Abstract
TP53 mutation is an independent marker of poor prognosis in patients with diffuse large B-cell lymphoma (DLBCL) treated with cyclophosphamide, hydroxydaunorubicin, vincristine, and prednisone (CHOP) therapy. However, its prognostic value in the rituximab immunochemotherapy era remains undefined. In the present study of a large cohort of DLBCL patients treated with rituximab plus CHOP (R-CHOP), we show that those with TP53 mutations had worse overall and progression-free survival compared with those without. Unlike earlier studies of patients treated with CHOP, TP53 mutation has predictive value for R-CHOP–treated patients with either the germinal center B-cell or activated B-cell DLBCL subtypes. Furthermore, we identified the loop-sheet-helix and L3 motifs in the DNA-binding domain to be the most critical structures for maintaining p53 function. In contrast, TP53 deletion and loss of heterozygosity did not confer worse survival. If gene mutation data are not available, immunohistochemical analysis showing > 50% cells expressing p53 protein is a useful surrogate and was able to stratify patients with significantly different prognoses. We conclude that assessment of TP53 mutation status is important for stratifying R-CHOP–treated patients into distinct prognostic subsets and has significant value in the design of future therapeutic strategies.
Introduction
Diffuse large B-cell lymphoma (DLBCL) is the most common (30%-40%) and perhaps the most heterogeneous type of nonHodgkin lymphoma and has aggressive clinical features. Using gene-expression profiling (GEP), DLBCLs can be classified into 2 molecularly distinctive types: germinal center B-cell like (GCB) and activated B-cell like (ABC) that resemble the GEPs of normal germinal center B cells and mitogenically activated blood B cells, respectively.1 Despite this heterogeneity, DLBCL patients are usually treated with the same combination chemotherapy regimen, traditionally cyclophosphamide, hydroxydaunorubicin, vincristine, and prednisone (CHOP), and in the past decade, rituximab plus CHOP (R-CHOP). R-CHOP has clearly improved the outcome of DLBCL patients. In a study of young patients, the 6-year event-free survival (EFS) was shown to be 74.3% and overall survival (OS) was 94.9%.2 In elderly patients, the rate of complete response (CR) has been shown to be 76%3 ; the 10-year progression-free survival (PFS) was shown to be 36.5% and the OS 43.5%.4 However, among patients with refractory disease or with early relapse, the CR rate was only 38%, and the 3-year EFS rate was only 31% after high-dose chemotherapy and autologous stem-cell transplantation.5 Therefore, the identification of prognostic biomarkers for high-risk DLBCL subgroups is essential for designing targeted chemotherapy.
The p53 (TP53) protein (393 aa) encoded by the TP53 gene is a crucial tumor suppressor that mediates cell-cycle arrest, DNA repair, apoptosis, senescence, and autophagy under cellular stress via transcription-dependent activity (TA) and transcription-independent activity (TIA) in the nucleus and cytoplasm, respectively.6,7 TP53 dysfunction is implicated in lymphomagenesis and disease progression,8,9 and normal function of TA and TIA of p53 is crucial for tumor suppression.10,11 Nonsynonymous TP53 mutations alter the p53 protein sequence and structure, disrupt function, and in many tumors are the most common mechanism that inactivates TP53. TP53 mutations could provide prognostic and predictive information to guide more targeted therapy for lymphoma patients.12
The DNA-binding domain (DBD) of p53 is the most important domain for its TA and TIA,13,14 so most TP53 mutations occur in the DBD encoded by exons 4-8 of the TP53 gene.15,16 In our previous study of TP53 mutations in 477 DLBCL patients treated with CHOP therapy,17 we detected exon 5-9 TP53 mutations in 21.4% of DLBCL patients, 66% of which occurred in the loop (L1)–sheet-helix (LSH) motif and 2 large loops L2 and L3, which are crucial for DNA binding.18,19 Functionally, TA of most p53 mutants have been characterized by a yeast-based functional assay20 and in some human and yeast lines, available in the International Agency for Research in Cancer (IARC) TP53 database (http://www.iarc.fr).15 Many p53 mutants (MUT-TP53) exhibit dominant-negative characteristics on wild-type p53 (WT-TP53) function by associating together as tetramers.
The prognostic significance of TP53 mutations has been inconsistent in several cancers, probably because of limitations of mutation detection methods, heterogeneity of TP53 mutations, inherent heterogeneity of disease, different therapies, diversity of p53 functions, and other mechanisms that inactivate the TP53 pathway.13,21,22 Our study and others have shown that TP53 mutation is an independent prognostic indicator of poor survival in DLBCL patients treated with CHOP chemotherapy.16,17,23-25 In our earlier study using a different DLBCL patient cohort treated with CHOP therapy, TP53 mutations were correlated with worse OS, mostly attributable to the poor prognostic impact of TP53 mutations in the DBD. When we subgrouped patients into GCB- and non–GCB-DLBCL based on GEP data, the predictive value of TP53 mutations was restricted to patients with GCB-DLBCL.17
The anti-CD20 Ab rituximab combined with CHOP has significantly improved survival of DLBCL patients. Rituximab acts through 3 mechanisms: direct signaling, complement-mediated cytotoxicity, and Ab-dependent cellular cytotoxicity.26 Influx of external calcium (Ca2+) is essential for signaling-induced apoptosis mediated by the BCR and CD20.27,28 Therapeutic (high) doses of rituximab induce intracellular Ca2+ mobilization independently of CD20 and inhibit tumor cell growth.29 The effects of rituximab are attributed to inhibition of the BCR-signaling cascade,30 p38, NF-κB, ERK, and Akt and down-regulation of cytokine IL-10 and BCL-2.26 The TA and function of p53 might differ after rituximab addition as a result of different stresses induced by R-CHOP and CHOP because the p53 transcriptional program is stimulus and cell-type specific.13,31 The TIA of p53 can also be affected by the p53-independent signaling pathways induced by rituximab. Therefore, the prognostic value of TP53 mutation in DLBCL patients treated with R-CHOP needs to be evaluated.
To address the prognostic significance of TP53 mutations in DLBCL, in the present study, we analyzed exons 2-11 of TP53 in a large cohort of de novo DLBCL patients treated with R-CHOP and compared mutation status with clinical outcome.
Methods
Patients
The study cohort consisted of 506 de novo DLBCL patients treated with R-CHOP therapy in 29 medical centers selected based on the eligibility and exclusion criteria of histology, treatments, availability of clinical data, and biologic results. All patients were diagnosed according to the World Health Organization (WHO) criteria and reviewed by a group of hematopathologists (participating center pathologists were S.M.-M., M.A.P., M.B.M., A.T., and K.H.Y.). Patients with primary mediastinal large B-cell lymphoma, cutaneous or CNS DLBCL, HIV infection, or a history of low-grade B-cell lymphoma with transformation to DLBCL were excluded.32,33 Eligible patients were diagnosed between June 15, 1998 and October 8, 2008 (14 patients diagnosed in 1998-1999 were enrolled in the R-CHOP clinical trials). Most patients (n = 416) underwent 6-8 cycles of R-CHOP-21; others (n = 90) received R-CHOP–like regimens. In 182 patients, radiotherapy (25-50 Gy) followed R-CHOP and in 29 patients, hematopoietic stem cell transplantation followed R-CHOP (7 patients received both radiotherapy and transplantation). Treatment responses (CR, partial response, stable disease, and progressive disease) and follow-up were evaluated according to the recommended criteria.34,35 Thirty-eight percent (193 of 506) of the patients had died by the last follow-up. The median follow-up interval for the 313 censored patients was 59.73 months (range, 12-180.26). The present study, conducted in accordance with the Declaration of Helsinki, was approved as being of minimal or no risk or as exempt by the institutional review boards of all participating centers, and the overall study was approved by the institutional review board of the University of Texas MD Anderson Cancer Center.
TP53 mutations by resequencing microarray
Genomic DNAs extracted from formalin-fixed, paraffin-embedded (FFPE) tissues were used for TP53 exon sequencing. Sequencing of the coding sequence (exons 2-11) and splicing sites was performed using p53 AmpliChip (Roche Molecular Systems).36 For data analysis and classification, TP53 sequences were generated and compared with the TP53 reference sequence (NC_000017.10) in the GenBank database.
GEP and subtype classification
Total RNAs extracted from FFPE tissues were subjected to GEP using Affymetrix GeneChip Human Genome HG-U133 Plus Version 2.0 to classify 506 DLBCL patients into GCB or ABC subtypes, as described previously.33 Immunohistochemistry (IHC) based on staining of biomarkers including CD10, BCL-6, GCET-1, FOXP-1, and MUM-133 was performed on tissue microarrays to classify the 506 patients according to the Visco-Young algorithm with high concordance with GEP analysis, as described previously.33
FISH for TP53 gene (chromosome 17p13.1) deletion
Among the 506 DLBCL patients studied by GEP, 440 FFPE tissue sections were available for evaluation of chromosome 17p13.1 deletions by interphase FISH using a LSI TP53 Spectrum Orange Probe (Vysis) and methods described previously.16 A CEP 18 Spectrum Aqua Probe (Vysis) was used to evaluate simultaneously for chromosome 17 copy number. The ratio of TP53 signals (orange) to centromere 17 signals (green) was counted in 200 tumor cells. A ratio lower than 0.81 was considered to support the presence of a TP53 deletion. Because no patients had a ratio below 0.5, it was assumed that no homozygous deletions occurred.
IHC for p53
IHC staining was performed for all patients using a method described previously and the DO-7 mAb (DAKO).16 The percentage of positive tumor cells for each DLBCL patient in 5% increments was evaluated independently by a group of 6 pathologists in addition to each contributing center pathologist, and disagreements were resolved by joint review on a multihead microscope. Receiver operating characteristic (ROC) curve analysis was performed using Prism Version 5 software (GraphPad) to assess the discriminatory accuracy of p53 protein overexpression in predicting TP53 mutation status.
Statistical analysis
A comparison of clinical and laboratory features at the time of presentation between different DLBCL subgroups was carried out using the χ2 test and the Spearman rank correlation. OS was calculated from time of diagnosis to time of death from any cause. PFS was calculated from the time of diagnosis to the time of progression or death from any cause.35 Patients who remained alive or progression free were censored at the time of last follow-up. Multivariate analysis for survival of the study cohort was performed with SAS Version 9.3 software for Windows (SAS Institute) using the Cox proportional hazards regression model. OS and PFS curves of different groups were analyzed by Prism 5 using the Kaplan-Meier method and differences were compared using the log-rank (Mantel-Cox) test. All differences with P ≤ .05 were considered to be statistically significant.
Results
Most TP53 mutations present in DLBCL are missense mutations in the DBD
Gene sequencing detected 133 TP53 gene mutations in 112 DLBCL patients, including 111 (21.9%) patients with nonsynonymous mutations or p53 mutants (MUT-TP53) and 17 patients with multiple (2-4) mutations. Mutation prevalence and patterns are shown in Figure 1A-B and Table 1. The 112 patients with 133 TP53 mutations included 92 with missense mutations (including 15 patients who carried 2 missense mutations); 14 with a nonsense mutation; 2 with a 2-bp deletion resulting in frameshift; 3 with a silent mutation; and 7 carrying a mutation in splicing sites. Missense mutations accounted for 81% of the total mutations present in this cohort.
Patterns of TP53 mutations in 506 DLBCL patients treated with R-CHOP. (A) Proportions of classified point mutations. (B) Proportions of mutations based on mutation impact on the p53 protein sequence. (C) Proportions of mutations based on mutation effect on the p53 function. (D) Distribution of mutation numbers according to TP53 exons. The numerals on top are numbers of mutations in exons, and the numerals at the bottom are numbers of mutations in splicing sites. (E) Codon distribution of TP53 mutations. Codons with mutations of high frequency in the cohort are marked.
Patterns of TP53 mutations in 506 DLBCL patients treated with R-CHOP. (A) Proportions of classified point mutations. (B) Proportions of mutations based on mutation impact on the p53 protein sequence. (C) Proportions of mutations based on mutation effect on the p53 function. (D) Distribution of mutation numbers according to TP53 exons. The numerals on top are numbers of mutations in exons, and the numerals at the bottom are numbers of mutations in splicing sites. (E) Codon distribution of TP53 mutations. Codons with mutations of high frequency in the cohort are marked.
Mutation prevalence in 506 de novo DLBCL patients treated with R-CHOP
| . | No. of patients . | Prevalence . | No. of mutations . | No. of mutation variants . |
|---|---|---|---|---|
| Total patients | 506 | 133 | 99 | |
| Patients with WT-TP53* | 395 | 78% | 1 | 1 |
| Patients with MUT-TP53† | 111 | 22% | 132 | 98 |
| Patients with missense mutations‡ | 92 | 18.2% | 107 | 79 |
| Patients with nonsense mutations | 14 | 2.8% | 14 | 9 |
| Patients with a 2-bp deletion causing reading frame shift | 2 | 0.4% | 2 | 2 |
| Patients with silent mutations§ | 2 | 0.6% | 2 | 2 |
| Patients with a mutation at the splicing sites¶ | 7 | 1.4% | 7 | 6 |
| . | No. of patients . | Prevalence . | No. of mutations . | No. of mutation variants . |
|---|---|---|---|---|
| Total patients | 506 | 133 | 99 | |
| Patients with WT-TP53* | 395 | 78% | 1 | 1 |
| Patients with MUT-TP53† | 111 | 22% | 132 | 98 |
| Patients with missense mutations‡ | 92 | 18.2% | 107 | 79 |
| Patients with nonsense mutations | 14 | 2.8% | 14 | 9 |
| Patients with a 2-bp deletion causing reading frame shift | 2 | 0.4% | 2 | 2 |
| Patients with silent mutations§ | 2 | 0.6% | 2 | 2 |
| Patients with a mutation at the splicing sites¶ | 7 | 1.4% | 7 | 6 |
One DLBCL patient with only 1 silent mutation is also counted as WT-TP53.
Seventeen DLBCL patients had multiple (2-4) mutations.
Fifteen DLBCL patients had 2 missense mutations.
These 2 patients also carried missense mutations.
Four patients carrying a mutation at the splicing site also carried missense mutations.
The exon and codon distributions of these mutations are shown in Figure 1D and E. Exons 5-8 were most often mutated. A total of 117 (88.6%) mutations occurred in the DBD, including 25 (18.9%) in L3, 24 (18.2%) in the LSH motif, and 20 (15.2%) in L2. Codons 248, 273, 175, 176, and 213 of the p53 protein had the highest mutation frequency. Codons 248, 273, and 175 are the TP53 mutation hot spots found in most human cancers.
Most missense and nonsense mutations are loss-of-function mutations
Functional analysis of TP53 mutations using bioinformatics models.
Functional prediction of the 121 missense or nonsense mutations was computed by SIFT (http://sift.jcvi.org/) and AGVGD (http://agvgd.iarc.fr/) methods based on protein sequence homology. By SIFT, 108 (90%) missense or nonsense mutations were predicted to be deleterious (disrupt the p53 function), and 12 (10%) missense or nonsense mutations were predicted to be tolerable (neutral). By AGVGD, 99 (81.8%) missense mutations were predicted to be deleterious and 8 (6.6%) missense mutations were predicted to be neutral.
Yeast-based functional assay data in IARC database.
Effect of mutations on p53 transcription function can be retrieved from the International Agency for Research on Cancer (IARC) TP53 database (http://www.iarc.fr; TP53MutFunction2R15 dataset) that was established by a p53 transactivation activity assay in 8 yeast promoters (WAF1, MDM2, BAX, 14-3-3δ, p53AIP1, GADD45, NOXA, and p53R2) that harbor p53 response elements. Of 133 detected mutations, there were 99 mutation variants (some patients carried the same TP53 mutation variant), including 79 missense mutation variants, 9 nonsense mutations resulting in truncated protein, 6 splicing mutation variants, 2 frameshift mutation variants in exon 6 or 7, and 3 silent mutations. Among the 88 missense and nonsense mutation variants, 66 (75%) were nonfunctional, 16 (18.2%) partially functional, and only 4 (4.5%) functional, as experimentally measured by the yeast-based functional assay (Figure 1C). These data correspond to 94 (77.7%) nonfunctional, 18 (15%) partially functional, and 5 (4%) functional mutations among the 121 detected missense and nonsense mutations, with 4 missense or nonsense mutations uncharacterized. Splicing mutations and frameshift mutations result in more alterations of protein sequence than missense mutations and therefore potentially have a more deleterious effect on p53 function. Altogether, more than 90% of mutations result in loss of p53 function. Most mutations at hot spots, including R248W, R248Q, R273C, R273H, R175H, C176R, R213X, and R213Q, are nonfunctional, whereas C176F is partially functional, and R175C is functional. Furthermore, according to the IARC TP53MutFunction1R15 dataset, 32 mutants detected have a dominant-negative effect on p53 transcription function, and 3 mutants have lost the ability to form tetramers.
Functional data for a limited number of missense and nonsense mutants regarding TIA are available in the IARC TP53MutFunction1R15 dataset. R175H, Y234C, G245S, R248Q, R248W, R273H, C277Y (partial), R280K, and R337C (partial) have lost the ability to induce apoptosis in human cells, whereas S121F, R175C, R181C, and S240R are functional or partially functional regarding transcriptional activities and have conserved apoptosis function. Altogether, either by bioinformatic computations or by laboratory-based evaluation, approximately 90% of TP53 mutations detected in this study cohort were accompanied by a loss of p53 function.
Clinical features of the cohort
The clinical features of the WT-TP53 and MUT-TP53 groups are compared and summarized in Table 2. There were no significant differences between these 2 groups regarding age, sex, stage, B symptoms, number of extranodal sites, performance status, tumor size, or International Prognostic Index (IPI). Serum lactate dehydrogenase level (P = .05), tumor size (P = .02), and therapeutic response (P < .0001) were the only variables that were significantly different between these 2 groups.
