Abstract
Abstract 2546
We recently identified an alternative splice variant for IL23R (SV-IL23R), a type I cytokine receptor that regulates proliferation and differentiation via the JAK/STAT pathway. This SV-IL23R transcript contained a cryptic exon (CE) within intron 6 of the reference sequence and showed no evidence of expressing the proximal exons 1–6; rather, transcription of the SV-IL23R started with the CE, which introduced a novel start codon and maintained the normal reading frame through exons 7–11. As a result of these changes, majority of the critical extracellular binding structures of the receptor would be lost. Thorough review of databases identified previous reports of this transcript in melanoma and chronic myeloid leukemia, but studies have not systematically examined the expression of SV-IL23R in normal or malignant hematopoietic cells. Therefore, we performed a large retrospective study to examine the expression and clinical significance of SV-IL23R in a cohort of previously untreated pediatric AML patients.
RNA from pretreatment bone marrow (BM) or peripheral blood (PB) was available in the Children's Oncology Group (COG) repository for 132 pediatric patients with AML (ages 0.1 – 21 yrs). Patients received double induction and intensification as per the single arm protocol COG-AAML03P1, which investigated the use gemtuzumab ozogamicin during induction and intensification. RNA was also available from 17 leukemic cell lines and the following subpopulations from healthy donors: mobilized PB CD34+ (N = 10), unsorted BM, (N = 10), unsorted PB (N =10), and T-cells (N = 10). SV-IL23R- specific primers were developed and optimized using standard RT-PCR conditions. The RT-PCR primers were developed to only amplify the SV-IL23R transcript.
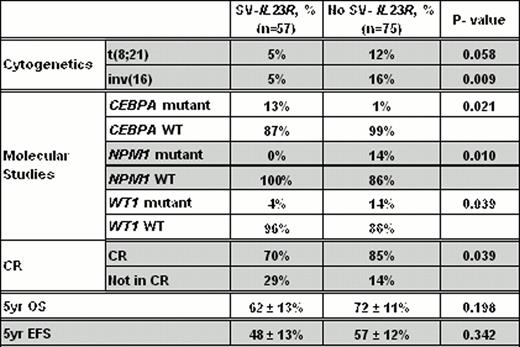
We found no evidence of SV-IL23R expression in the 40 normal samples tested; however, SV-IL23R was expressed in 10 of the 17 leukemic cell lines. Of the 132 samples from pediatric patients, 57 (43%) expressed SV-IL23R. The presence of SV-IL23R expression was not significantly associated with age, gender, race, WBC count, blast percentage or FLT3-ITD status. However, the SV-IL23R expression was inversely correlated with low-risk t(8;21) or inv16 (Table 1). Moreover, patients with a CEPBA mutation were more likely to express SV-IL23R, while those with NPM1 or WT1 mutations were less likely to express SV-IL23R (Table 1). Interestingly, SV-IL23R expression was associated with an increased risk for resistant disease (29% vs. 14%, P = 0.039). Although there was a modest reduction in the estimated 5-year overall survival (OS, 62% vs. 72%, P = 0.19) and event free survival (EFS, 48% vs. 57%, P = 0.34), but these were not statistically significant (Table 1).
This is the first study examining the prevalence and clinical significance of SV-IL23R in hematopoietic malignancies. We found that the blasts in 43% of evaluated pediatric AML patients expressed this transcript. Furthermore, SV-IL23R expression was significantly associated with resistant disease. Further studies examining samples from adult and additional pediatric AML patients are underway and will address the independent nature of this splice variant. If confirmed, SV-IL23R will become one of the most common biomarkers for AML and a potential target for future minimal residual disease assays. Moreover, its correlation with CR rates and potentially other clinical outcomes, if independent of other biomarkers, may enable us to add SV-IL23R to current portfolio of prognostic biomarkers, which may better enable us to more precisely risk-stratify future AML patients.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal