3).
Differentiation of reprogrammed α-thalassemia fibroblasts. (A) H&E staining demonstrating that iPSCs derived from α-thalassemia fibroblasts with transgene-free method can form teratoma in NSG mice, and generate cells from the 3 germ layers (bar = 50 μm). (B) HPLC analysis of globin expression and morphology of the erythroblasts obtained after differentiation of α0-thal-iPSCs and control H1 hESCs. (Top panels) Basophilic erythroblasts obtained after 14 days of coculture on FH-B-hTERT and 14 days of liquid culture. (Bottom panels) Orthochromatic erythroblasts obtained after an additional 10 days of liquid culture. (Left panels) HPLC profiles and Giemsa staining of cells obtained after differentiation of α0-thal-iPSCs. (Right panels) Same as left panels but for control H1 hESCs. α0-thal-iPSCs do not express any α-globin chains. Zeta-globin chains are silenced between the 14th and the 24th day of liquid culture.
Differentiation of reprogrammed α-thalassemia fibroblasts. (A) H&E staining demonstrating that iPSCs derived from α-thalassemia fibroblasts with transgene-free method can form teratoma in NSG mice, and generate cells from the 3 germ layers (bar = 50 μm). (B) HPLC analysis of globin expression and morphology of the erythroblasts obtained after differentiation of α0-thal-iPSCs and control H1 hESCs. (Top panels) Basophilic erythroblasts obtained after 14 days of coculture on FH-B-hTERT and 14 days of liquid culture. (Bottom panels) Orthochromatic erythroblasts obtained after an additional 10 days of liquid culture. (Left panels) HPLC profiles and Giemsa staining of cells obtained after differentiation of α0-thal-iPSCs. (Right panels) Same as left panels but for control H1 hESCs. α0-thal-iPSCs do not express any α-globin chains. Zeta-globin chains are silenced between the 14th and the 24th day of liquid culture.
Once we had shown that the α0-thal-iPSCs were pluripotent, we differentiated them and control H1 hESCs into erythroid cells to assess globin gene expression. Differentiation was induced as previously described9 by coculture with immortalized human fetal hepatocytes for 14 days, followed by 24 days of liquid culture (supplemental Figure 4A-B). In these conditions, the ζ-globin gene is expressed at a high level in the basophilic erythroblasts, which are predominant after 14 days of liquid culture, while the α-globin genes are expressed most highly in the mature orthochromatic erythroblasts which are the most abundant cells at day 24 of liquid culture.
HPLC analysis at day 14 and day 24 of liquid culture revealed that as expected, erythroid cells derived from α0-thal-iPSCs expressed no α-globin chains. Interestingly, we also observed that the ζ-globin gene on the chromosome carrying the SEA deletion was silenced normally during maturation of the α-thalassemic erythroid cells (Figure 2B). This univocally demonstrates that in these cells, ζ-globin silencing does not require the presence of α-globin genes and therefore that this globin switch is not dependent on competition between the α and ζ globin promoters for interaction with HS-40, the major regulatory element of the α-globin cluster.23
ZFN-mediated gene incorporation in AAVS1 site
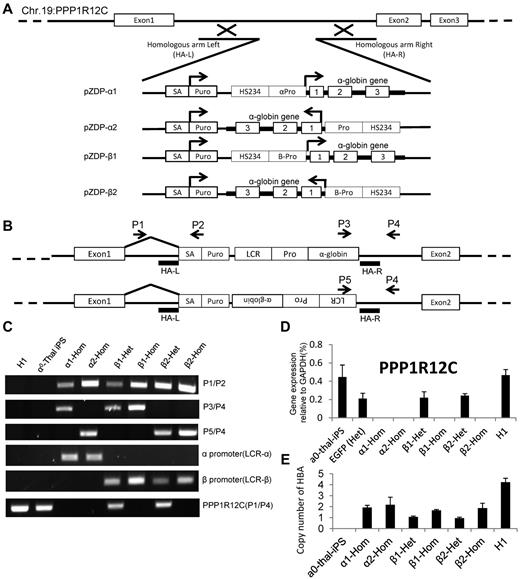
To correct the genetic defect in the α0-thal-iPSCs, we then integrated at AAVS1 constructs containing a gene trap based on the puromycin gene14 and a globin therapeutic cassette derived from a gene therapy vector developed by the Sadelain laboratory.18 The globin cassettes that we used contain the β-globin mini-LCR, and either the β or the α-globin promoter driving the α-globin gene (including all introns).
Four α-globin targeting cassettes that differ by the promoter and the orientation of the transgene relative to the PPP1R12C gene were constructed (Figure 3A). For validation purposes, the constructs were integrated at AAVS1 in K562 cells before testing in iPSCs. After transfection and isolation of puromycin-resistant clones, K562 cells were induced to express globin chains using hemin and globin expression was characterized by HPLC. This revealed that α-globin expression was increased 30% to 40% compared with the unmodified cells (supplemental Figure 5). Levels of α-globin mRNA were also elevated confirming that our vectors were functional (supplemental Figure 6).
ZFN-mediated integration of α-globin expression constructs in AAVS1 site. (A) Integration strategy. Four different α-globin constructs were tested. Therapeutic cassettes were inserted between exons 1 and 2 of the PPP1R12C by cotransfection of 2 ZFN-containing plasmids and of a targeting construct containing short homologies to part of intron 1 flanking the therapeutic transgenes. Successful targeting leads to expression of the puromycin gene under the control of the PPP1R12C promoter because of the presence of the splice acceptor (SA). (B) Location of the primer sets for demonstration of ZFN-mediated integration of α-globin constructs after selection of puromycin-resistant colonies. P1 hybridizes to a region just 5′ of the left homology arm. P2 hybridizes to the puromycin-resistance genes. P3 hybridizes to the 3′ part of the α-globin gene; P4 hybridizes to a region just 3′ of the right homology arm. P5 hybridizes to the 5′ region of the LCR. P1/P2, P3/P4, and P5/P6 only yield a PCR product if the transgenes are specifically inserted at AAVS1. P1/P4 detects the unmodified PPP1R12C gene. (C) PCR results demonstrating insertions of α-globin constructs at AAVS1. The PPP1R21C (P1/P4) PCR amplification can be used to identify heterozygous or homozygous insertions. α1-Hom indicates homozygous transgene in same orientation as PPP1R12C, driven by α-globin promoter; β1-Het, heterozygous transgene in same orientation as PPP1R12C, driven by β-globin promoter; β1-Hom, homozygous transgene in same orientation as PPP1R12C, driven by β-globin promoter; and β2-Het and β2-Hom, same as above but transgene is in opposite orientation. (D) Expression of PPP1R12C after insertion of the therapeutic transgenes. Heterozygous clones express PPP1R12C at approximately 50% the level of the unmodified locus. Homozygous clones do not express any detectable levels of PPP1R12C (n = 3). These results confirm the results of the analysis in panel C. (E) Histograms illustrating the number of copies of the corrective α-globin construct inserted in the genome. Q-PCR analyses were performed to compare the number of copies of α and β-globin present in the genome after. Y-axis = 2 × 2(Ctα-globin − Ctβ-globin). Together with the Southern blot analysis, these results demonstrate that these clones did not harbor any off-target integrations.
ZFN-mediated integration of α-globin expression constructs in AAVS1 site. (A) Integration strategy. Four different α-globin constructs were tested. Therapeutic cassettes were inserted between exons 1 and 2 of the PPP1R12C by cotransfection of 2 ZFN-containing plasmids and of a targeting construct containing short homologies to part of intron 1 flanking the therapeutic transgenes. Successful targeting leads to expression of the puromycin gene under the control of the PPP1R12C promoter because of the presence of the splice acceptor (SA). (B) Location of the primer sets for demonstration of ZFN-mediated integration of α-globin constructs after selection of puromycin-resistant colonies. P1 hybridizes to a region just 5′ of the left homology arm. P2 hybridizes to the puromycin-resistance genes. P3 hybridizes to the 3′ part of the α-globin gene; P4 hybridizes to a region just 3′ of the right homology arm. P5 hybridizes to the 5′ region of the LCR. P1/P2, P3/P4, and P5/P6 only yield a PCR product if the transgenes are specifically inserted at AAVS1. P1/P4 detects the unmodified PPP1R12C gene. (C) PCR results demonstrating insertions of α-globin constructs at AAVS1. The PPP1R21C (P1/P4) PCR amplification can be used to identify heterozygous or homozygous insertions. α1-Hom indicates homozygous transgene in same orientation as PPP1R12C, driven by α-globin promoter; β1-Het, heterozygous transgene in same orientation as PPP1R12C, driven by β-globin promoter; β1-Hom, homozygous transgene in same orientation as PPP1R12C, driven by β-globin promoter; and β2-Het and β2-Hom, same as above but transgene is in opposite orientation. (D) Expression of PPP1R12C after insertion of the therapeutic transgenes. Heterozygous clones express PPP1R12C at approximately 50% the level of the unmodified locus. Homozygous clones do not express any detectable levels of PPP1R12C (n = 3). These results confirm the results of the analysis in panel C. (E) Histograms illustrating the number of copies of the corrective α-globin construct inserted in the genome. Q-PCR analyses were performed to compare the number of copies of α and β-globin present in the genome after. Y-axis = 2 × 2(Ctα-globin − Ctβ-globin). Together with the Southern blot analysis, these results demonstrate that these clones did not harbor any off-target integrations.
Because the constructs and integration strategy had been validated, we integrated the same 4 cassettes as well as a GFP control cassette at the AAVS1 site in one selected α-thal-iPSC clone, as described in Figure 3. After puromycin selection, 5 clones for each construct were expanded and examined by PCR and RT-PCR to ascertain that the transgenes were indeed inserted at AAVS1. All clones had insertions at the AAVS1 site. Importantly, approximately 50% of the clones had homozygous insertions. This was demonstrated by the loss of the wild-type allele in the genomic DNA as detected by PCR and by complete loss of expression of PPP1R12C detected by quantitative RT-PCR (Figure 3B-D, supplemental Figure 7, supplemental Table 1). Insertion at AAVS1 was confirmed by Southern blots (supplemental Figure 8) which demonstrated the heterozygous and homozygous insertions. To ascertain that there was no off-target integration, we then performed quantitative PCR (Q-PCR) analysis to measure the number of α-globin gene copies in the cells using the β-globin gene copy number as a reference (Figure 3E). As expected, this analysis revealed that there were 4 α-globin genes in the H1 cells, none in the α0-thal-iPSCs, 1 in the heterozygous corrected α0-thal-iPSCs and 2 in the homozygous corrected α0-thal-iPSCs. Together with the Southern blots, these results demonstrate that there was no off-target integration in these clones.
Clones containing the 4 constructs in the heterozygous or homozygous forms were then further characterized to verify their pluripotency. All clones were similar to H1 and to the parental α0-thal-iPSCs in terms of surface markers, Nanog and Oct4 expression and teratoma formation (supplemental Figures 9-11). Karyotype analysis revealed that no translocation had occurred in the clones after ZFN-mediated insertion (supplemental Figure 12).
Effect of insertion of therapeutic transgenes on neighboring genes at AAVS1
The targeted iPSCs were then differentiated into erythroid cells and globin expression levels determined by HPLC as described in “Methods” using H1 hESC-derived erythroid cells as a control. Because erythroblasts derived from hESCs might be different from erythroblasts produced in vivo, we also produced erythroblasts from a 16-week-old fetal liver. These erythroblasts are the most appropriate controls because they express hemoglobin profiles that are very similar to those of erythroid cells derived from hESCs or iPSCs. Erythroblasts used for all analyses were at least 95% glycophorin A (CD235a) and CD71 positive (supplemental Figure 13).