Clinical characteristics of 506 de novo DLBCL with GCB or ABC subtypes treated with R-CHOP: comparison between patients with or without TP53 mutations
| . | Overall (N = 506) . | GCB (n = 258) . | ABC (n = 241) . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| WT-TP53 . | MUT-TP53 . | P . | WT-TP53 . | MUT-TP53 . | P . | WT-TP53 . | MUT-TP53 . | P . | |
| Patients, n | 395 | 111 | 190 | 68 | 198 | 43 | |||
| Age | |||||||||
| < 60 y | 163 | 43 | 94 | 33 | 64 | 10 | |||
| ≥ 60 y | 232 | 68 | .2073 | 96 | 35 | .8937 | 134 | 33 | .2427 |
| Sex | |||||||||
| F | 164 | 46 | 82 | 27 | 77 | 19 | |||
| M | 231 | 65 | .9883 | 108 | 41 | .6209 | 121 | 24 | .5201 |
| Stage | |||||||||
| I-II | 183 | 54 | 104 | 33 | 74 | 21 | |||
| III-IV | 192 | 57 | .9776 | 72 | 35 | .1361 | 118 | 22 | .2137 |
| B symptoms | |||||||||
| No | 232 | 66 | 119 | 41 | 108 | 25 | |||
| Yes | 112 | 29 | .7073 | 44 | 20 | .3929 | 67 | 9 | .1900 |
| LDH | |||||||||
| Normal | 129 | 27 | 71 | 14 | 57 | 13 | |||
| Elevated | 223 | 76 | .0497 | 95 | 48 | .0050 | 123 | 28 | .9906 |
| No. of extranodal sites | |||||||||
| 0-1 | 299 | 87 | 143 | 53 | 145 | 32 | |||
| ≥ 2 | 75 | 21 | .889 | 32 | 13 | .8020 | 47 | 10 | .9270 |
| Performance status | |||||||||
| 0-1 | 289 | 89 | 133 | 55 | 149 | 34 | |||
| ≥ 2 | 46 | 11 | .4775 | 17 | 6 | .7517 | 29 | 5 | .5891 |
| Size of largest tumor | |||||||||
| < 7.5 cm | 241 | 60 | 114 | 38 | 124 | 22 | |||
| ≥ 7.5 cm | 63 | 29 | .0201 | 27 | 21 | .0130 | 34 | 8 | .5349 |
| IPI risk group | |||||||||
| 0-2 | 225 | 66 | 118 | 41 | 100 | 25 | |||
| 3-5 | 122 | 36 | .9799 | 43 | 20 | .3698 | 79 | 16 | .5513 |
| Therapy response* | |||||||||
| CR | 316 | 67 | < .0001 | 153 | 41 | .0009 | 156 | 26 | .0113 |
| PR | 45 | 23 | 19 | 14 | 26 | 9 | |||
| SD | 11 | 7 | 6 | 4 | 5 | 3 | |||
| PD | 23 | 14 | 12 | 9 | 11 | 5 | |||
| . | Overall (N = 506) . | GCB (n = 258) . | ABC (n = 241) . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| WT-TP53 . | MUT-TP53 . | P . | WT-TP53 . | MUT-TP53 . | P . | WT-TP53 . | MUT-TP53 . | P . | |
| Patients, n | 395 | 111 | 190 | 68 | 198 | 43 | |||
| Age | |||||||||
| < 60 y | 163 | 43 | 94 | 33 | 64 | 10 | |||
| ≥ 60 y | 232 | 68 | .2073 | 96 | 35 | .8937 | 134 | 33 | .2427 |
| Sex | |||||||||
| F | 164 | 46 | 82 | 27 | 77 | 19 | |||
| M | 231 | 65 | .9883 | 108 | 41 | .6209 | 121 | 24 | .5201 |
| Stage | |||||||||
| I-II | 183 | 54 | 104 | 33 | 74 | 21 | |||
| III-IV | 192 | 57 | .9776 | 72 | 35 | .1361 | 118 | 22 | .2137 |
| B symptoms | |||||||||
| No | 232 | 66 | 119 | 41 | 108 | 25 | |||
| Yes | 112 | 29 | .7073 | 44 | 20 | .3929 | 67 | 9 | .1900 |
| LDH | |||||||||
| Normal | 129 | 27 | 71 | 14 | 57 | 13 | |||
| Elevated | 223 | 76 | .0497 | 95 | 48 | .0050 | 123 | 28 | .9906 |
| No. of extranodal sites | |||||||||
| 0-1 | 299 | 87 | 143 | 53 | 145 | 32 | |||
| ≥ 2 | 75 | 21 | .889 | 32 | 13 | .8020 | 47 | 10 | .9270 |
| Performance status | |||||||||
| 0-1 | 289 | 89 | 133 | 55 | 149 | 34 | |||
| ≥ 2 | 46 | 11 | .4775 | 17 | 6 | .7517 | 29 | 5 | .5891 |
| Size of largest tumor | |||||||||
| < 7.5 cm | 241 | 60 | 114 | 38 | 124 | 22 | |||
| ≥ 7.5 cm | 63 | 29 | .0201 | 27 | 21 | .0130 | 34 | 8 | .5349 |
| IPI risk group | |||||||||
| 0-2 | 225 | 66 | 118 | 41 | 100 | 25 | |||
| 3-5 | 122 | 36 | .9799 | 43 | 20 | .3698 | 79 | 16 | .5513 |
| Therapy response* | |||||||||
| CR | 316 | 67 | < .0001 | 153 | 41 | .0009 | 156 | 26 | .0113 |
| PR | 45 | 23 | 19 | 14 | 26 | 9 | |||
| SD | 11 | 7 | 6 | 4 | 5 | 3 | |||
| PD | 23 | 14 | 12 | 9 | 11 | 5 | |||
LDH indicates lactate dehydrogenase; PR, partial response; SD, stable disease; and PD, progressive disease.
For therapy response, we calculated P values as CR versus other responses.
Most patients were able to be classified into GCB- and ABC-DLBCL subtypes according to the GEP data (n = 441), supplemented by IHC assessment (n = 499). Of these, 258 patients were determined to be the GCB subtype, including 190 patients with WT-TP53 and 68 patients with MUT-TP53; 241 patients were determined to be the ABC subtype, including 198 patients with WT-TP53 and 43 patients with MUT-TP53. Serum lactate dehydrogenase (P = .005), tumor size (P = .013), and therapeutic response (P = .0009) differed significantly in patients with WT-TP53 versus MUT-TP53 in the GCB subtype, whereas only therapeutic response differed significantly in patients with WT-TP53 versus MUT-TP53 (P = .0113) in the ABC subtype.
Impact of TP53 mutations on survival in DLBCL patients treated with R-CHOP
TP53 mutations predict poor survival in all DLBCL patients.
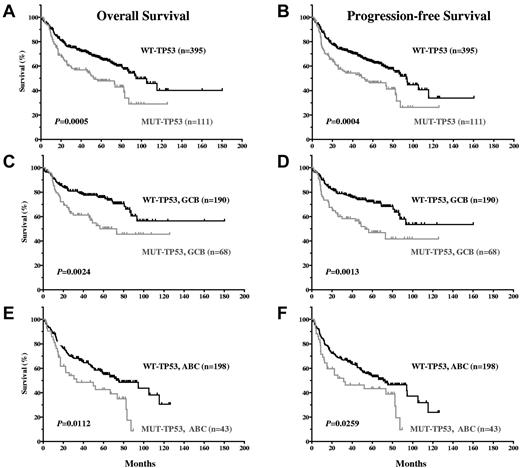
Our results show that TP53 mutation status is a significant prognostic factor that stratifies DLBCL patients treated with R-CHOP. DLBCL patients with WT-TP53 had significantly better OS and PFS compared with DLBCL with MUT-TP53 (P = .0005 and P = .0004, respectively; Figure 2A-B). The median OS of 395 patients with WT-TP53 DLBCL was 94.49 months, in contrast to 52.90 months for 111 patients with MUT-TP53 (hazard ratio [HR] = 0.5320; 95% confidence interval [CI], 0.37-0.76). Patients with MUT-TP53 were associated with a significantly higher, 1.9 times (95% CI, 1.32-2.68) hazard for OS (poorer survival) compared with patients with WT-TP53. The 5-year OS was 65.9% for patients with WT-TP53 versus 47.8% for those with MUT-TP53 DLBCL. The median PFS for patients with WT-TP53 DLBCL was 93.14 months versus 51.95 months for patients with MUT-TP53 DLBCL (HR = 0.5370; 95% CI, 0.38-0.76). The 5-year PFS was 63.5% for DLBCL patients with WT-TP53 versus 46.3% with MUT-TP53.
DLBCL patients with WT-TP53 have significantly better OS and PFS compared with DLBCL patients with MUT-TP53.
DLBCL patients with WT-TP53 have significantly better OS and PFS compared with DLBCL patients with MUT-TP53.
TP53 mutations predict poor survival in both the GCB- and ABC-DLBCL subtypes.
GCB-DLBCL patients had significantly better survival than ABC-DLBCL patients (supplemental Figure 1A-B, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). GCB-DLBCL patients with WT-TP53 had better OS (P = .0024; HR = 0.46; 95% CI, 0.28-0.76) and PFS (P = .0013; HR = 0.4483; 95% CI, 0.28-0.73) than GCB-DLBCL patients with MUT-TP53 (Figure 2C-D). Similarly, ABC-DLBCL patients with WT-TP53 had better survival than ABC-DLBCL patients with MUT-TP53 (P = .0112, HR = 0.5218, and 95% CI, 0.32-0.86 for OS; P = .0259, HR = 0.5665, and 95% CI, 0.34-0.93 for PFS; Figure 2E-F).
Multivariate analysis.
Multivariate analysis showed that an IPI score of > 2, TP53 mutation, ABC subtype, and B symptoms were the only prognostic factors that independently predicted worse OS and PFS in DLBCL patients treated with R-CHOP (Table 3). TP53 mutation is an important prognostic marker second to an IPI score of > 2. Patients with TP53 mutation had a 1.95 times hazard for OS (P = .0004) and a 1.84 times hazard (P = .001) for PFS compared with patients without TP53 mutation.
Multivariate analysis in terms of OS and PFS of patients with DLBCL treated with R-CHOP
| . | HR . | 95% CI . | P . |
|---|---|---|---|
| OS | |||
| IPI > 2 | 2.63 | 1.87-3.69 | < .0001 |
| TP53 mutation | 1.95 | 1.34-2.83 | .0004 |
| ABC subtype | 1.62 | 1.15-2.28 | .0062 |
| B symptoms | 1.43 | 1.02-2.01 | .0374 |
| PFS | |||
| IPI > 2 | 2.45 | 1.77-3.39 | < .0001 |
| TP53 mutation | 1.84 | 1.28-2.65 | .0010 |
| ABC subtype | 1.60 | 1.15-2.24 | .0052 |
| B symptoms | 1.43 | 1.03-1.99 | .0323 |
| . | HR . | 95% CI . | P . |
|---|---|---|---|
| OS | |||
| IPI > 2 | 2.63 | 1.87-3.69 | < .0001 |
| TP53 mutation | 1.95 | 1.34-2.83 | .0004 |
| ABC subtype | 1.62 | 1.15-2.28 | .0062 |
| B symptoms | 1.43 | 1.02-2.01 | .0374 |
| PFS | |||
| IPI > 2 | 2.45 | 1.77-3.39 | < .0001 |
| TP53 mutation | 1.84 | 1.28-2.65 | .0010 |
| ABC subtype | 1.60 | 1.15-2.24 | .0052 |
| B symptoms | 1.43 | 1.03-1.99 | .0323 |
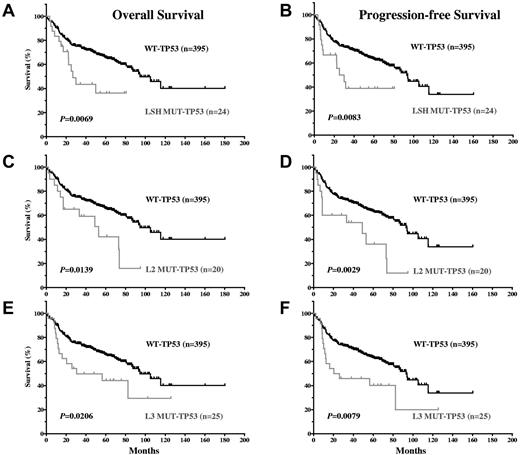
TP53 mutations in the LSH, L2, and L3 motifs predict poor survival in DLBCL patients.
Because most (105 of 111; 95%) MUT-TP53 DLBCL patients carried mutations in the DBD, survival curves for DLBCL patients with DBD TP53 mutations versus those with WT-TP53 closely mirrored the overall MUT-TP53 versus WT-TP53 data (figures not shown). Mutations in the 3 important motifs for DNA binding, LSH, L2, and L3, were all significantly associated with worse survival (Figure 3A-F). Mutations in LSH (including the codon 273 hot spot) and L3 (including the codon 248 hot spot) were particularly associated with decreased survival: for LSH, a median OS of 26.76 months and PFS of 27.64 months, and for L3, a median OS of 30.64 months and PFS of 23.23 months. The median OS and PFS for L2 mutations (52.90 and 48.99 months, respectively) were similar to those of DLBCL patients with MUT-TP53 overall.
DLBCL patients with TP53 mutations in the LSH, L2, and L3 motifs of the DNA binding domain have significantly inferior OS and PFS compared with DLBCL patients with WT-TP53.
DLBCL patients with TP53 mutations in the LSH, L2, and L3 motifs of the DNA binding domain have significantly inferior OS and PFS compared with DLBCL patients with WT-TP53.
Differentially expressed genes in MUT-TP53 and WT-TP53 patients
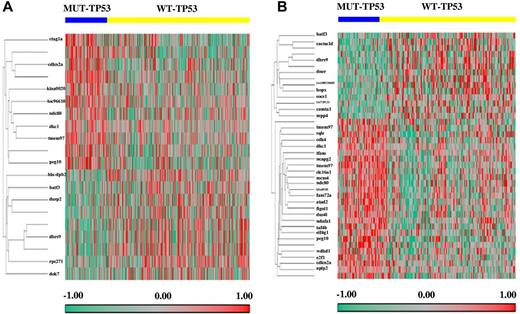
To identify the genes responsible for the different clinical outcomes of DLBCL patients with WT-TP53 versus MUT-TP53 and the different therapeutic response in DLBCL patients with GCB versus ABC subtypes, we profiled and compared the GEPs of WT-TP53 and MUT-TP53 DLBCLs (Figure 4A-B). Twenty probe sets were significantly differentially expressed between WT-TP53 and MUT-TP53 DLBCL patients. Genes up-regulated in WT-TP53 DLBCL included: DOK7, DHRS9, HLA-DPB2, BATF3, RPS27L, and DUSP2; genes down-regulated included: PEG10, CDKN2A, CTAG1A/CTAG1B, TMEM97, NDC80, LOC96610, KIAA0020, and DKC1.
Genes differentially expressed between DLBCL patients with MUT-TP53 and WT-TP53. (A) Fourteen genes significantly differentially expressed in DLBCL patients treated with R-CHOP. (B) Thirty-four genes significantly differentially expressed in the GCB-DLBCL subgroup.
Genes differentially expressed between DLBCL patients with MUT-TP53 and WT-TP53. (A) Fourteen genes significantly differentially expressed in DLBCL patients treated with R-CHOP. (B) Thirty-four genes significantly differentially expressed in the GCB-DLBCL subgroup.
The impact of p53 status on GEP was different in GCB- and ABC-DLBCL. In GCB-DLBCL (Figure 4B), 40 probe sets were differentially expressed between MUT-TP53 and WT-TP53. Genes up-regulated in WT-TP53 included: HOPX, DHRS9, LOC100510485, CACNA1D, BATF3, DNER, SOCS1, CAMTA1, LOC729121, and MPP4; genes down-regulated included KIAA0101, PEG10, TMEM97, GPR180, TAF4B, CDKN2A, FAM72A, FAM72B, FAM72C, FAM72D, MCM4, E2F1, ATAD2, SLC16A1, NDC80, FIGNL1, WDHD1, NCAPG2, DKC1, SQLE, NDUFA1, CDK4, EIF4G1, DUS4L, TFAM, PA2G4, PA2G4P4, and APLP2. In contrast, in ABC-DLBCL, only 1 gene (PSMD10) was significantly down-regulated in the WT-TP53 compared with the MUT-TP53 patients. The functions and subcellular locations of these genes (http://www.uniprot.org) are listed in supplemental Table 1.
Association of TP53 mutations and LOH
TP53 single-allele mutations are frequently followed by loss of heterozygosity (LOH), further promoting tumor development and disease progression.22,37 In the present study, we observed a weak association between TP53 mutations detected by p53 AmpliChip and TP53 deletions detected by FISH. Ninety-four MUT-TP53 and 346 WT-TP53 DLBCL patients were assessed by FISH (17 MUT-TP53 and 49 WT-TP53 patients had no tissue available or FISH failed). Overall, 12.3% of DLBCLs had allelic deletion, with a lower frequency than mutation (21.9%). In the MUT-TP53 group, there were 16 (17.0%) patients with TP53 allelic deletion. In comparison, 38 (11.0%) patients in the WT-TP53 group had TP53 allelic deletion. Therefore, TP53 deletion (LOH) was more common in DLBCL with MUT-TP53; however, the frequency difference did not reach statistical significance (P = .1136). Conversely, the TP53 mutation frequency was higher in DLBCLs with allelic deletions (29.6%) than in tumors without deletions (20.5%), but this was also not statistically significant (P = .1253). These results suggest a weak association between allelic deletion and TP53 mutations.
Others have reported that complete inactivation of TP53 by concomitant TP53 mutations and LOH is present most often in high-grade lymphomas and might be associated with high-grade transformation or disease progression.38 In the present DLBCL study cohort, a significantly higher frequency (P = .025) of allelic deletion was present in DLBCL patients with a high proliferation index (Ki-67 protein ≥ 70%; high Ki-67 = 7.4% vs low Ki-67 = 14.9%). TP53 mutations also occurred significantly more frequently (P = .0235) in DLBCLs with a high Ki-67 index (high = 25.7% vs low = 16.8%). In addition, 15 of 16 DLBCLs with LOH concurrent with TP53 mutation had a high Ki-67 index. These data suggest that TP53 mutations and LOH are associated with a high Ki-67 index.
Impact of TP53 allelic deletion on patient survival.
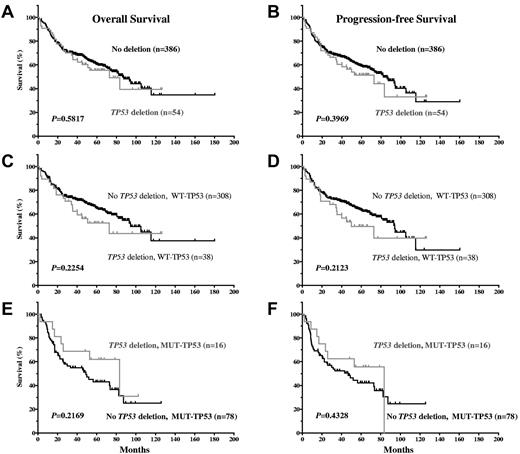
Our results show that TP53 allelic deletion was not significantly associated with decreased OS or PFS (Figure 5A-B). The impact of allelic deletion might have been attenuated by the poor survival potential of DLBCL patients with MUT-TP53 with or without allelic deletion. Therefore, we further examined the impact of TP53 allelic deletion after stratifying patients into the WT-TP53 and MUT-TP53 DLBCL subgroups. In DLBCL patients with either WT-TP53 or MUT-TP53, allelic deletion was not correlated with survival (Figure 5C-F.), although allelic deletion tended to be correlated with decreased survival in DLBCL patients with WT-TP53.
Impact of TP53 allelic deletion on OS and PFS in DLBCL patients treated with R-CHOP, with WT-TP53 or MUT-TP53.
Impact of TP53 allelic deletion on OS and PFS in DLBCL patients treated with R-CHOP, with WT-TP53 or MUT-TP53.
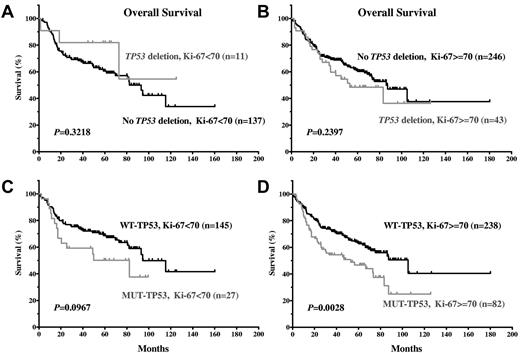
Because LOH and TP53 mutations are significantly associated with high Ki-67, we assessed the impact of LOH and TP53 mutations on survival in patients with a high or low Ki-67 index. TP53 mutations, but not allelic deletion, affected survival significantly in DLBCL patients with highly proliferative tumors (Ki-67 ≥ 70%; Figure 6A-D).
TP53 mutations and allelic deletions demonstrate distinct impacts on OS in DLBCL patients with a low or high proliferation index. (A-B) TP53 allelic deletions were not correlated with significantly inferior OS in DLBCL patients with low or high proliferation index. (C-D) TP53 mutations were correlated with significantly inferior OS in DLBCL patients with high proliferation index, but not in patients with low proliferation index.
TP53 mutations and allelic deletions demonstrate distinct impacts on OS in DLBCL patients with a low or high proliferation index. (A-B) TP53 allelic deletions were not correlated with significantly inferior OS in DLBCL patients with low or high proliferation index. (C-D) TP53 mutations were correlated with significantly inferior OS in DLBCL patients with high proliferation index, but not in patients with low proliferation index.
p53 overexpression by IHC in tissue sections.
The p53 protein expression level was determined in 483 DLBCL patients using IHC. Different cutoffs of positive cells yielded different sensitivity and specificity by ROC analysis (supplemental Table 2). Cutoffs between 10% and 60% stratified DLBCL patients into 2 groups with significantly different OS and PFS (P < .05; supplemental Table 2). However, p53 overexpression determined by IHC using cutoffs of 10%-40% is less discriminative than TP53 mutation status to stratify DLBCL patients into groups with different survival. Although a 30% cutoff of p53 expression by IHC had the biggest Youden index, a cutoff of 50% had a higher specificity, highest HR, and best P value in patient stratification. Therefore, we believe that > 50% is the best p53 expression cutoff with which to stratify DLBCL patients in diagnostic practice if sequencing of TP53 is not available.
Discussion
In the present study, we found a TP53 mutation prevalence of 21.9% in DLBCL patients, and the mutation patterns were similar to patterns in some other cancers. Most TP53 mutations were missense mutations, suggesting the importance of maintaining p53 structure in tumor suppression. In addition, most mutations occurred in the DBD, with hot spots at codons 248 (in L3), 273 (in LSH), and 175 and 176 (in L2), indicating that these motifs are critical for p53 tumor-suppression function. Bioinformatics tools based on alignments of TP53 sequences during evolution predicted that 90% of these missense mutations disrupt p53 function. Functional assays in human or yeast cells confirmed that most of these mutants lose transcriptional function. Impairment of transcription-independent apoptosis function by TP53 mutations has also been reported previously.39-41 The mutants R273H, L194F, R175H, and E285K have a low ability to translocate to mitochondria and induce apoptosis under stress conditions42 ; therefore, loss of p53 TA or TIA could be a driver for lymphomagenesis.
In this large cohort of de novo DLBCL patients treated with R-CHOP, we show that TP53 mutations, especially mutations in the L3 and LSH motifs, are correlated with disease progression and poor survival (Figures 2 and 3). To eliminate the possible impact of radiotherapy and transplantation on survival, we also performed survival analysis for patients who received standard R-CHOP therapy only and found that patients with WT-TP53 had significantly better OS (P = .0041) and PFS (P = .0084). Therefore, despite the addition of rituximab to CHOP, TP53 mutation is an independent prognostic factor that predicts poor survival in patients with DLBCL. This observation holds true in patients with the GCB or ABC subtypes of DLBCL (Figure 2C-F) and further supports the crucial role of p53 in death of tumor cells and tumor suppression. Nonetheless, the impact of TP53 mutation on survival of GCB-DLBCL patients is more pronounced than for ABC-DLBCL patients (Figure 2C-F). In fact, TP53 mutation status was not an independent prognostic factor in patients with ABC-DLBCL treated with CHOP.17 The distinctive molecular programs harbored by GCB- and ABC-DLBCL43 might explain the differences in treatment response and impact of TP53 mutations. Perhaps somatic hypermutation (SHM) in GCB-DLBCL under chemotherapy may trigger rapid apoptosis in WT-TP53 patients, whereas accumulation of more oncogenic events in MUT-TP53 makes these tumors resistant to apoptosis. In contrast, the SHM machinery is inactivated in ABC-DLBCL, which instead expresses a pre-plasma cell program. NF-κB and BCR signaling pathways are activated in ABC-DLBCL,43,44 causing resistance to apoptosis and chemotherapy, negatively affecting the survival of ABC-DLBCL patients with WT-TP53, and minimizing the prognostic value of MUT-TP53. Therefore, with CHOP treatment, TP53 status is an independent predictor of treatment response only in the GCB subtype. However, with R-CHOP therapy, rituximab inhibits the NF-κB and BCR signaling pathways, and ABC-DLBCL patients with MUT-TP53 have significantly worse survival than those with WT-TP53.
In addition, similar to CHOP regimen, ABC-DLBCL patients treated with R-CHOP have significantly inferior OS and PFS compared with GCB-DLBCL patients both in the entire DLBCL cohort (supplemental Figure 1A-B) and in the WT-TP53 subcohort (supplemental Figure 1C-D). Our multivariate analysis also suggested that ABC subtype independently predicted worse survival (Table 3). These data are in agreement with a recent study.45 Most of the clinical characteristics contributing to IPI score and IPI score itself were significantly different between GCB-DLBCL and ABC-DLBCL patients (supplemental Table 3) in overall DLBCL and DLBCL with WT-TP53, suggesting that molecularly distinctive types can be clinically reflected on IPI parameters. In DLBCL patients with MUT-TP53, IPI score was not significantly different between the GCB and ABC subtypes (supplemental Table 3); correspondingly, OS and PFS were not significantly different (supplemental Figure 1E-F). This might be because of the small cohort of DLBCL patients with MUT-TP53, but also might reflect the dominant adverse impact of p53 mutations on survival in both the GCB and ABC subtypes.
We also performed GEP in DLBCL patients with WT-TP53 and MUT-TP53, as well as according to GCB- and ABC-DLBCL subtypes (Figure 4). One critical feature between WT-TP53 and MUT-TP53 DLBCLs is that certain genes were differentially expressed in GCB-DLBCL, but only 1 gene (PSMD10, a negative regulator of RB1 and p53) was significantly down-regulated in ABC-DLBCL with WT-TP53. These results imply either absence or heterogeneity of the p53 transcription activities in ABC-DLBCL and probably indicate that p53 is less activated in ABC- compared with GCB-DLBCL, perhaps because of decreased activity of SHM. Similar phenomena were observed previously in DLBCL patients treated with CHOP.16,17 However, ABC-DLBCL patients with WT-TP53 still had better survival compared with patients with MUT-TP53 (Figure 2E-F), suggesting that transcription-independent p53 function is mainly responsible for the better survival of ABC-DLBCL patients with WT-TP53 treated with R-CHOP.
The GEP differences between WT-TP53 and MUT-TP53 DLBCLs (Figure 4) could be explained as loss or altered p53 function in MUT-TP53. From this perspective, loss of tumor-suppressor function of MUT-TP53 includes transactivation of several genes involved in signaling and transcription, including negative regulators of ERK, JNK, and cytokine signaling pathways; activators of the NOTCH1 pathway; and transrepression of several genes involved in antiapoptosis, cell-cycle progression, biosynthesis, proliferation, transcriptional activation, and G-protein signaling. Conversely, the difference in gene expression between the WT-TP53 and MUT-TP53 DLBCL patients may result from gain-of-function of some MUT-TP53 mutants. The tumor-suppressor gene CDKN2A (encoding p16INK4A and p14ARF) was down-regulated in WT-TP53 DLBCL patients (Figure 4A-B). p16INK4A can cause cell-cycle arrest by inhibiting CDK4; p14ARF sequesters the p53-negative regulator MDM2, thereby enhancing p53 level and activity, or induces cell-cycle arrest and apoptosis by p53-independent pathways. p14ARF also inhibits the function of BCL-6,46 which is expressed in B-cell germinal centers and activated in a subset of lymphomas.44 The repression of CDKN2A by p53 has also been reported in a cell line study,47 and might provide negative feedback to the TP53 pathway.48 The known p53 targets (eg, MDM2, BAX, and p21) did not appear to be differentially expressed (P = insignificant), reflecting the biologic heterogeneity of DLBCLs.
The prognostic value of p53 overexpression as a surrogate for TP53 mutations has been studied by others, but with inconsistent results. We used an IHC method to detect p53 overexpression and compared it with TP53 mutation status. We evaluated different cutoffs of p53 expression by ROC analysis and found that most cutoffs were able to stratify patients with significantly different OS and PFS. However, most stratification using IHC appears to be less discriminating than TP53 mutation status. In the present study, a > 50% cutoff was determined to be the best for diagnostic practice if gene-sequencing data are not available.
In contrast to TP53 mutations, we found that TP53 allelic deletion, which also may inactivate the TP53 pathway, had no significant impact on patient survival, either overall or within the WT-TP53 or MUT-TP53 subsets (Figure 5C-F). In fact, DLBCL patients with both LOH and MUT-TP53 had a slightly better survival than DLBCL patients with MUT-TP53 alone (Figure 5E-F). The higher frequency of TP53 mutations than allelic deletions in our cohort may imply that LOH occurred after TP53 mutation, which drives lymphomagenesis. LOH seemingly added no further selective growth advantage for tumor cells that carried TP53 mutation (Figure 5E-F), which probably reflects the dominant-negative effect exerted by MUT-TP53 on WT-TP53 expressed from the other intact allele and gain-of-function by MUT-TP53 resulting in a greater decrease in survival than loss-of-function by LOH. Alternatively, the slightly worse survival of MUT-TP53 patients (Figure 5E-F) could be caused by TP53 mutations present in the other allele. The AmpliChip we used for mutation detection cannot distinguish between homozygous and heterozygous mutations, but 13 of 78 MUT-TP53 patients without LOH carried 2 or 3 different TP53 mutations. The comparably weaker growth advantage provided by LOH compared with TP53 mutation explains the low percentage of allelic deletions (17%) in MUT-TP53 DLBCLs and also the absence of statistically significant association between TP53 mutations and LOH in this cohort. However, concomitant LOH and TP53 mutation were almost always in patients with a high proliferation index (Ki-67 ≥ 70%), suggesting an association of LOH with disease progression. Nonetheless, our results (Figure 6A-D) suggest that TP53 mutations and not LOH drive disease progression.
In conclusion, in the present study, we identified TP53 mutation patterns in a large cohort of patients with de novo DLBCL who were treated with R-CHOP and confirmed that TP53 mutation status is a valuable prognostic biomarker. TP53 mutations, especially the ones in the LSH, L3, and L2 motifs of the DBD, were significantly correlated with worse survival in patients with either GCB- or ABC-DLBCL. Therefore, therapeutic approaches targeting the inactivated TP53 pathway may further improve clinical outcomes of patients with DLBCL treated with R-CHOP.49,50
There is an Inside Blood commentary on this article in this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank their consortium program team of pathologists, hematologists, and clinicians; each of the contributing center principal physicians; their patients; and former and current hematopathology and hematology/oncology fellows for their support.
Z.Y.X.-M. is a recipient of the Shannon Timmins Leukemia Fellowship Award at The University of Texas MD Anderson Cancer Center. A.T. is a recipient of the Stiftung zur Krebsbekaempfung Zurich Grant 269 award. K.H.Y is supported by The University of Texas MD Anderson Cancer Center Institutional Research and Development Fund, Institutional Research Grant Award, MD Anderson Cancer Center SPORE Research Development Program Award, Gundersen Lutheran Medical Foundation Award, MD Anderson Cancer Center Collaborative Funds with Roche, HTG Molecular and Daiichi Sankyo, and the Forward Lymphoma Fund. This study was also partially supported by the National Cancer Institute/National Institutes of Health (R01CA138688, 1RC1CA146299, and P50CA142509).
National Institutes of Health
Authorship
Contribution: Z.Y.X.-M., L.W., and K.H.Y. designed and conducted the research, performed and supported the statistical analysis, and wrote the manuscript; Z.Y.X.-M., L.W., C.V., Y.C.T., A.T., W.L., S.M.-M., K.D., A.C., A.O., Y.Z., G.B., K.L.R., E.D.H., X.F.Z., W.W.L.C., X.Z., J.H.v.K., Q.H., J.H., W.A., M.P., A.J.M.F., F.Z., J.N.W., W.X., J.L., R.S.G., Y.L, M.A.P., M.B.M., R.N.M., L.V.A., L.J.M., and K.H.Y. contributed vital new reagents, resources, and analytical tools under approval by the institutional review boards and the material transfer agreement; Z.Y.X.-M., C.V., A.T., S.M.-M., K.D., A.C., A.O., Y.Z., G.B., K.L.R., E.D.H., X.F.Z., W.W.L.C., X.Z., J.H.v.K., Q.H., J.H., W.A., M.P., A.J.M.F., F.Z., J.N.W., R.S.G., M.A.P., M.B.M., R.N.M., L.V.A., L.J.M., and K.H.Y. collected clinical and follow-up data under approval by the institutional review boards and the material transfer agreement; Z.Y.X.-M., and K.H.Y. analyzed the data; and all authors contributed vital strategies, participated in discussions, and provided scientific input.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The DLBCL Rituximab-CHOP Consortium Program has the principal investigation center at The University of Texas MD Anderson Cancer Center in Houston, TX, and includes 29 participating centers. A material transfer agreement was established and approved by each of the participating centers of the DLBCL Rituximab-CHOP Consortium Program. For a complete list of DLBCL Rituximab-CHOP Consortium Program participating centers, please see the supplemental Appendix.
Correspondence: Ken H. Young, MD, PhD, The University of Texas MD Anderson Cancer Center, Department of Hematopathology, 1515 Holcombe Blvd, Houston, TX 77030-4009; e-mail: khyoung@mdanderson.org.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal