Abstract
Selection of a suitable graft for allogeneic hematopoietic stem cell transplantation involves consideration of both donor and recipient characteristics. Of primary importance is sufficient donor-recipient HLA matching to ensure engraftment and acceptable rates of GVHD. In this Perspective, the National Marrow Donor Program and the Center for International Blood and Marrow Transplant Research provide guidelines, based on large studies correlating graft characteristics with clinical transplantation outcomes, on appropriate typing strategies and matching criteria for unrelated adult donor and cord blood graft selection.
Introduction
The National Marrow Donor Program (NMDP) facilitates identification and procurement of hematopoietic stem cell grafts for transplantation. The Center for International Blood and Marrow Transplant Research (CIBMTR) is a research affiliation of the NMDP and the Medical College of Wisconsin. The guidelines herein, which update those previously published in 20031 and in 2008,2 are based on current and relevant data supporting optimal HLA donor-recipient matching criteria and other factors affecting graft selection.
HLA matching
What literature discusses the impact of HLA on hematopoietic cell transplantation outcome?
Many studies have evaluated the role of HLA matching and outcome in transplantation. Our recommendations are based primarily on large, contemporary studies from the NMDP and CIBMTR. Associations between HLA disparity and survival differ somewhat among published studies.3-7 These differences are detailed in previous publications1,2 and probably result from differences in study design (eg, sample size, recipient race/ethnicity, categorization of mismatches, and impact of other recipient variables, such as diagnosis and disease stage). However, taken together, these studies support 2 general concepts. First, there is a direct association between the number of donor-recipient HLA mismatches and the risk for mortality. Second, mismatching has a greater impact on absolute mortality differences in recipients with “low-risk” disease (ie, disease with a low risk of posttransplantation recurrence). One limitation of existing large studies is that they primarily evaluate the impact of HLA-matching on outcome of transplantation for malignant disease. Fewer data are available for transplantation of nonmalignant disorders, but the general principles are presumed to apply. One caveat to this is that graft-versus-tumor effects that offset some of the mortality associated with GVHD after transplantation for malignancies are of no benefit when treating nonmalignant diseases.
Which is the most important outcome to consider?
The outcome of primary importance after transplantation is survival. Survival is determined by multiple factors. Pretransplantation factors include donor-recipient HLA matching, graft cell-dose (particularly for umbilical cord blood grafts), recipient cytomegalovirus seropositivity, performance score, disease, and disease status. Posttransplantation factors include acute and chronic GVHD, infections, organ toxicity, and recurrent and second malignant neoplasms. When transplantation is being considered as a treatment option, early referral for transplantation, ensuring the recipient has an optimal graft, and using effective strategies to lower rates of acute and chronic GVHD and organ toxicity will maximize the likelihood of a good outcome.
What are the optimal match criteria for unrelated adult donors?
Our proposed guidelines are based on several studies that analyzed the effect of donor-recipient HLA match on survival.3,4,8,9 The study by Lee et al of 3857 transplantations for hematologic malignancies, using primarily marrow grafts, showed that high-resolution matching for HLA-A, -B, -C, and -DRB1 maximizes posttransplantation survival.9 This NMDP/CIBMTR study isolated the effect of each locus by comparing mismatches at a particular locus within recipients who were high-resolution matched for all other loci. Matching at all 4 loci was important, and there was a direct association between the number of HLA mismatches and the risk for mortality. This study also found that a high-resolution mismatch had an effect similar to an antigen-level mismatch. The possible exception was HLA-C where high-resolution mismatches appeared to be better tolerated than antigen-level mismatches. Recipient-related factors were also important, particularly disease stage at transplantation. The magnitude of the survival differences with HLA mismatching was greatest (∼ 10% lower with each mismatch) among recipients with “low-risk” disease (defined as chronic myelogenous leukemia in first chronic phase, myelodysplastic syndrome subtype refractory anemia, acute leukemia in first remission). Among recipients with “high-risk” disease, the higher mortality associated with HLA mismatching was statistically significant but of lesser magnitude (≤ 5% lower with each mismatch).
Thus, whenever possible, donors who are high-resolution matched at HLA-A, -B, -C, and -DRB1 should be sought, but unavailability of such a donor is not a contraindication for transplantation. If a mismatch is unavoidable, a single-locus mismatched donor (HLA-A, -B, -C, or -DRB1) can be used with acceptable risks of transplant-related mortality. The study suggested that mismatches at HLA-B and -C may be less detrimental than those at HLA-A and -DRB1, but the data supporting this difference were not conclusive.
Of importance is the observation in the Lee et al study that an isolated mismatch at HLA-DQ did not have the same impact as mismatching at the 4 other HLA loci,9 although other data indicate that HLA-DQ mismatches may be important in certain disease subsets or when coupled with mismatches at other loci.6,10 Similar to HLA-DQ, mismatches at HLA-DP did not seem to affect overall mortality in the Lee et al study.9 Several studies, including the Lee et al study,5,9,11 show an association between HLA-DP mismatches and acute GVHD; however, this negative impact is offset by a decreased risk for disease relapse with no net effect on survival.11 Additional work by Zino et al,12 subsequently confirmed in a large International Histocompatibility Working Group study,13 suggests that the nature of the DP mismatch may determine its effect. These studies characterized DP mismatches as permissive or nonpermissive based on whether they occurred within or between cross-reactive T-cell epitope groups. Nonpermissive mismatching was associated with higher risks of nonrelapse mortality, especially when there was additional mismatching at other loci.12,14 In further analysis of the dataset used by Lee et al,9 mismatching at HLA-DRB3, -DRB4, and -DRB5 did not appear to impact outcome when appearing in isolation, but multiple mismatches at secondary HLA loci (ie, HLA-DQ, -DP, and -DRB3/4/5) increased the risk associated with mismatching at HLA-A, -B, -C, or -DRB.15
Do HLA matching requirements differ in selection of adult peripheral blood stem cell donors?
Currently, most unrelated adult donor transplantations use G-CSF–mobilized peripheral blood stem cell (PBSC) grafts. The analyses of high-resolution HLA-matching discussed in the preceding paragraph mainly derive from studies of transplantations using myeloablative conditioning regimens and marrow grafts. PBSC and marrow grafts differ in both the number and relative proportion of cells, including CD3+ and CD34+ cells, which might influence the effects of HLA matching. In a separate NMDP/CIBMTR analysis of HLA matching in 1933 unrelated PBSC transplantations for hematologic malignancies,16 recipients of PBSC grafts with at least one HLA antigen level mismatch at HLA-A, -B, -C, or -DRB1 had worse disease-free and overall survival than those receiving an 8 of 8 matched graft. No significant effect was observed when the mismatch was at the allele-level only, but there were far fewer patients evaluable for these comparisons than in the Lee et al study,9 and the power to detect a difference was limited. As seen with marrow grafts, survival was not affected by mismatching at either HLA-DQ or -DP. Notably, HLA-C antigen mismatching conferred the greatest risk for mortality, grade 3 or 4 acute GVHD and chronic GVHD. The adverse effect of mismatching at the HLA-C locus was significant for recipients treated with either myeloablative or reduced intensity conditioning regimens.
In situations where HLA-C mismatching cannot be avoided, one might wonder whether a marrow graft would be better tolerated than PBSCs. In an exploratory analysis that compared the PBSC dataset with the marrow dataset used for the Lee et al analysis,16 no advantage was found to using marrow versus PBSCs as the cell source for transplantation when the donor had an isolated HLA-C antigen mismatch.
What HLA matching is required for umbilical cord blood?
Many studies have established the utility of umbilical cord blood transplantation, particularly as a treatment for childhood and adult leukemia,17-20 but also for other indications.21,22 The impact of HLA matching on outcomes after unrelated donor umbilical cord blood transplantation was summarized by the NMDP in 2008.23 A more recent report of more than 1000 recipients24 focused on the combined effects of HLA matching and cell dose, the other most recognized donor-dependent factor influencing prognosis after cord blood transplantation. This report demonstrated a significant effect of both better matching and higher cell doses.
HLA matching for unrelated cord blood transplantation generally focuses on 3 loci (HLA-A, -B, and -DRB1). Although selection currently is done to maximize matching at the antigen level for HLA-A and -B, and at the allele-level for DRB1, all 3 loci and HLA-C are being typed by many centers at high-resolution. In a recent analysis from NMDP/CIBMTR and Eurocord,25 transplants mismatched at HLA-C were associated with higher transplant-related mortality compared with transplants matched at HLA-C; among transplants mismatched at 2 loci, mismatching at HLA-C and -DRB1 was associated with the highest risk of mortality. This study suggests that extended HLA matching may yield better outcomes after cord blood transplantation. This study defined matching at HLA-A, -B, and -C at the antigen level because there were insufficient numbers of transplantations with high-resolution donor-recipient typing to allow analysis. The NMDP encourages extended high-resolution typing of umbilical cord blood units to facilitate further study of the impact of HLA on outcome.
Some centers are addressing the limitations in cell dose by combining 2 cord blood units for transplantation. There are no studies that evaluate the matching criteria for the 2 units related to one another, but current practice is to maximize matching of the 2 units at the antigen level for HLA-A and -B and at the allele level for DRB1 with a minimum of 4 of 6 match.26
Donor search
How do I search for the best donor?
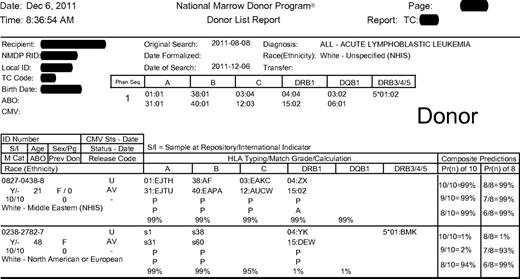
Search should be based on high-resolution HLA assignments of the patient.27 HLA-A, -B, -C, and -DRB1 loci should be characterized because they are important in matching; others (eg, DQB1, DRB3/4/5, and DPB1) may assist in designing an efficient search strategy for the patient and, when necessary, for selecting among more than one mismatched donor. Most listed donors do not have extended high-resolution typing of all of these loci available. The NMDP search algorithm HapLogic uses data on the frequencies of alleles and haplotypes in human populations to predict the probability of high-resolution matches at individual HLA loci and at all key loci simultaneously (Figure 1) for the patient and each potential donor. This is especially helpful when there are many potential adult donors or cord blood units but only sufficient resources and/or time to type a few of the potential donors/units at higher resolution.
Example of an NMDP search report. The columns labeled “HLA Typing/Match Grade/Calculation” include a letter indicating the match status of each allele at the locus indicated (A indicates allele match; P, potential allele match; and M, mismatch), the probability of matching both alleles at the locus (99% for the first donor at each locus). The columns labeled “Composite Predictions” display the probability of a 10 of 10 HLA-A, -B, -C, -DRB1, -DQB1 allele match (99% for first donor) and 8 of 8 HLA-A, -B, -C, -DRB1 allele match (99% for the first donor).
Example of an NMDP search report. The columns labeled “HLA Typing/Match Grade/Calculation” include a letter indicating the match status of each allele at the locus indicated (A indicates allele match; P, potential allele match; and M, mismatch), the probability of matching both alleles at the locus (99% for the first donor at each locus). The columns labeled “Composite Predictions” display the probability of a 10 of 10 HLA-A, -B, -C, -DRB1, -DQB1 allele match (99% for first donor) and 8 of 8 HLA-A, -B, -C, -DRB1 allele match (99% for the first donor).
In the United States, the NMDP serves as the single point of access for unrelated adult donors and umbilical cord blood units under the US Stem Cell Therapeutic and Research Act. The NMDP donor file includes volunteers from the United States as well as Germany, Israel, The Netherlands, Norway, and Sweden. The NMDP cord blood inventory contains units from US banks as well as Germany, Israel, Singapore, and Taiwan. An NMDP search includes a general search of Bone Marrow Donors Worldwide (BMDW),28 as well as an automatic detailed search of certain international registries using the European Marrow Donor Information System network. The BMDW report is particularly helpful to set an optimal, but realistic, target for an international donor search. Searches can also be submitted directly to BMDW to view potentially matched donors/units in differing formats. However, the decision on the overall search strategy and the usefulness of an extended international search must also take into account the variation of allele and haplotype frequencies in different geographic, racial, or ethnic groups and the time and resources available for a particular patient. The NMDP will assist transplant centers with requests to worldwide registries and cord blood banks not included in its own file.
The optimal number of potential donors to select from the search report for additional HLA typing should be individualized for each patient because many factors influence the likelihood of finding a compatible donor. Factors to be considered include the patient's alleles and haplotypes (eg, rare vs common) as well as clinical urgency. Multiple donors should always be selected because donors may be unavailable, mistyped, or not matched once high-resolution testing is complete. For searches listing numerous potential donors with a high probability of matching, per HapLogic or other advanced search algorithms, high-resolution typing of a small number (ie, 3-5) is usually sufficient. However, in the case of patients with rare alleles and haplotypes, where the likelihood of matching is low, 10 or more donors may be required to find the best match. Whenever deemed useful, the NMDP can provide specifically filtered match lists for searches with many donor candidates or with relaxed matching criteria for difficult cases. In the latter situation, help should be immediately sought from a histocompatibility expert (available through the NMDP) to design an effective search strategy that includes evaluation of worldwide donor registries.
How long do I search for adult donors?
For patients with common HLA phenotypes, a suitably matched adult donor can usually be identified on the first match run. For patients with uncommon phenotypes, a well-matched donor may not be readily apparent on the initial match run. For these patients, it is recommended that one request help from a local HLA expert or NMDP consultant to assist in identifying the best potential match.
If one is not able to identify an available, acceptably matched volunteer donor in a worldwide search, it is very unlikely that newly recruited donors will match the patient in a useful time frame. The NMDP donor file contains nearly 9.5 million donors (∼ 87% typed for HLA-A, -B, and -DR) and the NMDP search also provides a match report of an additional ∼ 8.5 million donors listed in BMDW, so patients who are not able to find a suitably matched donor in this pool have uncommon HLA phenotypes. The NMDP adds an average of 30 000 new donors to the file monthly. The likelihood that a patient's type will be represented in those new recruits is low. Therefore, it is recommended that one reevaluate alternative treatment options for those patients and decide whether to reduce the matching requirements or select another graft source (eg, unrelated cord blood transplantation, partially matched related donor transplantation). The high cost of extensive donor screening must also be recognized. Enlisting the assistance of an HLA expert can help maximize available resources by focusing selection of donors for screening to those most likely to match the patient.
How should the clinical status of my patient influence the selection of the donor?
The clinical status of the patient may affect graft selection. Almost all studies of HLA matching find a greater effect of mismatching on mortality among recipients with early-stage or low-risk diseases compared with those with intermediate-stage or advanced diseases, so more mismatching may be acceptable in the latter cases. In addition, patients with diseases that are likely to progress rapidly may need transplantation urgently, favoring the selection of a readily available cord unit over an unrelated donor, if a suitable unit is found. However, use of cord blood will limit access to subsequent donations for relapse control that would be available from an unrelated adult donor.
HLA typing
How should potentially matched donors and cord blood units be HLA typed?
Donors identified on the NMDP search report with the highest likelihood of matching the patient should undergo complete HLA-A, -B, -C, and -DRB1 high-resolution testing to select the best HLA match. DPB1 typing may be performed if a DPB1 permissive mismatching strategy is to be used. If the search is unlikely to identify a donor who is HLA-A, -B, -C, and -DRB1 matched, or if such a donor is not identified after initial screening, -DQB1 and -DRB3/4/5 should be typed to allow selection of the optimal mismatched donor. Testing of loci other than HLA-A, -B, -C, and -DRB1 will also support donor selection in the context of an HLA-sensitized patient to avoid the potential risk of graft failure.29,30 An HLA expert might recommend a strategy that initially targets selected loci for higher resolution typing to rapidly screen several donors and reduce the typing costs; however, this approach should be balanced against the patient's medical condition so as not to unduly delay an urgent transplantation.
Cord blood units should be typed by DNA-based methods for HLA-A and -B at a minimum of antigen-level resolution and for DRB1 at high resolution. Inclusion of HLA-C is strongly recommended. High-resolution typing for all loci helps to ensure that potential allele-level mismatches are well characterized and will allow better evaluation of the impact of HLA matching in the future.
How “high” does high-resolution typing have to be?
High-resolution DNA typing may not always be able to completely distinguish among similar HLA alleles (allele-level resolution). Current high-resolution techniques focus only on alleles that code for proteins that are found on the cell surface (and so are immunologically “active”) and on genes encoding the antigen recognition site of HLA molecules.31 The antigen recognition site is the “active” portion of the HLA molecule that binds peptide antigens and interacts with T-cell receptors. Available data indicate that alleles that are identical in the antigen recognition site domain do not have immunologic differences. Consequently, HLA reports may designate a donor or recipient as having one of several possible alleles, all with the same antigen recognition site, for a given locus, and it is standard practice to accept identity of these donor and recipient assignments as a match.32,33
Selection of HLA-mismatched donors or cord blood units
How do I select the best partially matched unrelated donor or cord blood unit?
For marrow recipients, Lee et al showed that a single HLA mismatch, antigen-level or high-resolution, at HLA- A, -B, -C, or -DRB1 loci was associated with a higher mortality and decreased survival9 ; however, the reduction in survival may be acceptable compared with the survival rates for currently available alternative treatments. Because not all patients will have a fully matched donor, it is important to optimize selection among mismatched donors and cord blood units. In the Lee et al study, mismatches at HLA-B and/or -C seemed to be better tolerated than mismatches at HLA-A and -DRB1.9 In contrast, for PBSC recipients, allele-level mismatches at HLA-A, -B, and/or -C seemed to be better tolerated than antigen-level mismatches; the most disadvantageous situation appeared to be with a mismatch for an HLA-C antigen.16 Data from the NMDP8,9,16 and others suggest that risks accompanying multiple mismatches may be cumulative or even synergistic. Although single mismatches at HLA-DRB3/4/5, -DQ, or -DP were not associated with increased mortality,9 a study by Fernandez-Vin̈a et al found an impact on survival when several HLA-DRB3/4/5, -DQ, or -DP mismatches occurred in combination with HLA-A, -B, -C, or -DRB1 mismatches.15 Therefore, in the setting of a mismatch at the latter loci, donors with the lowest cumulative number of HLA-DRB3/4/5, DQ, and DP mismatches should be favored if other matching criteria are equal. DP mismatches may be selected to be permissive. For searches that seek to optimize donor selection among multiple mismatched donors, we recommend that one request help from a local HLA expert or NMDP consultant.
It is hoped that in the future it may be possible to identify “permissible” mismatches at loci other than DP; however, currently there are currently insufficient data to support this as a standard of practice. Although several schematics for selecting permissive mismatches have been proposed, most have failed to be validated in large datasets. For example, in an analysis of NMDP data, HLA mismatching within a serologic cross-reactive group was not associated with a survival benefit compared with mismatches outside a cross-reactive group.34 Likewise, algorithms for selecting less immunogenic mismatches based on protein sequence and the location and characteristics of amino acid mismatches have not predicted improved outcome.35-37 However, few studies provide information on the potential for identifying permissible mismatches. Studies published by Morishima et al, on behalf of the International Histocompatibility Working Group, evaluated specific HLA-A2 allele-mismatched pairs from the Japanese Marrow Donor Program.38 The data of Morishima et al suggested that certain A2 allele mismatches (A*02:01 vs A*02:06) had a higher chance for mortality compared with A*02:01 versus A*02:05 or A*02:07.38 However, the challenges of evaluating specific permissive mismatches, such as described for the A2 alleles, are formidable. A paper assessing the likelihood of retrospectively analyzing permissive mismatches at the HLA-A locus in United States recipients estimated that to achieve 80% power to detect an effect of the A2 and several other common mismatches on survival would require a retrospective study of 11 000 to more than 1 million donor-recipient pairs.39 Consequently, although one may avoid the specific mismatches identified by Morishima et al as nonpermissive,38 there are few to no data indicating whether the majority of other mismatches are indeed permissive.
Some data regarding permissible mismatching in the context of umbilical cord blood transplantation are promising. These involve considering the HLA type of the cord blood donor's parents. Prior work suggests that the maternal-fetal experience may convey tolerance to the maternal HLA that was not inherited by the fetus.40 These noninherited maternal antigens (NIMAs) may define permissible HLA mismatches and could be used to extend the genotypes that are suitable matches for particular donors or umbilical cord blood units. van Rood et al demonstrated that umbilical cord bloods matched for NIMA were associated with lower treatment-related mortality and overall mortality and decreased relapse.41 A study by the CIBMTR, NMDP, and Eurocord42 found that NIMA-matched umbilical cord blood transplantation resulted in superior overall survival and disease-free survival compared with equivalent NIMA-mismatched transplantations. NIMA matches were relatively rare in both study populations, ranging from 7% to 10%. At present, there is only limited maternal typing available for evaluation of NIMA matching at the time of search. When prospectively searching for a NIMA match, one should keep in mind that the relative frequency of the mismatched antigen(s) will have a strong influence on the potential to identify a NIMA match. Searching for a unit matched for the lower-frequency recipient allele(s) will increase the probability of finding a NIMA match for the mismatched allele when the maternal sample is typed.43 Consulting an HLA expert for guidance could maximize the chances to find a NIMA match; however, searching for a NIMA match may delay transplantation.
Should patient sensitization be considered when selecting an HLA-mismatched donor or umbilical cord blood unit?
An evaluation of antibodies directed to HLA antigens is important for all patients receiving unrelated allogeneic transplantations. Many patients will be sensitized to HLA antigens, as demonstrated by the presence of circulating antibodies. Solid-phase assays make it very easy to assess presensitization in transplant recipients. Studies in both animals and humans29,30,44,45 show the association of preformed HLA-directed antibodies with failed engraftment. In a recent NMDP/CIBMTR study,29 approximately one-third of patients possessed antibodies to HLA antigens. Among recipients with a failed graft, approximately 24% possessed donor-specific HLA antibody, compared with 1% in appropriately matched controls without failed engraftment. These findings were recently replicated in a single-center study by Ciurea et al.30 HLA-directed antibodies have also been demonstrated as a barrier to engraftment in umbilical cord blood transplantation. In a recent study by the Japanese Red Cross,46 the presence of umbilical cord blood–specific HLA-directed antibodies was associated with a significantly decreased incidence of neutrophil and platelet recovery. Cutler et al also found that the presence of umbilical cord blood–specific HLA-directed antibodies led to increased graft failure, delayed neutrophil engraftment, and decreased overall survival in double umbilical cord transplantation.47 Thus, for patients with HLA-specific antibodies and a potentially mismatched allograft, careful antibody specificity analysis and/or testing of the patient's sera for reactivity with cells from potential donors (ie, cross-matching) should be done before transplantation.
In summary, although selection of a donor matched for HLA-A, -B, -C and -DRB1 at high resolution is preferred, the inability to identify a matched donor is not a contraindication for transplantation. Several strategies exist for selection of a mismatched donor or cord blood unit that will optimize the likelihood of a successful outcome. In many instances, survival rates with a well-selected HLA-mismatched graft are equivalent or nearly equivalent to survival rates after fully matched transplantation. Figure 2 summarizes HLA-related and cell dose factors to consider during the selection of an optimal donor.
Chart illustrating the HLA- and cell-dose–related factors to be considered in selection of unrelated donors and umbilical cord blood units. Consideration may also be given to HLA-DP permissive versus nonpermissive mismatching when choosing among donors who are equivalent by other criteria.
Chart illustrating the HLA- and cell-dose–related factors to be considered in selection of unrelated donors and umbilical cord blood units. Consideration may also be given to HLA-DP permissive versus nonpermissive mismatching when choosing among donors who are equivalent by other criteria.
Donor selection based on non-HLA factors
What non-HLA donor characteristics should I consider?
Other non-HLA factors are often considered when selecting donors including CMV negative serology (for CMV-negative patients), male sex, younger age, ABO compatibility, prior pregnancies, and larger body weight. To date, few reports have focused on donor characteristics as the primary objective. In a large study that specifically addressed donor characteristics by the NMDP, the only donor characteristic other than HLA match to be associated with survival was the age of the donor; mortality risks were higher with increasing donor age.48 Among cord blood units, the primary non-HLA factor to be considered is cell dose.23,24
Does the race/ethnicity of the donor need to be the same as the race/ethnicity of the recipient?
Some HLA alleles and haplotypes are distributed at different frequencies among different racial/ethnic groups. When searching for a donor, for some alleles, a high-resolution match is more likely to be found among persons of the same ancestry as the patient. HapLogic takes the race/ethnicity into account when predicting the likelihood of a high-resolution match. Once high-resolution HLA matches are identified, the ancestry of the matched donor does not appear to affect the outcome of the transplant.8,9,48 It should be recognized that the number of racially/ethnically mismatched donor/recipient pairs in these studies was small and further studies are needed to confirm these data.
How does donor availability affect my search?
More than 18 million people have registered worldwide as potential donors. Most are not seriously pursued as potential matches until months or years after initially volunteering. At that time, these potential donors may be unavailable because of changes in their personal circumstances. Overall, the NMDP finds that nearly 50% of all registrants are unavailable when called on. This means that a searching strategy should routinely include alternatives, such as equivalent acceptable adult donors or suitable cord blood units. Operational issues that may impact the timeliness of the search and donation process may also cause delays and impact outcome.
Should targets of NK cell alloreactivity be considered?
Identifying an HLA-matched donor should be the first priority. For an umbilical cord blood unit, HLA match and cell dose are top priorities. There are currently no data to unequivocally indicate that unrelated donors with mismatches at HLA class I loci (ie, the ligands for natural killer cell immunoglobulin-like receptors) should be preferred in any clinical circumstance. An early report from Ruggeri et al49 indicated a strong antileukemic effect and survival advantage with haploidentical related donor transplants with particular HLA class I mismatches that generate donor killer cell reactivity directed toward recipient's tissues. This association was observed only for recipients with acute myeloid leukemia. Several subsequent studies analyzing the impact of killer cell immunoglobulin-like receptors on outcome have had varied conclusions, which have been reviewed and put into context by others.50-52 At this time, more information is needed to understand the role of this complex system in transplantation outcome. Donor selection based on killer cell immunoglobulin-like receptors should only be considered within the context of a clinical trial.
Where can I find additional information and get help with an NMDP search?
NMDP provides extensive information online at: http://www.marrow.org/ and http://bioinformatics.nmdp.org/. Search strategy assistance can be requested by contacting the NMDP Search Strategy team at search-strategies@nmdp.org.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
CIBMTR is supported by the National Cancer Institute (Public Health Service Grant/Cooperative Agreement U24-CA76518), the National Heart, Lung, and Blood Institute, and the National Institute of Allergy and Infectious Diseases; Health Resources and Services Administration (HRSA/DHHS contract HHSH234200637015C); the Office of Naval Research (grants N00014-10-1-0204 and N00014-1-1-0339); and the following: Allos Inc, Amgen Inc, Angioblast, anonymous donation to the Medical College of Wisconsin, Ariad, Be the Match Foundation, Blue Cross and Blue Shield Association, Buchanan Family Foundation, CaridianBCT, Celgene Corporation, CellGenix GmbH, Children's Leukemia Research Association, Fresenius-Biotech North America Inc, Gamida Cell Teva Joint Venture Ltd, Genentech Inc, Genzyme Corporation, GlaxoSmithKline, Kiadis Pharma, Leukemia & Lymphoma Society, Medical College of Wisconsin, Millennium Pharmaceuticals Inc, Milliman USA Inc, Miltenyi Biotec Inc, National Marrow Donor Program, Optum Healthcare Solutions Inc, Otsuka America Pharmaceutical Inc, Seattle Genetics, Sigma-Tau Pharmaceuticals, Soligenix Inc, Swedish Orphan Biovitrum, THERAKOS Inc, and Wellpoint Inc.
The views expressed in this article do not reflect the official policy or position of the National Institutes of Health, the Department of the Navy, the Department of Defense, or any other agency of the US government.
National Institutes of Health
Authorship
Contribution: S.R.S. and C.K.H. had primary responsibility for drafting and preparing the manuscript; M.E., B.R.L., C.M., P.R., M.I.S., and A.E.W. contributed sections to the manuscript and approved the final report; and M.E., B.R.L., C.M., P.R., M.I.S., A.E.W., D.L.C., and M.M.H. critically revised the manuscript and approved the final report.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Mary M. Horowitz, Center for International Blood and Marrow Transplant Research, Division of Hematology and Oncology, Department of Medicine, Medical College of Wisconsin, 9200 West Wisconsin Ave, Milwaukee, WI 53226; e-mail: marymh@mcw.edu.