Abstract
The coding single nucleotide polymorphism GFI136N in the human gene growth factor independence 1 (GFI1) is present in 3%-7% of whites and increases the risk for acute myeloid leukemia (AML) by 60%. We show here that GFI136N, in contrast to GFI136S, lacks the ability to bind to the Gfi1 target gene that encodes the leukemia-associated transcription factor Hoxa9 and fails to initiate histone modifications that regulate HoxA9 expression. Consistent with this, AML patients heterozygous for the GFI136N variant show increased HOXA9 expression compared with normal controls. Using ChipSeq, we demonstrate that GFI136N specific epigenetic changes are also present in other genes involved in the development of AML. Moreover, granulomonocytic progenitors, a bone marrow subset from which AML can arise in humans and mice, show a proliferative expansion in the presence of the GFI136N variant. In addition, granulomonocytic progenitors carrying the GFI136N variant allele have altered gene expression patterns and differ in their ability to grow after transplantation. Finally, GFI136N can accelerate a K-RAS driven fatal myeloproliferative disease in mice. Our data suggest that the presence of a GFI136N variant allele induces a preleukemic state in myeloid precursors by deregulating the expression of Hoxa9 and other AML-related genes.
Introduction
Gfi1 is a DNA binding transcriptional repressor that recognizes specific target genes via its C-terminal C2H2 zinc finger domains. Gfi1 plays an important role in hematopoietic stem cell (HSC) function and B- and T-cell differentiation.1,2 It is also an important factor for myeloid differentiation because Gfi1−/− mice have increased numbers of myeloid precursors, such as granulomonocytic progenitors (GMPs) and common myeloid progenitors (CMPs), accumulate aberrant monocytes and lack neutrophil granulocytes,3-7 and in humans loss of function mutations in GFI1 cause severe congenital neutropenia.8 A number of studies suggest that acute myeloid leukemia (AML) cells originate both in human and mice from the CMP, GMP, or lymphoid primed multipotential progenitors (LMPP) fractions.9-13 Recently, we reported that a single nucleotide polymorphism (SNP) in the GFI1 gene, which leads to the exchange of a serine to an asparagine at position 36, is associated with AML.14 The GFI136N variant allele is present in 3% to 7% of healthy whites and in 11% of AML patients. Persons carrying this SNP have a 1.6-fold increased risk to develop AML compared with persons not carrying this variant. It remains unclear, however, how the AML-associated GFI136N variant can be responsible for this predisposition. To better understand the effects of GFI136N, we have made knock-in mice carrying the human GFI136N variant or the more common GFI136S form at the endogenous murine Gfi1 gene locus. This enabled a more detailed investigation of the pathways that are normally regulated by GFI1 in cells expressing the GFI1 variant. Here we present the results that we obtained studying these engineered mice and offer a first mechanistic insight into the function of the GFI136N variant.
Methods
Generation of KI mice
Gfi136Nneo/+ or Gfi136Sneo/+ mice were generated following a previously described strategy.15 All mice were backcrossed to C57Bl/6 background (verified by PCR). All mice were housed under SPF conditions, and the institutional animal ethics committee approved all animal experiments. The following primers were used for genotyping mice: mGfi11, 5′-cccttctctcagaactcagag-3′; mGfi24, 5′-ctggcaagctcagcaaatctg-3′; and R9, 5′-GTTCACAGAAGAGGCCCAGG-3′. The expected knock-in band is at 520 bp, and the control wild-type band at 350 bp. Mice were backcrossed to B57L/6J background, and this was verified by background specific satellite PCR.
K-RAS activation and bone marrow transplantation
Expression of a mutated form of K-RAS was induced by injecting the various MxCre tg K-RASflstoplfK12D mice with 500 mg polyriboinosinic acid/polyribocytidylic acid (poly(I:C); Sigma-Aldrich) 3 times every other day.
In vitro colony assay
Approximately 450-500 GMPs were seeded on methylcelluose (Stem Cells, M3434) and expanded further. At day 10, 17, and 24, colonies were counted and replated, and pictures were taken using the Axiovert s100TV microscope.
ChIP
For ChIP with Lin−, Sca1−, ckit+ 5 × 105 cells were used, otherwise 5-50 × 106 cells were used. Cells were washed, cross-linked with formaldehyde, and lysed. The collected lysate was sonicated to an average size of 600 bp. Samples were immunoprecipitated with anti-Gfi1 antibody (σ-2.5D) or control antibody and collected with protein G/salmon sperm DNA. Beads were washed and samples were eluted. After reverse-crosslinking, DNA was purified with the Qiaquick PCR Purification Kit (QIAGEN); ∼ 5 μL of diluted DNA was used for each real-time PCR reaction. An α-Gfi1 antibody (2.5D; Sigma-Aldrich) was used for examining binding of Gfi1 to the target genes, as control an α-GFP antibody (sc9996; Santa Cruz Biotechnology) was used. For the H3K4 dimethyl ChIP, a kit (GAM-3203) from SAB-Bioscience was used. For the examination of the Hoxa9 locus, the following primer pairs from SAB Bioscience were used: GPM1052016(+)02A, +1A, −1A, −2A, −3A, and −13A. The primers for the ID2 and Gfi1 have been previously described.16,17
Chip seq
ChIP-Seq ChIP assays were performed as previously described18 using polyclonal anti–dimethyl-histone H3 (Lys4; Millipore 07-030) and anti-Gfi1 (Abcam ab21061) antibodies. Briefly, 5 × 105 Lin−Sca1−cKit+ cells and 2 × 107 MLL-ENL–immortalized bone marrow progenitor cells were cross-linked in 1% formaldehyde, lysed and sonicated to fragments of ∼ 150-400 bp. Each sample was amplified using the Illumina kit following the manufacturer's instructions and sequenced using the Illumina 2G Genome Analyzer. Sequencing reads were mapped to the mouse reference genome using Bowtie19 converted to a density plot as described20 and displayed in the UCSC genome browser. In a first step, reads were normalized as reads per indicated region divided through number of total reads of the genome. In order to take account in different efficiencies of the ChIP, the curves were normalized to genes, which are not Gi1 target genes such as hypoxanthin-phosphoribosyl-transferase (HPRT) or beta-actin. After normalization, the different curves were superimposed and the area under each curve was determined using the TL 100 software from Non-Linear Dynamics. Sequence data have been submitted to the NCBI archive under accession no. GSE31657.
Gene expression array and real-time PCR
Gene expression array was performed according to published procedures.21 Array data are accessible under GEO Omnibus accession no. 25551. Real-time PCR for Hoxa9 expression in human and murine samples was performed with specific predesigned primers from ABI (human HOXA9 HS00365956_m1 and murine Hoxa9 Mm00439364-m1) according to the manufacturer's instructions.
Statistical analysis
The log-rank test was used for comparing survival rates of mice and the unpaired Student t test for analyzing the differences in the number of the different populations. All P values were calculated 2-sided. Statistical analysis was done with Prism Version 4 software (GraphPad).
Flow cytometry analysis and sorting of populations
Cells were analyzed and sorted as previously described.22-24 Lineage-negative cells were defined as the 1% of the live-gated BM cells with the lowest fluorescence of the lineage antibodies.
Reporter assay, methylcellulose assay, and immunoprecipitation
Patient cohorts
The characteristics of the cohort of patient cohorts from Nijmegen and Rotterdam were previously described.14 All patient samples were obtained after informed consent according to the Declaration of Helsinki. The respective local ethic committees have approved the use of all patient samples.
Results
Generation of mice carrying the human GFI136N and GFI136S cDNA in the murine Gfi1 locus
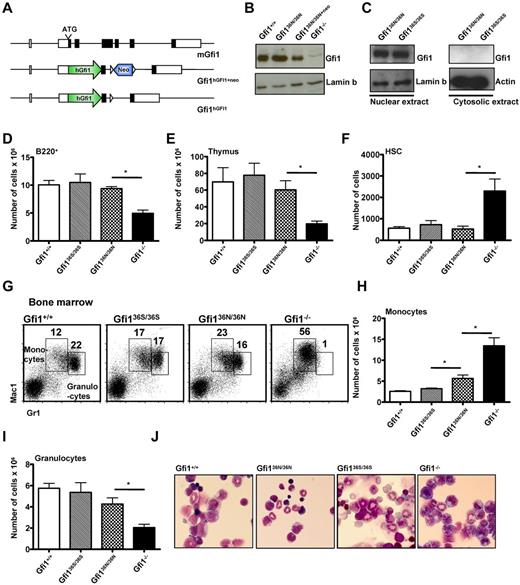
To understand how the GFI136N variant contributes to AML development, we used gene targeting to knock-in either the more common GFI136S form (Gfi136S/36S) or the GFI136N variant (Gfi136N/36N) into the murine Gfi1 locus (Figure 1A; supplemental Figure 1A-C, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). The 2 knock-in strains expressed GFI1 at levels similar to the endogenous murine Gfi1 (Figure 1B-C) and showed no significant difference to wt mice with regard to lymphoid differentiation in bone marrow, spleen, and thymus or the number of HSCs (Figure 1D-F; and data not shown). In addition, the cellular composition of the bone marrow and peripheral blood and the number of colony forming cells were also similar to wt mice, indicating a functionally intact hematopoiesis in both types of knock-in mice (Tables 1 and 2; supplemental Figure 2).
Human GFI1 is equivalent to murine Gfi1 in hematopoiesis. (A) Schematic representation of the murine Gfi1 locus and the targeted alleles. (B) Western blot of nuclear extracts of Gfi136N/36N, Gfi1+/+, Gfi1−/−, and Gfi136N/36N+neo thymocytes. (C) Western blot of nuclear and cytosolic extracts of Gfi136N/36N and Gfi136S/36S thymocytes. (D) Total number of B220+ cells in the bone marrow (per both hind limbs; n = 23 Gfi1+/+, n = 13 Gfi1−/−, n = 13 Gfi136N/36N, and n = 9 Gfi136S/36S). *P ≤ .05. (E) Total number of thymocytes (n = 9 Gfi1+/+, n = 5 Gfi1−/−, n = 5 Gfi136N/36N, and n = 6 Gfi136S/36S). *P ≤ .05. (F) Total number of HSCs (Lin−, Kit+, Sca-1+, CD150+, CD48− bone marrow cells) in both hind limbs (n = 15 Gfi1+/+, n = 7 Gfi1−/−, n = 3 Gfi136N/36N, and n = 4 Gfi136S/36S). *P ≤ .05. (G) Flow cytometric analysis showing the staining of bone marrow cells for Gr1 and Mac1 surface markers. (H) Total number of monocytes in the bone marrow (per both hindlimb; n = 19 Gfi1+/+, n = 6 Gfi1−/−, n = 6 Gfi136N/36N, and n = 4 Gfi136S/36S). *P ≤ .05. (I) Total number of granulocytes in the bone marrow of the indicated mouse strains per hindlimb (n = 19 Gfi1+/+, n = 6 Gfi1−/−, n = 6 Gfi136N/36N, and n = 4 Gfi136S/36S). *P ≤ .05. (J) Wright-Giemsa staining of bone marrow cytospins (original magnification ×100, Leitz DMRB from Leica, Micropublisher digital color camera, QImaging, Northern Eclipse Version 7.0 software).
Human GFI1 is equivalent to murine Gfi1 in hematopoiesis. (A) Schematic representation of the murine Gfi1 locus and the targeted alleles. (B) Western blot of nuclear extracts of Gfi136N/36N, Gfi1+/+, Gfi1−/−, and Gfi136N/36N+neo thymocytes. (C) Western blot of nuclear and cytosolic extracts of Gfi136N/36N and Gfi136S/36S thymocytes. (D) Total number of B220+ cells in the bone marrow (per both hind limbs; n = 23 Gfi1+/+, n = 13 Gfi1−/−, n = 13 Gfi136N/36N, and n = 9 Gfi136S/36S). *P ≤ .05. (E) Total number of thymocytes (n = 9 Gfi1+/+, n = 5 Gfi1−/−, n = 5 Gfi136N/36N, and n = 6 Gfi136S/36S). *P ≤ .05. (F) Total number of HSCs (Lin−, Kit+, Sca-1+, CD150+, CD48− bone marrow cells) in both hind limbs (n = 15 Gfi1+/+, n = 7 Gfi1−/−, n = 3 Gfi136N/36N, and n = 4 Gfi136S/36S). *P ≤ .05. (G) Flow cytometric analysis showing the staining of bone marrow cells for Gr1 and Mac1 surface markers. (H) Total number of monocytes in the bone marrow (per both hindlimb; n = 19 Gfi1+/+, n = 6 Gfi1−/−, n = 6 Gfi136N/36N, and n = 4 Gfi136S/36S). *P ≤ .05. (I) Total number of granulocytes in the bone marrow of the indicated mouse strains per hindlimb (n = 19 Gfi1+/+, n = 6 Gfi1−/−, n = 6 Gfi136N/36N, and n = 4 Gfi136S/36S). *P ≤ .05. (J) Wright-Giemsa staining of bone marrow cytospins (original magnification ×100, Leitz DMRB from Leica, Micropublisher digital color camera, QImaging, Northern Eclipse Version 7.0 software).
Myeloid maturation in bone marrow
| . | Myeloblast . | Neutrophil maturation, % of all counted cells . | Monocyte . | Lymphocyte . | |||
|---|---|---|---|---|---|---|---|
| Promyelocyte . | Myelocyte . | Metamyelocyte . | Segmented . | ||||
| Wt | 4 ± 1 | 2 ± 0.3 | 12 ± 1 | 12 ± 2 | 31 ± 4 | 6 ± 1 | 21 ± 5 |
| Gfi1−/− | 4 ± 1 | 0.2 ± 0.2 | 4 ± 1* | 2 ± 1* | 2 ± 2* | 54 ± 12* | 10 ± 2* |
| Gfi136N/36N | 2 ± 2 | 3 ± 1 | 6 ± 2 | 10 ± 2 | 25 ± 3 | 19 ± 3 | 31 ± 3 |
| Gfi136S/36S | 2 ± 1 | 1 ± 1 | 14 ± 7 | 10 ± 1 | 16 ± 5 | 14 ± 3 | 37 ± 3 |
| . | Myeloblast . | Neutrophil maturation, % of all counted cells . | Monocyte . | Lymphocyte . | |||
|---|---|---|---|---|---|---|---|
| Promyelocyte . | Myelocyte . | Metamyelocyte . | Segmented . | ||||
| Wt | 4 ± 1 | 2 ± 0.3 | 12 ± 1 | 12 ± 2 | 31 ± 4 | 6 ± 1 | 21 ± 5 |
| Gfi1−/− | 4 ± 1 | 0.2 ± 0.2 | 4 ± 1* | 2 ± 1* | 2 ± 2* | 54 ± 12* | 10 ± 2* |
| Gfi136N/36N | 2 ± 2 | 3 ± 1 | 6 ± 2 | 10 ± 2 | 25 ± 3 | 19 ± 3 | 31 ± 3 |
| Gfi136S/36S | 2 ± 1 | 1 ± 1 | 14 ± 7 | 10 ± 1 | 16 ± 5 | 14 ± 3 | 37 ± 3 |
Quantification of the different cell types according to their morphology enumerated from bone marrow cytospins of the indicated mouse strains (n = 3 for all genotypes).
Significant difference to wt mouse strain.
Cellular composition of blood of different mouse strains
| . | Lymphocytes, % . | Neutrophils, % . | Monocytes, % . | Eosinophils, % . | Basophils, % . |
|---|---|---|---|---|---|
| Wt | 62 ± 10 | 26 ± 7 | 11 ± 4 | 1 ± 1 | 0 |
| Gfi1−/− | 60 ± 17 | 1 ± 1* | 36 ± 14 | 2 ± 3 | 0 |
| Gfi136N/36N | 78 ± 5 | 10 ± 1 | 9 ± 3 | 2 ± 1 | 0 |
| Gfi136S/36S | 80 ± 5 | 16 ± 5 | 3 ± 1 | 3 ± 1 | 1 ± 1 |
| . | Lymphocytes, % . | Neutrophils, % . | Monocytes, % . | Eosinophils, % . | Basophils, % . |
|---|---|---|---|---|---|
| Wt | 62 ± 10 | 26 ± 7 | 11 ± 4 | 1 ± 1 | 0 |
| Gfi1−/− | 60 ± 17 | 1 ± 1* | 36 ± 14 | 2 ± 3 | 0 |
| Gfi136N/36N | 78 ± 5 | 10 ± 1 | 9 ± 3 | 2 ± 1 | 0 |
| Gfi136S/36S | 80 ± 5 | 16 ± 5 | 3 ± 1 | 3 ± 1 | 1 ± 1 |
Quantification of the different cell types according to their morphology enumerated from bone marrow cytospins of the indicated mouse strains (n = 3 for all genotypes).
Significant difference to wt mouse strain.
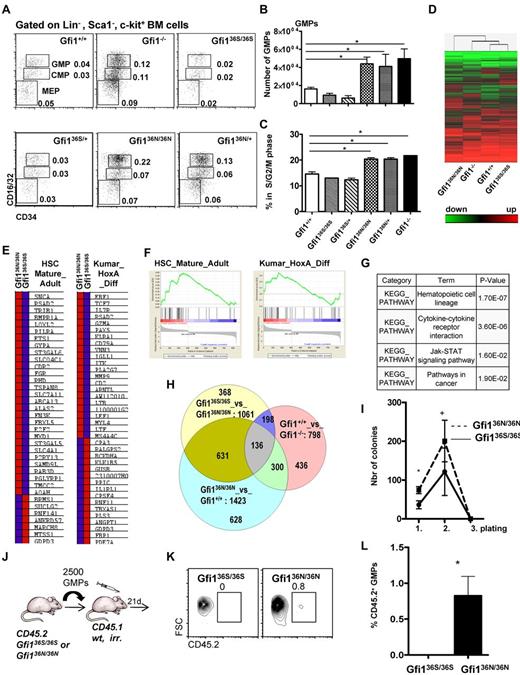
However, we observed a higher number of monocytes and a tendency toward lower numbers of granulocytes in the Gfi136N/36N mice compared with Gfi136S/36S or wt mice, but no obvious difference in cell morphology (Figure 1G-J; Tables 1 and 2). Moreover, significantly increased numbers of GMPs were evident in Gfi136N/36N mice compared with Gfi136S/36S mice or wt mice (Figure 2A-B), a phenotype that was similar to the expansion of GMPs previously described in Gfi1−/− mice,4,25 where a higher proportion of GMPs are in S/G2/M phase compared with wt controls because Gfi1 restricts cell cycle progression of these cells.25,26 Similarly, a higher percentage of Gfi136N/36N GMPs were undergoing cell cycling than in Gfi136S/36S or wt mice, indicating that the GFI136N variant also causes a proliferative expansion of the GMP progenitor fraction (Figure 2C). Interestingly, this proliferative expansion of GMPs is also observable in Gfi136N/+ mice, which resembles the situation found in human AML patients that are heterozygous for this variant.14 The fact that the presence of GFI136N affects specifically GMPs is of particular interest because it has been shown that, in humans as well as in mice, leukemic stem cells arise in the CMP and GMP fractions and that human AML stem cells can exhibit expression patterns similar to normal LMPPs and GMPs.9-12
The presence of GFI136N affects the GMP bone marrow fraction in Gfi136N/36N knock-in mice. (A) Representative flow cytometric analysis of CMPs, GMPs, and MEPs of the indicated mouse strains. Numbers indicate percentage of the different populations with regard to the total bone marrow cells. (B) Total number of GMPs in the different mouse strains (both hind limbs; n = 24 for Gfi1+/+, n = 15 for Gfi1−/−, n = 15 for Gfi136N/36N, n = 7 for Gfi136S/36S, n = 4 for Gfi136N/+, n = 3 for Gfi136N/+). *P ≤ .05. (C) Cell cycle progression of the GMPs from the indicated strains (n = 5 for Gfi1+/+, n = 3 for Gfi1−/−, n = 5 for Gfi136N/36N, n = 5 for Gfi136S/36S,, n = 5 for Gfi136N/+, n = 3 for Gfi136N/+). *P ≤ .05. (D) Heat map representing the different mRNA expression profiles in GMPs from the indicated mouse strains. Genes expressed in GMPs at more than a 2-fold difference in the comparisons, Gfi1−/− vs Gfi1+/+, Gfi136N/36N vs Gfi136S/36S, or Gfi136N/36N vs Gfi1+/+, were analyzed by hierarchical clustering using Wards method (similarity measure: half square eucledian distance). (E-F) Gene set enrichment analysis showed that sets of genes differentially expressed in mature adult hematopoietic cells compared with stem cells (HSC_Mature_Adult) and in MLL-AF9 leukemia (Kumar_HoxA_Diff) are enriched in the list of genes that are differentially expressed between Gfi136S/36S and Gfi136N/36N GMPs. (G) Table of results of KEGG pathway analysis showing that genes differentially expressed between Gfi136S/36S and Gfi136N/36N GMPs fall into the indicated pathways, the one composing hematopoietic cell lineage genes being the most significant. (H) Venn diagram comparing the number of overlapping genes differentially expressed in GMPs from the indicated mouse strains. (I) A total of 450 GMPs from Gfi136S/36S and Gfi136N/36N mice were sorted directly on methylcellulose, and 20 000 cells were replated from the emerging colonies every 10 days (see also “In vitro colony assay”). *P = .05. n = 3 for all genotypes. (J) Schematic outline of transplantation experiment. A total of 2500 GMPs were sorted from Gfi136S/36S or Gfi136N/36N mice and transplanted together with 105 bone marrow cells from CD45.1+ mice into lethally irradiated CD45.1+ mice. (K) At 24 days after transplantation, the number of CD45.1+ (all wt) and CD45.2+ (either Gfi136S/36S or Gfi136N/36N) GMPs was determined. (L-M) At 24 days after transplantation of either Gfi136S/36S or Gfi136N/36N CD45.2+ GMPs, the percentage of CD45.2+ GMPs was determined in the recipient mice. Whereas no CD45.2+, Gfi136S/36S GMPs were detectable, ∼ 1% of the GMPs were CD45.2+Gfi136N/36N GMPs.
The presence of GFI136N affects the GMP bone marrow fraction in Gfi136N/36N knock-in mice. (A) Representative flow cytometric analysis of CMPs, GMPs, and MEPs of the indicated mouse strains. Numbers indicate percentage of the different populations with regard to the total bone marrow cells. (B) Total number of GMPs in the different mouse strains (both hind limbs; n = 24 for Gfi1+/+, n = 15 for Gfi1−/−, n = 15 for Gfi136N/36N, n = 7 for Gfi136S/36S, n = 4 for Gfi136N/+, n = 3 for Gfi136N/+). *P ≤ .05. (C) Cell cycle progression of the GMPs from the indicated strains (n = 5 for Gfi1+/+, n = 3 for Gfi1−/−, n = 5 for Gfi136N/36N, n = 5 for Gfi136S/36S,, n = 5 for Gfi136N/+, n = 3 for Gfi136N/+). *P ≤ .05. (D) Heat map representing the different mRNA expression profiles in GMPs from the indicated mouse strains. Genes expressed in GMPs at more than a 2-fold difference in the comparisons, Gfi1−/− vs Gfi1+/+, Gfi136N/36N vs Gfi136S/36S, or Gfi136N/36N vs Gfi1+/+, were analyzed by hierarchical clustering using Wards method (similarity measure: half square eucledian distance). (E-F) Gene set enrichment analysis showed that sets of genes differentially expressed in mature adult hematopoietic cells compared with stem cells (HSC_Mature_Adult) and in MLL-AF9 leukemia (Kumar_HoxA_Diff) are enriched in the list of genes that are differentially expressed between Gfi136S/36S and Gfi136N/36N GMPs. (G) Table of results of KEGG pathway analysis showing that genes differentially expressed between Gfi136S/36S and Gfi136N/36N GMPs fall into the indicated pathways, the one composing hematopoietic cell lineage genes being the most significant. (H) Venn diagram comparing the number of overlapping genes differentially expressed in GMPs from the indicated mouse strains. (I) A total of 450 GMPs from Gfi136S/36S and Gfi136N/36N mice were sorted directly on methylcellulose, and 20 000 cells were replated from the emerging colonies every 10 days (see also “In vitro colony assay”). *P = .05. n = 3 for all genotypes. (J) Schematic outline of transplantation experiment. A total of 2500 GMPs were sorted from Gfi136S/36S or Gfi136N/36N mice and transplanted together with 105 bone marrow cells from CD45.1+ mice into lethally irradiated CD45.1+ mice. (K) At 24 days after transplantation, the number of CD45.1+ (all wt) and CD45.2+ (either Gfi136S/36S or Gfi136N/36N) GMPs was determined. (L-M) At 24 days after transplantation of either Gfi136S/36S or Gfi136N/36N CD45.2+ GMPs, the percentage of CD45.2+ GMPs was determined in the recipient mice. Whereas no CD45.2+, Gfi136S/36S GMPs were detectable, ∼ 1% of the GMPs were CD45.2+Gfi136N/36N GMPs.
GFI136N affects gene expression and function of GMPs
To understand why GFI136N but not GFI136S causes an expansion of GMPs, we compared genome-wide mRNA expression patterns of GMPs from Gfi136N/36N and Gfi136S/36S animals with those from wt and Gfi1−/− mice. Unsupervised hierarchical clustering of the expression levels of individual genes demonstrated a high similarity between GMPs from Gfi136S/36S and wt mice (Figure 2D; see also “Gene expression array and real-time PCR”) consistent with the notion that GFI136S is functionally equivalent to the endogenous murine Gfi1. In contrast, Gfi136N/36N GMPs exhibited a genome-wide mRNA expression pattern that differed from that obtained with mRNA of wt, Gfi136S/36S and even Gfi1−/− GMPs (Figure 2D). Using gene set enrichment analysis, we observed that the genes differentially expressed in GMPs from Gfi136S/36S and Gfi136N/36N mice showed a pattern reminiscent of the features of adult HSCs and contained a HoxA specific gene signature (Figure 2E-F). In addition, these differentially expressed genes were found to be part of KEGG pathways describing hematopoietic cell lineage determination, cytokine receptor interactions, Jak-STAT signaling, and cancer signaling (Figure 2G).
When we compared the lists of genes that were differentially expressed between GMPs from different mouse mutants (Figure 2H), the largest overlap was found when genes differentially expressed in GMPs from Gfi136N/36N and Gfi136S/36S mice were compared with those differentially expressed in GMPs from Gfi136N/36N and wt mice (631 genes, Figure 2H). This indicated that Gfi136S/36S and wt GMPs were similar in their expression pattern and that GMPs from Gfi136N/36N mice clearly differed from wt, Gfi136S/36S, and even Gfi1−/− GMPs. This also suggested that the presence of GFI136N is not simply recapitulating Gfi1 deficiency but exerts a distinctly aberrant function
A gene set enrichment analysis indicated that Gfi136N cells have characteristics also found in HSCs. Hence, we tested the self-renewal capacity of GFI136N GMPs in colony replating assays and found that GFI136N-expressing GMPs generated more colonies than GFI136S expressing GMPs during the initial plating and the first replating (Figure 2I). However, the replating efficiency of GFI136N-expressing GMPs was exhausted after 3 platings similar to GFI136S GMPs (Figure 2I). Next, we transplanted GMPs sorted from Gfi136S/36S or Gfi136N/36N mice (both CD45.2+) along with CD45.1+ bone marrow carriers into sublethally irradiated CD45.1+ animals and determined the frequency of CD45.2+ GMPs in the recipients 24 days later. Consistent with published findings, no CD45.2+ GFI136S expressing GMPs were detectable, but we found that ∼ 1% of all GMPs of mice transplanted with GMPs from Gfi136N/36N mice were CD45.2+ (Figure 2K-L). This suggested that GMPs with GFI136N variant alleles not only expand because of higher proliferation but have gained a certain capacity of self-renewal that is not found in GMPs expressing the more common GFI136S form.
GFI136N derepresses Hoxa9 expression in GMPs by altering histone modifications
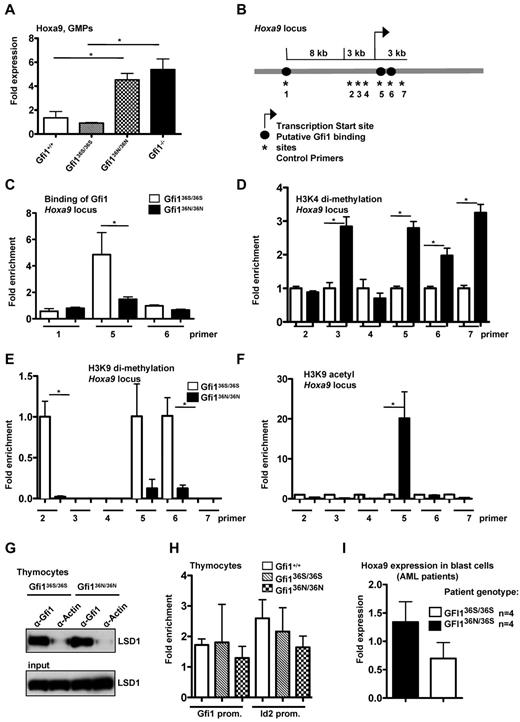
Expression of the Gfi1 target and known oncogene HoxA9 was significantly up-regulated in Gfi136N/36N versus wt or Gfi136S/36S GMPs (Figure 3A). Hoxa9 is important for AML pathogenesis, and high levels of Hoxa9 or Hoxa9 fusion proteins are sufficient to induce AML in mice.27,28 High Hoxa9 expression levels are characteristic for human AML stem cells9 and correlate in AML patients with a poor prognosis.29 Loss of Gfi1 leads to up-regulation of Hoxa9 expression in GMPs (Figure 3A), probably contributing to the numeric expansion of these cells in Gfi1-deficient mice.4 Although high Hoxa9 expression and GMP expansion are detectable in Gfi1−/− mice, these animals do not develop an AML. This is probably because Gfi1 also protects against apoptosis because the introduction of a Bcl-2 transgene leads to the development of a myeloproliferative disease in Gfi1−/− mice that resembles AML.22
Deregulation of Hoxa9 expression and epigenetic modification in myeloid cells from Gfi136N/36N knock-in mice. (A) Hoxa9 mRNA expression was determined in GMPs by RT-PCR. One representative experiment (with triplicates for each experiment) from 2 independent experiments is shown. (B) Schematic representation of the Hoxa9 locus. Numbers indicate the position of the primer pairs used for ChIP-PCR. (C) ChIP-PCR with sorted Lin−, c-kit+, Sca1− cells from the indicated mouse strains using an α-Gfi1 antibody. Shown is the representative result of 2 independent experiments, each done in triplicate (for location of primers see panel B). (D) ChIP-PCR with sorted Lin−, c-kit+, Sca1− cells from the indicated mouse strains using an α-H3K4 dimethyl-antibody (for locations of primers see panel B). Enrichment for Gfi136S is set at 1. (E) ChIP-PCR with sorted Lin−, c-kit+, Sca1− cells from the indicated mouse strains using an α-H3K9dimethyl-antibody (for locations of primers see panel B; for enrichment see panel D). (F) ChIP-PCR with sorted Lin−, c-kit+, Sca1− cells from the indicated mouse strains using an α-H3K9acetyl antibody (for locations of primers see panels B and D). (G) Immunoprecipitations (IP) using thymocyte extracts from the indicated mouse strains with an α-Gfi1 antibody. As control, an α-actin antibody was used. (H) ChIP-PCR with thymocytes using an anti-Gfi1 antibody (from left to right: Gfi1+/+, Gfi136S/36S, Gfi136N/36N). Relative enrichment was determined by amplification with previously described primers (see “ChIP”). (I) Hoxa9 mRNA expression level was determined in blast cells of 4 GFI136S/36S and 4 GFI136N/36S AML patients (P ≤ .10).
Deregulation of Hoxa9 expression and epigenetic modification in myeloid cells from Gfi136N/36N knock-in mice. (A) Hoxa9 mRNA expression was determined in GMPs by RT-PCR. One representative experiment (with triplicates for each experiment) from 2 independent experiments is shown. (B) Schematic representation of the Hoxa9 locus. Numbers indicate the position of the primer pairs used for ChIP-PCR. (C) ChIP-PCR with sorted Lin−, c-kit+, Sca1− cells from the indicated mouse strains using an α-Gfi1 antibody. Shown is the representative result of 2 independent experiments, each done in triplicate (for location of primers see panel B). (D) ChIP-PCR with sorted Lin−, c-kit+, Sca1− cells from the indicated mouse strains using an α-H3K4 dimethyl-antibody (for locations of primers see panel B). Enrichment for Gfi136S is set at 1. (E) ChIP-PCR with sorted Lin−, c-kit+, Sca1− cells from the indicated mouse strains using an α-H3K9dimethyl-antibody (for locations of primers see panel B; for enrichment see panel D). (F) ChIP-PCR with sorted Lin−, c-kit+, Sca1− cells from the indicated mouse strains using an α-H3K9acetyl antibody (for locations of primers see panels B and D). (G) Immunoprecipitations (IP) using thymocyte extracts from the indicated mouse strains with an α-Gfi1 antibody. As control, an α-actin antibody was used. (H) ChIP-PCR with thymocytes using an anti-Gfi1 antibody (from left to right: Gfi1+/+, Gfi136S/36S, Gfi136N/36N). Relative enrichment was determined by amplification with previously described primers (see “ChIP”). (I) Hoxa9 mRNA expression level was determined in blast cells of 4 GFI136S/36S and 4 GFI136N/36S AML patients (P ≤ .10).
It is known that Gfi1 represses transcription by recruiting histone-modifying enzymes, such as LSD1, G9a, and HDACs, to target gene promoters.30,31 LSD1 and HDAC enable the demethylation of H3K4 and deacetylation of H3K9 residues, respectively, both markers associated with gene activation. In contrast, G9a catalyzes the methylation of H3K9 residues, which is associated with gene repression.31 To test whether the GFI136N variant can affect histone modifications at the Hoxa9 locus, we sorted Lin−, Sca1−, ckit+ cells (which contain GMPs) from bone marrow of Gfi136S/36S and Gfi136N/36N mice and performed ChIP with antibodies either recognizing the human GFI1 or methylated or acetylated H3K4 or H3K9 residues. We found that GFI136N bound to a lesser degree to the Hoxa9 locus than GFI136S and that this correlated with a higher degree of H3K4 dimethylation, H3K9 acetylation, and lower H3K9 dimethylation at GFI1 binding sites at the Hoxa9 locus in GFI136N-expressing cells (Figure 3B-F). A similar finding could be found for GFI136N heterozygous mice (supplemental Figure 3). These data strongly suggest that the GFI136N variant is unable to regulate the expression of Hoxa9, which is a validated GFI1 target gene with high relevance for AML. As a result, the transcription of Hoxa9 is not properly silenced in GMPs that carry a Gfi136N variant allele.
Albeit this inability of GFI136N to bind to the HoxA9 promoter, both GFI1 variants were still able to interact with LSD1 (Figure 3G) and other known cofactors, such as PU.1 (supplemental Figure 4). In addition, both GFI1 variants could bind to other direct Gfi1 target genes, such as Id2 or Gfi1, itself at similar rates in thymocytes (Figure 3H).16,17 In addition, GFI136S and GFI136N were both able to repress transcription of the human Hoxa9 gene in a reporter gene assay (supplemental Figure 5). This suggested that the observed differences of GFI136S and GFI136N in their ability to regulate the Hoxa9 locus are specific for GMPs and do not occur in other cell types. Hence, a cell type specific factor or pathway must exist that affects the 2 GFI1 variants differentially in GMPs.
Consistent with our observation with GMPs from Gfi136N/36N mice, blast cells from GFI136N/36S AML patients featured higher Hoxa9 expression levels than blast cells originating from GFI136S/36S patients (Figure 3G), although the level of up-regulation was less pronounced in humans than in mice. In addition, in an independent cohort of 350 patients, Hoxa9 was among the 10 most up-regulated genes between GFI136N/36S and GFI136S/36S AML patients as determined by large-scale gene expression analyses (Table 3).21 These results and previous reports, which demonstrated that Hoxa9 up-regulation is indeed responsible for the expansion of the GMPs in Gfi1-deficient mice,4 indicates that elevated Hoxa9 expression could play a major role in the pathogenesis of AML in GFI136N heterozygous patients. Based on these data and our findings with Gfi136N/36N knock-in mice and in particular because Hoxa9 expression is directly linked to the development of AML,4,27,32,33 we reasoned that the inability of GFI136N to repress Hoxa9 in GMPs may explain the association of the variant GFI136N allele with AML in human patients.
Up-regulated genes in human Gfi136N/36S AML blast cells
| Gene . | Fold change in Gfi136S/36N versus GFI136S/36S patients . |
|---|---|
| SOCS2 | 2.05 |
| MARCKS | 2.02 |
| HOXA9* | 1.87 |
| CYBB | 1.84 |
| BASP1 | 1.84 |
| SOCS2 | 1.82 |
| RNASE6 | 1.8 |
| DACH1 | 1.8 |
| SERPINB2 | 1.79 |
| MPEG1 | 1.79 |
| Gene . | Fold change in Gfi136S/36N versus GFI136S/36S patients . |
|---|---|
| SOCS2 | 2.05 |
| MARCKS | 2.02 |
| HOXA9* | 1.87 |
| CYBB | 1.84 |
| BASP1 | 1.84 |
| SOCS2 | 1.82 |
| RNASE6 | 1.8 |
| DACH1 | 1.8 |
| SERPINB2 | 1.79 |
| MPEG1 | 1.79 |
List of the 10 most up-regulated genes comparing gene expression arrays of AML blast from patients heterozygous for GFI136N (n = 25) to AML blast from patients homozygous for GFI136S (n = 325).
Presence of GFI136N induces epigenetic changes at a genome-wide level
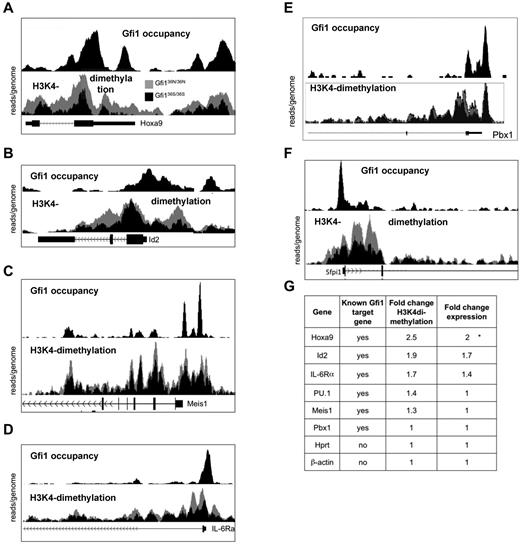
We next tested whether the epigenetic changes associated with GFI136N were restricted to the Hoxa9 locus or whether they could be found at GFI1 binding sites throughout the genome. To determine genome-wide occupancy by Gfi1, we performed a ChIP-Seq experiment using Gfi1 antibodies and cells from murine AML induced by the MLL-Enl onco-fusion gene because large numbers of cells are required to efficiently immune-precipitate murine Gfi1 with currently available antibodies. We also performed ChIP-Seq experiments on sorted Lin−, Sca1−, ckit+ cells from both Gfi136N/36N and Gfi136S/36S mice using antibodies recognizing H3K4 dimethylation.
These experiments demonstrated that Gfi1 indeed occupies the genomic regions of Hoxa9 and a number of other validated target genes, such as Id2, Meis1, IL-6Ra, PU.1, and Pbx1 (Figure 4A-G). In addition, GFI136N-expressing cells showed a much higher degree of H3K4 dimethylation at the Hoxa9 promoter, but also in the 5′region and within the Hoxa9 gene than cells that express the GFI136S form (Figure 4A), which is consistent with the ChIP-PCR results obtained with sorted GMPs (Figure 3B,D). Interestingly, similar epigenetic changes were also evident, albeit to a lesser degree, at other Gfi1 target genes, such as the ID2 and IL-6RA locus, whereas other Gfi1 target genes, such as Meis1, PU.1, and Pbx1, showed no noticeable differences in H3K4 dimethylation when GFI136N was present (Figure 4B-F). A quantification of expression levels (fold up-regulation calculated using values from the GMP expression arrays) and H3K4 dimethylation (fold change calculated using peak areas) confirmed that the effect of the GFI136N variant was most pronounced at the Hoxa9 locus.
Presence of Gfi136N induces epigenetic changes at several loci. Result of the ChIP-Seq experiments indicating Gfi1 occupancy and histone H3 lysine 4 9H3K4) at selected gene loci. For the detection of H3K4 dimethylation, ChIP was performed with sorted Lin−, c-kit+, Sca1− cells from the indicated mouse strains using a α-H3K4 dimethyl antibody. For the detection of Gfi1, ChIP was performed with an α-Gfi1 antibody using MLL-ENL-transduced AML cells. After immunoprecipitation, DNA-protein complexes were subjected to high throughput sequencing to determine the genome-wide distribution of H3K4 dimethyl as well as of Gfi1 binding. In each figure, the top half represents the enrichment for Gfi1 binding and the lower half the distribution of H3K4dimethylation in cells from either Gfi136N/36N or Gfi136S/36S mice. The results of the H3K4dimethyl ChIP-Seq were normalized (see “ChIP seq”), and the curves of obtained with cells from the 2 different mouse strains were superimposed. The curves representing Gfi136N/336N cells appear in gray, the curves of Gfi136S/36S in black. The following gene loci have been analyzed: Hoxa9 (A), Id2 (B), Meis1 (C), IL-6Ra (D), PU.1 (E), and Pbx1 (F). (G) Summary of changes in H3K4 dimethylation and expression of the indicated Gfi1 target and control genes. The fold change in H3K4dimethylation between cells from Gfi136N/36N and Gfi136S/36S mice was calculated by dividing the areas under the respective curves. The area under the curves was determined using the TL-100 Version 2008 software from Non-Linear Dynamics. Fold change expression of the indicated genes between cells from Gfi136N/36N and Gfi136S/36S mice was calculated by dividing the absolute expression levels obtained from the DNA microarray expression analysis (Figure 2D).
Presence of Gfi136N induces epigenetic changes at several loci. Result of the ChIP-Seq experiments indicating Gfi1 occupancy and histone H3 lysine 4 9H3K4) at selected gene loci. For the detection of H3K4 dimethylation, ChIP was performed with sorted Lin−, c-kit+, Sca1− cells from the indicated mouse strains using a α-H3K4 dimethyl antibody. For the detection of Gfi1, ChIP was performed with an α-Gfi1 antibody using MLL-ENL-transduced AML cells. After immunoprecipitation, DNA-protein complexes were subjected to high throughput sequencing to determine the genome-wide distribution of H3K4 dimethyl as well as of Gfi1 binding. In each figure, the top half represents the enrichment for Gfi1 binding and the lower half the distribution of H3K4dimethylation in cells from either Gfi136N/36N or Gfi136S/36S mice. The results of the H3K4dimethyl ChIP-Seq were normalized (see “ChIP seq”), and the curves of obtained with cells from the 2 different mouse strains were superimposed. The curves representing Gfi136N/336N cells appear in gray, the curves of Gfi136S/36S in black. The following gene loci have been analyzed: Hoxa9 (A), Id2 (B), Meis1 (C), IL-6Ra (D), PU.1 (E), and Pbx1 (F). (G) Summary of changes in H3K4 dimethylation and expression of the indicated Gfi1 target and control genes. The fold change in H3K4dimethylation between cells from Gfi136N/36N and Gfi136S/36S mice was calculated by dividing the areas under the respective curves. The area under the curves was determined using the TL-100 Version 2008 software from Non-Linear Dynamics. Fold change expression of the indicated genes between cells from Gfi136N/36N and Gfi136S/36S mice was calculated by dividing the absolute expression levels obtained from the DNA microarray expression analysis (Figure 2D).
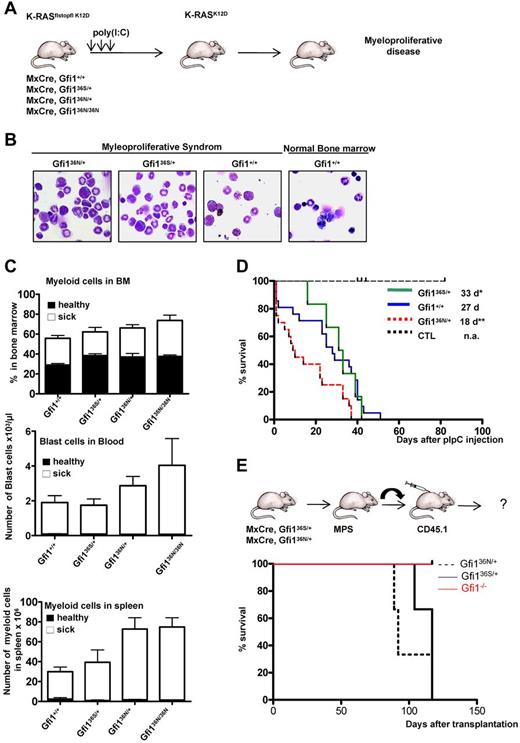
GFI136N accelerates the onset of a myeloproliferative disorder
We reasoned that the up-regulation of Hoxa9 and the epigenetic changes in other genes, which play a role in AML, might render GFI136N-expressing GMPs more susceptible for a progression to a myeloid malignancy. To verify this hypothesis, we tested whether the presence of GFI136N could affect the malignancy of a myeloproliferative-like disease or its transformation into an AML in vivo. It is known that constitutive activation of K-RAS is a recurring oncogenic event in AML,34 and transgenic mice carrying a conditional mutant K-RASfsfK12D allele (K12D floxstopflox K-RAS) allow develop a malignant myeloproliferative disease.35-37 In this model, the K-RAS oncogene is activated after removal of a stop codon, which can be achieved by injecting MxCre, K-RASfsfK12D mice with poly(I:C; Figure 5A). We used this model and crossed wt, Gfi136N/+, Gfi136N/36N, and Gfi136S/+ mouse strains with MxCre, K-RASfsfK12D mice and injected offspring with poly(I:C) to induce expression of the mutated form of K-RAS (Figure 5A). All combinatorial mouse mutants died as expected from a fatal myeloproliferative disorder that was accompanied by an expansion of myeloid cells in the bone marrow, an infiltration of myeloid cells in the spleen, and the appearance of blast cells in the blood (Figure 5B-C). As control mice, we treated Mx Cre tg Gfi1flox/flox mice or wt mice with polyriboinosinic acid/polyribocytidylic acid and monitored their survival to exclude that a higher mortality might be related to Cre activation or to a toxic effect of poly(I:C). However, none of the control mice died of a myeloproliferative disease or any other disease and survived well beyond the observation period (Figure 5D). This excludes that polyriboinosinic acid/polyribocytidylic acid treatment or Cre activation interferes with the K-RAS induced disease. Whereas Gfi1+/+ and Gfi136S/+ mice developed a myeloproliferative disease within a similar median latency of 27 and 33 days, respectively, Gfi136N/+ and Gfi136N/36N mice died significantly faster from this disease (18 for Gfi136N/+ and 16 days for Gfi136N/36N mice, respectively, P ≤ .01; Figure 5D; only data for Gfi136N/+ mice are shown). This indicates that heterozygosity of GFI136N suffices to accelerate a fatal myeloproliferative disorder in mice and further suggests that a causal relationship might exist between the variant GFI136N allele and fatal myeloproliferative diseases in humans.
Accelerated onset of a myeloproliferative disorder in the presence of GFI136N. (A) Experimental setup to induce expression of a mutated K-RASK12D transgene. (B) Representative bone marrow cytospins of moribund mice with the indicated genotypes (original magnification ×100, Leitz DMRB from Leica, Micropublisher digital color camera, QImaging). (C) Frequency of myeloid and blast cells in healthy (dark bar) and sick mice (white bar). Top panel: Percentage of myeloid cells in the bone marrow (healthy mice: n = 18 Gfi1+/+, n = 6 Gfi136N/36N, n = 3 Gfi136S/+, n = 3 Gfi136N/+; sick mice: n = 15 Gfi1+/+, n = 8 Gfi136N/36N, n = 5 Gfi136S/+, n = 15 Gfi136N/+). Middle panel: Number of blast cells per microliter of blood, healthy mice: n = 21 Gfi1+/+, n = 5 Gfi136N/36N, n = 3 Gfi136S/+, n = 4 Gfi136N/+; sick mice: n = 17 Gfi1+/+, n = 5 Gfi136N/36N, n = 5 Gfi136S/+, n = 7 Gfi136N/+). Bottom panel: Total number of myeloid cells in the spleen (healthy mice: n = 21 for Gfi1+/+, n = 3 for Gfi136N/36N, n = 3 for Gfi136S/+, n = 3 for Gfi136N/+; sick mice: n = 16 Gfi1+/+, n = 8 Gfi136N/36N, n = 5 Gfi136S/+, n = 14 Gfi136N/+). All sick mice carried both the MxCre transgene and the K-RASflstopfl K12D allele. (D) Kaplan-Meier survival curve of different strains (n = 12 for control mice, n = 22 for Gfi1+/+, n = 7 for Gfi136S/+, n = 22 for Gfi136N/+). P ≤ .01 between Gfi136N/+ and Gfi1+/+. P ≤ .04 between Gfi136N/+ and Gfi136S/+. The cohort of control mice (CTL) was composed of 2 different subgroups. One group (n = 8) consisted of Mx Cre tg Gfi1flox/flox mice, which were injected with poly(I:C) to exclude that a higher mortality of mice might be related to Cre activation or toxicity of poly(I:C). The second subgroup (n = 6) consisted of wt mice, which were injected with poly(I:C) also to monitor toxic effects of poly(I:C). With the exception of the control mice, all other mice carried an MxCre transgene and a K-RASflstopfl K12D allele. (E) Approximately 2 × 106 bone marrow cells of moribund mice (see panel D) with the indicated genotypes were transplanted alongside 105 CD45.1+ carrier bone marrow cells into sublethally irradiated CD45.1+ mice. Mice were then subsequently observed for emergence of disease; n = 3 for all genotypes. *P ≤ .05.
Accelerated onset of a myeloproliferative disorder in the presence of GFI136N. (A) Experimental setup to induce expression of a mutated K-RASK12D transgene. (B) Representative bone marrow cytospins of moribund mice with the indicated genotypes (original magnification ×100, Leitz DMRB from Leica, Micropublisher digital color camera, QImaging). (C) Frequency of myeloid and blast cells in healthy (dark bar) and sick mice (white bar). Top panel: Percentage of myeloid cells in the bone marrow (healthy mice: n = 18 Gfi1+/+, n = 6 Gfi136N/36N, n = 3 Gfi136S/+, n = 3 Gfi136N/+; sick mice: n = 15 Gfi1+/+, n = 8 Gfi136N/36N, n = 5 Gfi136S/+, n = 15 Gfi136N/+). Middle panel: Number of blast cells per microliter of blood, healthy mice: n = 21 Gfi1+/+, n = 5 Gfi136N/36N, n = 3 Gfi136S/+, n = 4 Gfi136N/+; sick mice: n = 17 Gfi1+/+, n = 5 Gfi136N/36N, n = 5 Gfi136S/+, n = 7 Gfi136N/+). Bottom panel: Total number of myeloid cells in the spleen (healthy mice: n = 21 for Gfi1+/+, n = 3 for Gfi136N/36N, n = 3 for Gfi136S/+, n = 3 for Gfi136N/+; sick mice: n = 16 Gfi1+/+, n = 8 Gfi136N/36N, n = 5 Gfi136S/+, n = 14 Gfi136N/+). All sick mice carried both the MxCre transgene and the K-RASflstopfl K12D allele. (D) Kaplan-Meier survival curve of different strains (n = 12 for control mice, n = 22 for Gfi1+/+, n = 7 for Gfi136S/+, n = 22 for Gfi136N/+). P ≤ .01 between Gfi136N/+ and Gfi1+/+. P ≤ .04 between Gfi136N/+ and Gfi136S/+. The cohort of control mice (CTL) was composed of 2 different subgroups. One group (n = 8) consisted of Mx Cre tg Gfi1flox/flox mice, which were injected with poly(I:C) to exclude that a higher mortality of mice might be related to Cre activation or toxicity of poly(I:C). The second subgroup (n = 6) consisted of wt mice, which were injected with poly(I:C) also to monitor toxic effects of poly(I:C). With the exception of the control mice, all other mice carried an MxCre transgene and a K-RASflstopfl K12D allele. (E) Approximately 2 × 106 bone marrow cells of moribund mice (see panel D) with the indicated genotypes were transplanted alongside 105 CD45.1+ carrier bone marrow cells into sublethally irradiated CD45.1+ mice. Mice were then subsequently observed for emergence of disease; n = 3 for all genotypes. *P ≤ .05.
Discussion
Our experiments provide evidence that the GFI136N variant affects the GMP bone marrow fraction, which has been recognized to be one of the original progenitors (besides CMPs and LMPPs) from which AML leukemic stem cells originate in humans and mice.9-12 Our findings suggest that the presence of GFI136N changes the histone code in GMPs by retaining epigenetic activation marks at the Hoxa9 locus, causing a continued, high level of Hoxa9 expression. In contrast, in wild type GMPs and in those expressing the more common GFI1 form (GFI136S), Hoxa9 is down-regulated in comparison. It has been shown that high levels of Hoxa9 increase the number of GMPs and confer an enhanced capacity for self-renewal to these cells.4,9,27,32,33,38 GMPs expressing the GFI136N variant show, as predicted from their high Hoxa9 expression, a proliferative expansion, higher ability to form colonies in semisolid medium and a higher self-renewal capacity as their wt or GFI136S counterparts. It is thus conceivable that the presence of GFI136N enhances the risk that GMPs develop into a leukemic stem cell and eventually give rise to myeloid malignancies. Our experiments with the K-RAS model of a myeloproliferative disorder support this view, but additional future work with other models in particular those that elicit AML is required to confirm such a role of GFI136N. However, our results confirm the findings of Horman et al4 that Gfi1 regulates the expression of Hoxa9 and that altered function of Gfi1, such as in case of Gfi136N or loss of Gfi1, leads to an accelerated onset of a myeloproliferative disease. We can also observe the emergence of T-cell leukemia after transplanting bone marrow cells from mice expressing a mutant K-RAS oncogene, but not when Gfi1 is absent. In addition, Horman et al show experiments suggesting that Gfi1 deficiency converts a K-RAS–induced myeloproliferative disease into a transplantable AML.4 The differences with regard to transplantation of Gfi1-deficient cells could be because of the fact, that in the experimental setting used here, constitutively and not conditionally Gfi1 deficient mice were used. It is thus possible that conditional ablation is less efficient and that a different Gfi1 dosage might have a different effect on KRA-transformed cells than a full, constitutive deletion of Gfi1.
The epigenetic changes observed at the Hoxa9 locus also occur at other Gfi1 target genes in GMPs carrying variant GFI136N alleles, albeit at a lower level. It is possible that these changes are also critical for the accelerated development of a myeloproliferative disease in Gfi136N/36N or Gfi136N/+ mice. The extent by which H3K4 dimethylation is retained in GFI136N cells is however striking and suggests that the aberrant function of the GFI136N variant is either specifically targeted to the Hoxa9 locus or is most visible at this locus for yet unknown reasons. The potential relevance of elevated Hoxa9 levels in our mouse model with regard to the development of human AML is also underscored by our finding that we observed in 2 independent cohorts of AML patients a tendency for higher Hoxa9 expression levels in blast cells carrying one GFI136N allele compared with blasts homozygous for GFI136S. Finally, the causative role of Hoxa9 overexpression in the pathogenesis of human and murine myeloid leukemia has been demonstrated in many different experiments and animal models and high Hoxa9 expression levels have been documented at many occasions in human AML blasts.4,9,22,27,29,32,33,38 Based on these data and our own findings, it is conceivable that the presence of the GFI136N variant causes elevated levels of Hoxa9, which is responsible for the proliferative expansion and higher self-renewal capacity of GMPs, setting the stage for a myeloproliferative disease and eventually for AML, if other cooperating events occur.
The analysis of 6 different Gfi1 target genes (Hoxa9, Id2, IL-6Ra, PU.1, Meis1, and Pbx1) with regard to expression changes and altered H3K4 dimethylation pattern in GFI136N cells showed that the best correlation was found for the Hoxa9 locus. Hoxa9 expression was up-regulated and had higher H3K4 dimethylation in GFI136N cells. Only Id2 and IL-6Ra showed a similar correlation, but not the other target genes. Although the exact reasons for this remain to be elucidated, it is possible that these genes are subject to control by other histone modifiers and transcription factors. This may also explain why the GFI136N variant does not cause AML but rather accelerates the development of an oncogene-driven nonmalignant myeloproliferative disease and that additional events leading to the deregulation of other AML-related genes must take place before an AML develops, even in GFI136N-expressing cells. In addition, no disease phenotype could be observed in cohorts of mice homozygous or heterozygous for the Gfi136N variant allele after 1 year. Hence, the presence of Gfi136N alone without any additional mutations is not sufficient to cause AML in mice. This is also consistent with the fact that 3%-5% of all healthy persons carry this variant, but the overall incidence rate for AML with 3.6 per 100 000 men and women per year is much lower.
It is important to note that the association between the GFI136N variant and AML was found with patients heterozygous for the variant allele (ie, GFI136N/GFI136S) because only 1 homozygous patient was found in a large cohort of >1000 individuals.14 Similarly, in our present study, we find that the proliferative expansion of GMPs and the faster onset of a myeloproliferative disease already occurs in mice heterozygous for the GFI136N allele (ie, in Gfi136N/wt knock-in mice) and that the epigenetic changes at the HoxA9 locus can also be found in GFI136N heterozygous mice. One possible explanation would be that the GFI136N variant form may cause a loss of function as a dominant negative mutant. Two findings would argue against such a role: first, our gene expression profiling suggests that the Gfi136N variant does not cause a loss of function, but Gfi136N homozygous GMP mice have a distinct expression pattern different from Gfi1 deficient GMPs. Second, although Gfi1−/− mice feature increased expression of Hoxa9 in GMPs,4,22 as a result of this proliferative expansion of GMPs, they never develop an AML or an AML-like disease. This can be explained by the fact that Gfi1 also inhibits apoptosis and, as a result of this, loss of Gfi1 is associated with increased cell death, preventing emergence of a full-blown AML from the GMP fraction.22 In line with this, constitutive overexpression of the antiapoptotic factor Bcl-2 in Gfi1-deficient mice causes a fatal myeloproliferative disorder resembling AML.4,22
Although Gfi136N is unable to repress Hoxa9 expression in GMPs, it can still protect against apoptosis (data not shown), and GMPs from Gfi136N/36N mice are in this respect different from Gfi1−/− GMPs. Because the GFI136N variant has lost the function to repress Hoxa9 in GMPs but retains its capacity to protect GMPs against apoptosis, it is possible that the deregulation of Hoxa9 and possibly other genes that remain to be identified confer an increased self-renewal capacity to GFI136N GMPs as is evident from colony replating and transplantation experiments. This and the increased number of GMPs may increase the likelihood of malignant transformation because the higher number of GMPs expands the pool of target cells. Similar findings have been reported for other mutations, which have been shown to induce AML by increasing the number of the cells, from which AML can arise.39 The presence of GFI136N could increase the likelihood of for a premalignant disease, such as a myeloproliferative disease to occur, which then might develop into an AML, but only if additional mutations occur (Figure 6).
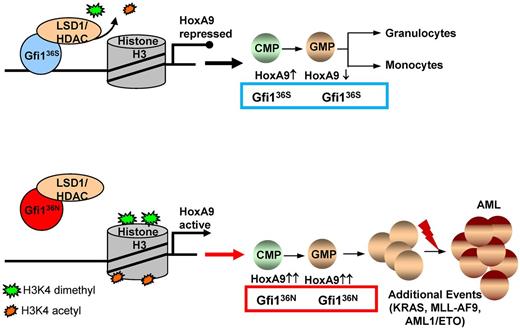
The role of the GFI136N variant in the development of a myeloproliferative disorder that may lead to AML. Proposed model for the function of the GFI136N variant: diminished binding of GFI136N to the Hoxa9 locus as well as to other gene promoters results in epigenetic changes leading to deregulation of expression, in particular of the Hoxa9 gene and subsequently to a proliferative expansion of the GMPs. This effect of the GFI136N variant can increase the likelihood that an AML develops, if other cooperating events occur.
The role of the GFI136N variant in the development of a myeloproliferative disorder that may lead to AML. Proposed model for the function of the GFI136N variant: diminished binding of GFI136N to the Hoxa9 locus as well as to other gene promoters results in epigenetic changes leading to deregulation of expression, in particular of the Hoxa9 gene and subsequently to a proliferative expansion of the GMPs. This effect of the GFI136N variant can increase the likelihood that an AML develops, if other cooperating events occur.
Our observations support an emerging concept that epigenetic changes and, as a result of this, reprogramming of gene expression patterns significantly contribute to the development of diseases and to malignant transformation.40-50 As an example, we show here how an SNP that causes the expression of a variant form of a transcription factor (GFI136N) leads to specific epigenetic alterations and as a result of these changes might render myeloid precursors more susceptible to develop more quickly into preleukemic cells when an oncogenic event has occurred. To our knowledge, this is the first report of a mouse model, in which the introduction of a human coding SNP accelerates a myeloproliferative disease by inducing epigenetic changes. Our experiments support the notion that the epigenetic changes that occur in the presence of GFI136N lead to Hoxa9 up-regulation and potentially to the altered expression of other genes. This changes the overall gene expression pattern causing an altered function of GMPs, which is a cell population, from which in human and mice myeloproliferative disorders can arise. More research is needed to fully clarify the apparent locus specific function of the GFI136N variant. Our GFI136N or 36S knock-in mice already offer the possibility for further investigation and for the testing new therapeutic strategies targeting epigenetic changes in preleukemic cells.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Mathieu Lapointe, Saskia Grunwald, and Rachel Bastien for technical assistance; Nancy Laverriere, Marlène Bernier, Marie-Claude Lavallée, and Mélanie St-Germain for excellent animal care; Eric Massicotte and Julie Lord for FACS and cell sorting; Odile Neyret for performing ChIPs; and Polly Chen for MLL-ENL transduced bone-marrow cells.
This work was supported by the Cancer Research Society, Canada (T.M.). J.S. and F.J.C.-N. were supported by Leukemia and Lymphoma Research United Kingdom. E.D. was supported by the Cambridge Stem Cell Center. R.H. was supported by the NIHR Cambridge Biomedical Research Center. C.K. was supported by the Cole Foundation (fellowship), the University Clinic of Essen (IFZ fellowship), and the German Cancer fund (Max-Eder-fellowship). M.-C.G. was supported by the CIHR (fellowship). T.M. was supported by a Tier 1 Canada Research Chair, the CIHR (MOP-84328), and the Cancer Research Society (Canada).
Authorship
Contribution: C.K. designed and performed research, analyzed data, and wrote the manuscript; J.K., L.V., M.-C.G., J.S., F.B., F.J.C.-N., E.D., R.H., and R.C. performed research, analyzed data, and edited the manuscript; B.A.v.d.R., J.H.J., B.L., J.K.P., H.L.G., S.E.M., and C.V.P. provided vital reagents; U.D. performed research, analyzed data, and provided funding; and B.G. and T.M. designed research, analyzed data, wrote the manuscript, and provided funding.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Bertie Göttgens, Cambridge University, Department of Haematology, Cambridge Institute for Medical Research, Hills Road, Cambridge CB2 0XY, United Kingdom; e-mail: bg200@cam.ac.uk; and Tarik Möröy, Institut de recherches cliniques de Montréal, 110, Avenue des Pins Ouest, Montréal, QC, Canada H2W 1R7; e-mail: tarik.moroy@ircm.qc.ca.
References
Author notes
J.K. and J.S. contributed equally to this study as second authors.