As illustrated in Figure 4A and supplemental Figure 14, and summarized in Table 1, the expression of α-globin in basophilic erythroblasts derived from clones in which the therapeutic α-globin gene was driven by the β-globin promoter was 4.7% to 5.4% of total globin in the heterozygous states and 8.2% to 12.9% in the homozygous states. In the orthochromatic erythroblasts, α-globin expression was 13.3% of total globin in heterozygous and 25.8% in the homozygous clones. Importantly, in the clones in which the α-globin promoter was driving the transgenes, the levels of expression were higher, reaching, in the homozygous state, 14.5% of total globin expression in basophilic erythroblasts and 42.6% in orthochromatic erythroblasts. In the control H1 and fetal liver–derived cells, the α-globin gene represented, respectively, 46% and 45.4% of all globins in orthochromatic erythroblasts. To determine whether that level of expression was sufficient to correct the chain imbalance, we calculated the α-like/β-like ratios (Table 1). Introduction of homozygous transgenes driven by the α-globin promoter led to a complete correction of chain imbalance in basophilic erythroblasts and to an almost complete correction in the mature erythroblasts (α-like/β-like ratio = 0.86 in the corrected iPSCs compare with 0.02 in the untargeted α0-thal-iPSCs) in the corrected iPSCs. α-like/β-like ratio in the control H1 and fetal liver cells were, respectively, 101.2 and 91.4 in orthochromatic erythroblasts. Correction of chain imbalance was higher in the more immature cells because of higher level expression of the ζ-globin gene. These results were confirmed by quantitative RT-PCR analysis of globin mRNA levels in basophilic erythroblasts, a stage where globin expression is rapidly increasing. The levels of globin expression expressed as ratio to GAPDH (Figure 4C) were in excellent agreement with the results at the protein levels with the homozygous corrected iPSCs expressing approximately twice as much α-globin than the heterozygous iPSCs.
Erythroid cell differentiation of corrected α0-thal-iPSCs. (A) Chromatograms illustrating the results of HPLC analyses of globin chain expression performed after 14 days (14 FHB 14LC) or 24 days (14FHB 24 LC) of liquid culture of corrected iPSCs differentiated into erythroid cells. The 3 chromatograms on the left illustrate the globin expression pattern in orthochromatic erythroblasts obtained from culture of CD34+ cells from cord blood, fetal liver, and peripheral blood from a transfusion dependent adult with α0-thalassemia. β1-Het indicates heterozygous transgene integrated in same orientation as PPP1R12C, driven by β-globin promoter; β1-Hom, homozygous transgene in same orientation as PPP1R12C, driven by β-globin promoter; α1-Hom, homozygous transgene in same orientation as PPP1R12C, driven by α-globin promoter. Almost complete correction of chain imbalance in mature erythroid cells was obtained when homozygous transgenes driven by the α-globin promoter were inserted at AAVS1. The β-globin promoter was less effective than the α-globin promoter. Orientation of the transgene had no major effect on expression. (B) Q-RT-PCR analysis of globin expression in hESCs and iPSCs after differentiation into basophilic erythroblasts (14 days of liquid culture). The y-axis indicates the fold-difference compared with GAPDH. (C) IEF electrophoresis on lysates of orthochromatic erythroblasts illustrating the hemoglobin tetramers expressed in controls, in H1 and in corrected iPSCs. Globin content of each tetramer was determined by HPLC after cutting-out each major band from the gel. Corrected iPSCs express Hb F and Hb Gower II in addition to embryonic globins. A vertical line has been inserted to indicate a repositioned gel lane.
Erythroid cell differentiation of corrected α0-thal-iPSCs. (A) Chromatograms illustrating the results of HPLC analyses of globin chain expression performed after 14 days (14 FHB 14LC) or 24 days (14FHB 24 LC) of liquid culture of corrected iPSCs differentiated into erythroid cells. The 3 chromatograms on the left illustrate the globin expression pattern in orthochromatic erythroblasts obtained from culture of CD34+ cells from cord blood, fetal liver, and peripheral blood from a transfusion dependent adult with α0-thalassemia. β1-Het indicates heterozygous transgene integrated in same orientation as PPP1R12C, driven by β-globin promoter; β1-Hom, homozygous transgene in same orientation as PPP1R12C, driven by β-globin promoter; α1-Hom, homozygous transgene in same orientation as PPP1R12C, driven by α-globin promoter. Almost complete correction of chain imbalance in mature erythroid cells was obtained when homozygous transgenes driven by the α-globin promoter were inserted at AAVS1. The β-globin promoter was less effective than the α-globin promoter. Orientation of the transgene had no major effect on expression. (B) Q-RT-PCR analysis of globin expression in hESCs and iPSCs after differentiation into basophilic erythroblasts (14 days of liquid culture). The y-axis indicates the fold-difference compared with GAPDH. (C) IEF electrophoresis on lysates of orthochromatic erythroblasts illustrating the hemoglobin tetramers expressed in controls, in H1 and in corrected iPSCs. Globin content of each tetramer was determined by HPLC after cutting-out each major band from the gel. Corrected iPSCs express Hb F and Hb Gower II in addition to embryonic globins. A vertical line has been inserted to indicate a repositioned gel lane.
Quantitation of HPLC analysis in controls and corrected iPSCs
| . | . | α0 Thal iPS . | β1* . | β2* . | α1† . | α2† . | H1 . | FL . | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Het . | Hom . | Het . | Hom . | Hom . | Hom . | |||||
| Baso-E, 14-day liquid culture | ||||||||||
| α/all globins | % α globin | 0.0 | 4.7 | 8.2 | 5.4 | 12.9 | 14.5 | 13.6 | 9.8 | |
| α/α+ζ | αζ ratio | 0.0 | 12.1 | 18.8 | 11.8 | 24.6 | 31.8 | 28.8 | 19.0 | |
| α+ζ/γ+ϵ+β | Chain imbalance | 73.2 | 63.6 | 76.6 | 83.9 | 110.9 | 83.8 | 89.3 | 106.2 | |
| Ortho-E, 24-day liquid culture | ||||||||||
| α/all globins | % α globin | 0.0 | 13.3 | 25.8 | ND | 26.0 | 42.6 | 37.8 | 46 | 45.4 |
| α/α+ζ | αζ ratio | 0.0 | 79.5 | 75.4 | ND | 94.5 | 98.4 | 81.9 | 91.5 | 95.0 |
| α+ζ/γ+ϵ+β | Chain imbalance | 2.3 | 20.1 | 52.0 | ND | 38.0 | 76.5 | 85.8 | 101.2 | 91.4 |
| . | . | α0 Thal iPS . | β1* . | β2* . | α1† . | α2† . | H1 . | FL . | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Het . | Hom . | Het . | Hom . | Hom . | Hom . | |||||
| Baso-E, 14-day liquid culture | ||||||||||
| α/all globins | % α globin | 0.0 | 4.7 | 8.2 | 5.4 | 12.9 | 14.5 | 13.6 | 9.8 | |
| α/α+ζ | αζ ratio | 0.0 | 12.1 | 18.8 | 11.8 | 24.6 | 31.8 | 28.8 | 19.0 | |
| α+ζ/γ+ϵ+β | Chain imbalance | 73.2 | 63.6 | 76.6 | 83.9 | 110.9 | 83.8 | 89.3 | 106.2 | |
| Ortho-E, 24-day liquid culture | ||||||||||
| α/all globins | % α globin | 0.0 | 13.3 | 25.8 | ND | 26.0 | 42.6 | 37.8 | 46 | 45.4 |
| α/α+ζ | αζ ratio | 0.0 | 79.5 | 75.4 | ND | 94.5 | 98.4 | 81.9 | 91.5 | 95.0 |
| α+ζ/γ+ϵ+β | Chain imbalance | 2.3 | 20.1 | 52.0 | ND | 38.0 | 76.5 | 85.8 | 101.2 | 91.4 |
Summary of HPLC analyses of globin chain expression performed after 14 days or 24 days of liquid culture.
iPSC indicates induced pluripotent stem cell; Thal, thalassemia; FL, fetal liver; Het, heterozygous insertion; Hom, homozygous insertion; and ND, not determined.
Columns β1 and β2 summarize the results for the iPSCs in which the α-globin therapeutic gene is driven by the β-globin promoter. In iPSCs, the α1 and β1 transgenes are in the same orientation as the PPP1R12C genes. In iPSCs, the α2 and β2 transgenes are in opposite orientation (n = 3).
Columns α1 and α2 summarize the results for the iPSCs in which the α-globin therapeutic gene is driven by the α-globin promoter.
To further characterize the hemoglobin produced by the corrected cells, we quantified the total level of globin expression per cell using spectrophotometry (Table 2). Concentration of hemoglobin per cells was 22.6 pg/cell for the fetal liver–derived cells, 6.2 pg/cells for the uncorrected α0-thal-iPSCs and 21.1 pg/cell for the homozygous corrected iPSCs where the transgene was driven by the α-globin promoter.
Hemoglobin concentration
| Sample . | MCH, pg/cell . |
|---|---|
| Fetal liver | 22.6 ± 0.3 |
| α0-Thal iPS | 6.2 ± 0.0 |
| β1-Het | 16.6 ± 0.1 |
| β1-Hom | 19.9 ± 0.2 |
| β2-Het | 17.9 ± 0.3 |
| α1-Hom | 21.1 ± 0.4 |
| α2-Hom | 19.5 ± 0.5 |
| Sample . | MCH, pg/cell . |
|---|---|
| Fetal liver | 22.6 ± 0.3 |
| α0-Thal iPS | 6.2 ± 0.0 |
| β1-Het | 16.6 ± 0.1 |
| β1-Hom | 19.9 ± 0.2 |
| β2-Het | 17.9 ± 0.3 |
| α1-Hom | 21.1 ± 0.4 |
| α2-Hom | 19.5 ± 0.5 |
Hemoglobin concentrations (in pictogram per cell) in orthochromatic erythroblasts produced from fetal liver and uncorrected and corrected α0-thal iPSCs. Hemoglobin concentrations (in pictograms/cell) determined by the TMB method in orthochromatic erythroblasts derived from fetal liver and uncorrected and corrected iPSCs with the various constructs integrated (see Table 1 legend).
MCH indicates mean corpuscular hemoglobin; Thal, thalassemia; iPS, induced pluripotent stem; Het, heterozygous insertion; Hom, homozygous insertion; and TMB, tetramethylbenzidine.
Finally, to obtain a qualitative view of the tetrameric hemoglobins present in the cells, we performed IEF electrophoresis on lysates of orthochromatic erythroblasts (Figure 4C). As expected, the results revealed that the α0-thal-iPSCs express mostly Hb Bart (γ4), Hb Gower I (ζ2ϵ2), and Hb Portland (ζ2γ2). As expected, we did not detect any hemoglobin tetramers that contain α-globin. By contrast, the corrected iPSCs expressed the same hemoglobins but also express Hb F (α2γ2) and Hb Gower II (α2ϵ2). The control H1 and fetal liver–derived erythroblasts expressed hemoglobins that were very similar to the corrected α0-thal-iPSCs. Control adult orthochromatic erythroblasts with α0-thalassemia expressed predominantly Hb H which is a tetramer of the adult β-globin chains, a chain not expressed in erythroblasts derived from pluripotent cells.
Whether the AAVS1 site is a safe harbor is unclear at the current time because the site is located in a gene-rich region that contains several important genes.15 To start addressing this question, we measured by Q-RT-PCR expression of 10 neighboring genes (Figure 5A) in undifferentiated iPSCs and in erythroid cells obtained after iPSC differentiation. To maximize sensitivity, the analysis was performed in clones with homozygous insertions. Insertion of the LCR at AAVS1 in undifferentiated cells had remarkably little effect on the neighboring genes (supplemental Figure 15). In differentiated cells, insertion of the β-globin LCR-containing constructs at AAVS1 led to activation of 4 genes (Figure 5B). The most highly deregulated genes (up to 10-fold) were TNNT1 and TNNI3 2 subunits of troponin, which are regulatory components of the thin filament in sarcomere and GP6 (glycophorin VI), a platelet membrane glycoprotein of the immunoglobulin superfamily. Activation was generally more pronounced in the presence of the β-globin promoter containing cassette than in the presence of the α-globin promoter cassettes. The orientation of the therapeutic transgenes inserted at AAVS1 also had an effect on the activation of the neighboring genes with the weakest activation observed when the α-globin promoter was intercalated between the neighboring genes and the LCR. Importantly, when the results were compared in absolute terms with GAPDH expression (Figure 5C), the levels of expression of all the neighboring genes were extremely low, virtually undetectable in most cases, and lower than 0.5% of GAPDH in the others cases. When the cassette containing the α-promoter was inserted in the reverse orientation relative to PPP1R12C, the most highly activated gene was TNNI3 which reached 0.06% of GAPDH.
Effect of insertion of therapeutic transgenes on neighboring genes at AAVS1. (A) Top panel shows a map of 10 neighboring genes on chromosome 19 around AAVS1 site. (B) Bar graph illustrates Q-RT-PCR analysis of 10 neighbor genes after differentiation of corrected α0-thal-iPSCs into basophilic erythroblasts (normalized to parental iPSCs). All iPSCs analyzed carried homozygous insertions at AAVS1. mRNA expression levels were calculated using the Delta (δ Ct) method using differentiated uncorrected α0-thal-iPSCs as the controls (n = 3). Four genes were activated. The smallest activation was obtained when the α-globin promoter construct was inserted in the same orientation as the PPP1R12C gene. (C) Same as above but data are expressed relative to GAPDH. Expression of the neighboring genes is low compared with GAPDH.
Effect of insertion of therapeutic transgenes on neighboring genes at AAVS1. (A) Top panel shows a map of 10 neighboring genes on chromosome 19 around AAVS1 site. (B) Bar graph illustrates Q-RT-PCR analysis of 10 neighbor genes after differentiation of corrected α0-thal-iPSCs into basophilic erythroblasts (normalized to parental iPSCs). All iPSCs analyzed carried homozygous insertions at AAVS1. mRNA expression levels were calculated using the Delta (δ Ct) method using differentiated uncorrected α0-thal-iPSCs as the controls (n = 3). Four genes were activated. The smallest activation was obtained when the α-globin promoter construct was inserted in the same orientation as the PPP1R12C gene. (C) Same as above but data are expressed relative to GAPDH. Expression of the neighboring genes is low compared with GAPDH.
Discussion
We have produced transgene-free iPSCs carrying homozygous α-thalassemia deletions and have inserted 2 different therapeutic constructs at AAVS1 using zinc-finger nucleases. These results showed that introduction of homozygous transgenes at AAVS1 driven by the α-globin promoter correct the chain imbalance, leads to the formation of Hb F and Hb Gower II tetramer and leads to the production of cells containing a hemoglobin concentration very similar to control pluripotent cells or fetal liver–derived erythroblasts.
We also observed that the orientation of the therapeutic construct relative to the PPP12R1C gene had no major effects on transgene expression and that the α-globin promoter yields higher levels of expression than the β-globin promoter. Generally, hemoglobin concentration and chain balance were proportional to the level of expression of the transgene as determined by HPLC.
Achieving quasinormal expression levels of globin transgenes is of importance because it has remained a difficult goal to achieve for the gene therapy field. For instance, it was recently reported that a hemoglobinopathy patient treated by gene therapy was almost cured because he became transfusion independent and his hemoglobin levels were raised very significantly.24 However, even in this successful trial, the patient would have likely benefited from higher levels of expression of the transgenic globin because the final levels obtained were below normal levels.
The observation that the α-globin promoter is stronger than the β-globin promoter when inserted at AAVS1 in pluripotent stem cells is potentially important for the gene therapy field because most studies have focused on the β-globin promoter. However, whether this will also hold true in adult erythroid cells which express their endogenous β-globin genes will have to be tested.
To determine whether the AAVS1 site was a safe harbor, we measured expression of 10 neighboring genes before and after insertions of the LCR-containing therapeutic transgenes. No effects were detected in undifferentiated iPSCs but a few genes were slightly activated in the erythroid lineage probably because of the presence of the LCR. Importantly, activation of the neighboring genes was dependent on the promoter used and the orientation of the cassette and was lowest when the transgenes were most highly expressed, maybe because the α-globin promoter competes more effectively than the β-globin promoter for interaction with the LCR. It will be interesting to determine whether insulators can be used to completely eliminate activation of the neighboring genes.25
Lombardo et al have recently reported that transgenes inserted at AAVS1 were expressed without any effect on the neighboring genes.26 The discrepancy between this report and our findings might be because of the fact that globin transgenes are expressed at extremely high levels in fully differentiated erythroid cells. It is unclear whether activation of the neighboring genes could have any deleterious effects in the erythroid or any other hematopoietic cells because the absolute levels of activation observed in the presence of the α-promoter cassettes are very small.
An additional potential problem with the AAVS1 site is that complete correction of chain imbalance was only achieved when the insertion was homozygous which led to a complete knock-out of PPP1R12C expression. Lombardo et al have shown that it was possible to insert genes at AAVS1 without affecting PPP1R12C by removing introns from the cassette.26 It will be important to test whether this is true for genes expressed at very high levels such as the globin genes. In any case, these findings clearly suggest that other possible integration sites should be thoroughly evaluated.
Two other major issues will have to be solved before clinical trials can be considered. The first is the lack of a method to efficiently differentiate human pluripotent cells into transplantable HSCs. Mouse transplantable HSCs are also difficult to produce from mouse embryonic stem cells, but the problem has been solved by overexpressing transcription factors such as Bcr-abl or HoxB4 in mouse embryonic stem cells.4,27,28 To the best of our knowledge, this method does not work in human cells, but other factors might achieve the same result. The second issue that must be resolved is linked to the genomic integrity of the iPSCs. It has been shown that iPSCs carry genetic and epigenetic mutations that are in part carried-over from the donor cells, and in part acquired during the reprogramming process.29-33 In addition, the use of ZFNs has been reported to be associated with genotoxicity.34,35 Many of these genetic and epigenetic abnormalities might be eliminated by carefully choosing the donor cells and by optimizing the iPSC derivation and the genetic correction procedures. Endogenous HSCs also acquire genomic and epigenomic abnormalities throughout life. The number and type of genomic abnormalities that is safe and therefore tolerable is currently unknown but the rapid technological progress in sequencing might provide an answer in the future.
In any case, our study demonstrates that quasinormal protein levels of α-globin can be obtained by introducing 2 transgenes at the AAVS1 in α0-thal-iPSCs. This provides an important method to correct genetic defect such as large deletions that cannot be easily repaired by homologous recombination.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr M. Sadelain for generously providing plasmid pLM18-TNS9.3-HBA1 and for useful discussions. They also thank Ms S. Suzuka for helping with IEF electrophoresis, and Dr J. Locker for help with the Southern blots.
C.-J.C. and E.E.B. are supported in part by National Institutes of Health grant HL08846 and by grants C024405 and C024172 from NYSTEM, the funding agency of the New York State Empire Stem Cell Board.
National Institutes of Health
Authorship
Contribution: C.-J.C. performed most experiments and contributed to the experimental design and interpretation of the results; E.E.B. contributed to the experimental design and interpretation of the results.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Eric E. Bouhassira, PhD, Department of Medicine, Division of Hematology, Department of Cell Biology, Albert Einstein College of Medicine, Ullamnn 903, 1300 Morris Park Ave, Bronx, NY 10461; e-mail: eric.bouhassira@einstein.yu.edu.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal