Abstract
Cellular and interpatient heterogeneity and the involvement of different stem and progenitor compartments in leukemogenesis are challenges for the identification of common pathways contributing to the initiation and maintenance of acute myeloid leukemia (AML). Here we used a strategy of parallel transcriptional analysis of phenotypic long-term hematopoietic stem cells (HSCs), short-term HSCs, and granulocyte-monocyte progenitors from individuals with high-risk (−7/7q−) AML and compared them with the corresponding cell populations from healthy controls. This analysis revealed dysregulated expression of 11 genes, including IL-1 receptor accessory protein (IL1RAP), in all leukemic stem and progenitor cell compartments. IL1RAP protein was found to be overexpressed on the surface of HSCs of AML patients, and marked cells with the −7/7q− anomaly. IL1RAP was also overexpressed on HSCs of patients with normal karyotype AML and high-risk myelodysplastic syndrome, suggesting a pervasive role in different disease subtypes. High IL1RAP expression was independently associated with poor overall survival in 3 independent cohorts of AML patients (P = 2.2 × 10−7). Knockdown of IL1RAP decreased clonogenicity and increased cell death of AML cells. Our study identified genes dysregulated in stem and progenitor cells in −7/7q− AML, and suggests that IL1RAP may be a promising therapeutic and prognostic target in AML and high-risk myelodysplastic syndrome.
Introduction
Acute myeloid leukemia (AML) and myelodysplastic syndromes (MDSs) are heterogeneous neoplastic diseases, and most subtypes have poor clinical outcomes. Despite the established use of poly-chemotherapy and the development of new agents that transiently reduce the tumor burden, relapse or failure to achieve durable remission continues to be the most common causes of death in most subtypes of AML and MDS. Recent experimental evidence suggests that AML arises from transformed immature hematopoietic cells after the accumulation of multiple stepwise genetic and epigenetic changes in hematopoietic stem cells (HSCs) and committed progenitors.1 The series of transforming events are thought to initially give rise to preleukemia stem cells (pre-LSCs), preceding the formation of fully transformed LSCs. Defining the characteristics of LSCs, and also of pre-LSCs, is critical to understanding the genesis of leukemia and to developing strategies by which these cells can be eradicated. AML is characterized by a cellular heterogeneous tumor bulk, with LSCs at the top of the hierarchy and a differentiation block at various stages during myeloid maturation.2 To address the problem of cellular heterogeneity within the tumor and to identify relevant molecular pathways effective in LSCs and pre-LSCs, novel experimental approaches other than the examination of bulk tumor cells need to be established. Recent findings have suggested that human LSCs are contained within different phenotypic compartments and at relatively low frequencies.3-5 Several surface molecules were reported to permit enrichment of LSCs in AML.4,6-11 However, reliable markers for human LSCs at the single-cell level have yet to be identified; and because of the challenges associated with the use of xenograft models, the search for such markers remains difficult. Moreover, although there is clear evidence for the involvement of HSCs in AML pathogenesis, studies from murine models suggest that fully transformed and transplantable LSCs may reside at a committed progenitor stage.12-15 Here we applied a novel approach of parallel transcriptional analysis of multiple, highly fractionated stem and progenitor populations in individual patients. We isolated phenotypic long-term HSCs (LT-HSCs), short-term HSCs (ST-HSCs), and committed granulocyte-monocyte progenitors (GMP) from individual patients with AML, and compared gene expression profiles of each population with their phenotypic counterparts from age-matched healthy controls (HCs). Subsequent intersection of differentially expressed genes in the different cellular compartments allowed us to identify candidate genes that are consistently dysregulated at multiple immature stem and progenitor cell stages. Therapeutic targeting of these commonly dysregulated genes may be efficient at relevant pre-LSC stages as well as LSCs. To reduce experimental variation for transcriptional analysis and candidate target identification because of interpatient heterogeneity, we initially focused our study on a genetically defined subset of AML; AML with complete loss (−7) or deletions of the long arm of chromosome 7 (7q−) as the sole cytogenetic aberration. Monosomy 7 is the most common numerical chromosomal aberration found as a sole abnormality in AML16 and the second most frequent in MDS,17 and displays poor response to chemotherapy and an adverse prognosis. Moreover, the molecular pathogenesis of AML with −7/7q− is largely unknown.
Using our novel strategy, we report the identification of 11 genes that are commonly dysregulated in LT-HSCs, ST-HSCs, and GMP of patients with AML with −7/7q−. We show that one of the top differentially expressed genes, IL-1 receptor accessory protein (IL1RAP), is aberrantly expressed on the surface of clonotypic stem and progenitor cells of AML patients with −7/7q−. We also found IL1RAP overexpression in stem cells of a subset of AML patients with normal karyotype, and of patients with high-risk MDS, and show that high levels of IL1RAP are independently associated with poor overall survival in 3 independent clinical cohorts. Functional studies showed that down-regulation of IL1RAP inhibited the clonogenic capacity of AML cells and led to increased apoptosis. Our study suggests that IL1RAP is dysregulated in stem and progenitor cells in AML and high-risk MDS and that it may be a promising novel therapeutic and prognostic target.
Methods
Patient samples and cell lines
BM samples from 16 patients with AML at initial diagnosis were obtained from the Eastern Cooperative Oncology Group (ECOG) within the framework of routine diagnostic bone marrow aspirations after written informed consent in accordance with the Declaration of Helsinki (for patients' characteristics, see supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). BM samples from 7 MDS patients were obtained from Einstein/Montefiore Medical Center (supplemental Table 3). All experiments were approved by the Institutional Review Board of the Albert Einstein College of Medicine (CCI#2008-942). Healthy BM specimens were obtained from Stem Cell Technologies (supplemental Table 4). The AML cell lines THP-1, HL-60, and HEL were grown in RPMI medium supplemented with 10% FBS and 1% penicillin/streptomycin. OCI-AML3 cells were grown in α-MEM medium supplemented with 20% FBS and 1% penicillin/ streptomycin.
Multiparameter high-speed FACS of stem and progenitor cells
To purify the stem and progenitor compartments from total BM of AML or MDS patients, samples were processed as follows: Frozen BM aspirates were quickly thawed in a water bath at 37°C and resuspended in IMDM supplemented with 2% FBS. After repeated washes with IMDM 2% FBS, cells were resuspended in MACS buffer (PBS supplemented with 0.5% BSA and 2mM EDTA, pH 7.2). CD34+ cells were immunomagnetically selected from mononuclear cells from AML patients and healthy donors using Miltenyi MACS technology (130-046-702, Miltenyi Biotec) according to the manufacturer's protocol. Afterward, cells were stained for 30 minutes on ice with PE-Cy5–conjugated antibodies against lineage antigens (CD2[RPA-2.10], CD3[UCHT1], CD4[S3.5], CD7[6B7], CD8[3B5], CD10[CB-CALLA], CD11b[VIM12], CD14[TueK4], CD19[HIB19], CD20[2H7], CD56[MEM-188], Glycophorin A[CLB-ery-1(AME-1)]), and hematopoietic stem and progenitor markers (APC-conjugated CD34[581/CD34(class III epitope)], PE-CY7–conjugated CD38[HIT-2], FITC-conjugated CD45RA[MEM-56], PE-conjugated CD123[6H6] and APC-Cy7–conjugated CD90[5E10]) to distinguish LT-HSCs (Lin−/CD34+/CD38−/CD90+), ST-HSC (Lin−/CD34+/CD38−/CD90−), and GMP (Lin−/CD34+/CD38+/CD123+/CD45RA+). After staining, cells were washed with MACS buffer and subjected to 7-color sorting using a FACSAria II Special Order System (BD Biosciences) as previously described.18 Cells were sorted directly into RLT plus buffer (QIAGEN) for RNA extraction. For some experiments, we sorted and used total myeloid “progenitors” (where indicated) defined as Lin−CD34+CD38+.
RNA amplification and genome-wide transcriptional analysis
Total RNA was extracted from sorted LT-HSC, ST-HSC, and GMP populations from AML patients and HCs using a denaturing buffer containing guanidine isothiocyanate (ALLPrep Micro Kit, QIAGEN). After checking the quality of RNA with an Agilent 2100 Bioanalyzer, total RNA was amplified using the Single Primer Isothermal Amplification (SPIANugen Ovation pico WTA) system according to the manufacturer's instructions. After labeling with the GeneChip WT terminal labeling kit (Affymetrix), labeled cRNA of each individual sample was hybridized to GeneChip Human Gene 1.0 ST microarrays (Affymetrix), stained, and scanned by GeneChip Scanner 3000 7G system (Affymetrix) according to standard protocols. The complete array data are deposited in the gene expression omnibus database (www.ncbi.nlm.nih.gov/geo/; accession no. GSE35008 and GSE35010) according to MIAME standards. Array data were normalized in Expression Console 1.2 software from Affymetrix using Robust Multichip Average. Normalized data were analyzed in MeV Version 4.7.3 software,19 for differential gene expression between groups using Welch t test with a significance level of P < .05. Genes with an absolute value of the group mean difference equal or greater than 1.5 (log2 scale) and P values < .05 were called as differentially expressed between groups. Gene expression differences between LT-HSC, ST-HSC, and GMP groups in −7/7q− AML samples were assessed with 1-way ANOVA (P value distribution based on 1000 permutations, P value cut-off of .01). Principal component analysis based on differentially expressed genes between the −7 AML stem and progenitor populations was carried out in MeV using default parameters. A view of these 3 populations within the −7 AML group as well as within both the −7 AML group and healthy LT-HSC, ST-HSC, and GMP populations is shown in supplemental Figure 3.
Quantitative real-time RT-PCR and detection of IL1RAP protein by flow cytometry
IL1RAP mRNA expression was corroborated by quantitative RT-PCR (forward, 5′-TGCATCTTTGACCGAGACAG-3′; reverse, 5′-CGGCTGAAAATGCAGAAAA-3′) using complementary DNA (cDNA) amplified from total RNA with the WT-Ovation RNA Amplification System (Nugen) and unamplified cDNA with SYBR Green on iQ5real-time PCR instrument (Bio-Rad). Abundance of each transcript was calculated using the Pfaffl method.20 IL1RAP expression was validated at the protein level by flow cytometry in a FACSAria II Special Order System (BD Biosciences) using a biotinylated IL1RAP antibody (R&D Systems) and streptavidin APC-AlexaFluor-750 conjugate (SA1027, Invitrogen) as a secondary antibody. For flow cytometry, AML patient and HC samples were processed as described in “Multiparameter high-speed FACS of stem and progenitor cells” with the addition of IL1RAP antibody. Biotinylated normal goat IgG antibody (BAF108, R&D Systems) was used as isotype control.
FISH
IL1RAP-positive and -negative populations were sorted directly onto polylysine-coated slides. The cells on the slides were fixed in Carnoy solution. The following dual-color probe (Abbott Molecular) was used to detect monosomy 7/7q deletions: LSI D7S522 (7q31) SpectrumOrange/CEP7 SpectrumGreen. FISH was performed according to the manufacturer's instructions. Cells were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) and examined at room temperature on a Zeiss Axioplan 2 fluorescence microscope with a Plan-APOCHROMAT 100×/1.4 oil objective. The images were acquired using a CV−M9+CL camera (JAI) and MetaSystems Isis software (Version 5.4.9). The frequency of false-positive signal loss for the used FISH probe was established by hybridization to cytospin preparations of normal hematopoietic stem and progenitor populations and myeloblasts. For the purpose of this study, the cut-off value for true signal loss, corresponding to monosomy 7/7q deletion, was set at > 10%, counting 100 nuclei per slide.
Lentiviral vectors and transduction
For knockdown studies, shRNA template oligonucleotides (supplemental Table 5) were cloned into the pSIH1-H1-copGFP shRNA vector (System Biosciences). For production of lentiviral particles, lentiviral shRNA expression constructs were transfected together with packaging vectors into 293T producer cells using Fugene HD transfection reagent (Roche Diagnostics), supernatants were harvested after 48 and 72 hours, and concentrated by ultracentrifugation. The THP1, OCI-AML3, HL-60, and HEL cell lines were transduced with the short-hairpin–containing lentivirus (MOI = 10-15). After culture with fresh medium, GFP-positive cells were sorted 48 to 72 hours after infection using a FACSAria II sorter (BD Biosciences) and used for experiments. IL1RAP knockdown efficiency was determined by flow cytometry and quantified as described elsewhere.21
Flow cytometric determination of apoptosis and clonogenic assays
To determine viability after IL1RAP knockdown, 1 × 106 AML cells were washed with PBS and mixed with prediluted PE-conjugated annexin V (BD Pharmingen) and DAPI. Cells were stained at room temperature for 15 minutes and resuspended in 0.5 mL of annexin V–FLUOS incubation buffer (Roche Diagnostics) for analysis. Annexin V−/DAPI− cells were sorted and plated in methylcellulose (StemCell Technologies H4434, or R&D Systems HSC002SF) at 1000 cells/mL in 6-well plates. Cells were incubated at 37°C and 5% CO2. Colonies were scored after 7 days in culture.
Cell cycle analysis
Cell cycle analysis was performed as previously described.22 In brief, 1 × 106 THP-1 cells were rinsed with ice-cold PBS, incubated 45 minutes at 37°C in the dark with 0.5 mL Hoechst buffer (20μg/mL Hoechst 33342 in Hanks balanced salt solution containing 10% FBS, 20mM HEPES, pH 7.2, 1 g/L glucose, and 50 μg/mL verapamil (Sigma-Aldrich). Pyronin Y (Sigma-Aldrich) was added at 1μg/mL, and cells were incubated for 15 minutes at 37°C in the dark, washed with PBS, and analyzed by flow cytometry using a FACSAria II Special Order System (BD Biosciences).
Gene expression and survival analysis
We analyzed publicly available gene expression datasets from human AML studies with the accession numbers GSE12417 (training set hybridized to Affymetrix U133A and U133B microarrays; test set to U133plus2.0) and GSE10358 (U133plus2.0). CEL files were downloaded from GEO and processed using GenePattern (Broad Institute) for normalization (ExpressionFileCreator algorithm) according to the preset parameters of the software (Robust Multichip Average method, with quintile normalization, background correction, median scale normalization method) and collapsing (using CollapseDatast with default parameters). All aforementioned datasets were then analyzed separately to dichotomize the population of patients of each dataset into subsets with high versus low expression of IL1RAP transcript, using the 75th percentile of normalized IL1RAP expression in each dataset as the cut-off point. Publicly available clinical annotation accompanying each one of these datasets was then used to perform Kaplan-Meier survival analysis (GraphPad Prism Version 5.0) comparing clinical outcomes of patients with high versus low IL1RAP expression. Multivariate analysis with stepwise forward and backward model selection by Aikake Information Criterion was performed in R/Bioconductor using the survival and MASS packages. The model parameters included IL1RAP status, French-American-British subclass, FLT3 mutation status, age, sex, and cytogenetic risk. IL1RAP gene expression comparisons between CD34+ cells from healthy donors and patients with chromosome 7 deletions were carried out using the published GSE14468 patient data. CEL files from del(7) patients (n = 18) and HCs (n = 11) were Robust Multichip Average-normalized, and log2-transformed IL1RAP expression was plotted. We analyzed gene expression data and clinical information from 183 MDS CD34+ cells and 17 controls.23 IL1RAP expression was represented as scatter plots using GraphPad Prism Version 5.0 software.
Results
Identification of genes consistently dysregulated in multiple distinct stem and progenitor cell compartments in patients with AML
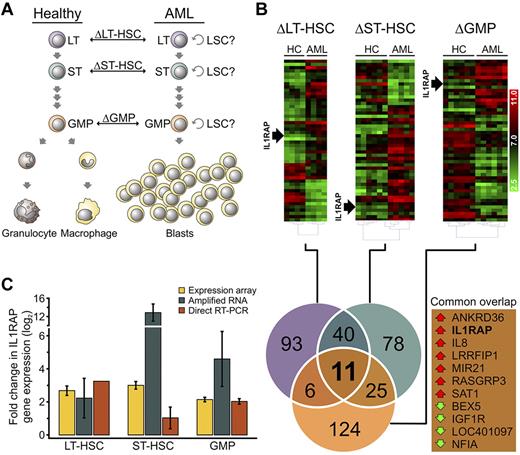
In this study, we carried out gene expression analysis of highly fractionated stem and progenitor cell compartments from individual patients with AML with monosomy 7 (−7) or deletions of the long arm of chromosome 7 (7q−) as the sole cytogenetic aberration compared with phenotypically identical cell populations from age-matched HCs, in an attempt to reduce interpatient as well as cellular heterogeneity. Specifically, we sorted and analyzed Lin−CD34+CD38−CD90+ cells (referred to as LT-HSCs), Lin−CD34+CD38−CD90− cells (referred to as ST-HSCs), and Lin−CD34+CD38+CD123+CD45RA+ cells (referred to as GMPs), using previously established marker schemes18,24-26 (supplemental Figure 1; supplemental Table 2). We included rigorous lineage depletion in our strategy to avoid analysis of the leukemic bulk (blast) population and to focus on the earliest known stem cell and committed myeloid progenitor populations in humans. Lineage depletion eliminated an additional 70% to 90% of cells from the stem and progenitor gates (supplemental Figure 2). To identify genes dysregulated within the distinct stem and progenitor cell compartments of AML patients, we used Affymetrix GeneST 1.0 arrays to compare gene expression with the respective HCs at each cellular level (AML-LT-HSCs vs HC-LT-HSCs, AML-ST-HSCs vs HC-ST-HSCs, and AML-GMPs vs HC-GMPs; Figure 1A). We found significant differences in gene expression (> 1.5-fold change; P < .05) in all examined compartments (Figure 1B). In LT-HSCs, 150 genes were differentially expressed (115 up-regulated, 35 down-regulated); in ST-HSCs, 154 genes were differentially expressed (90 up-regulated, 64 down-regulated); and in GMPs, 166 genes were differentially expressed (35 up-regulated, 131 down-regulated; supplemental Table 6). We then compared differentially expressed genes in each individual compartment (ΔLT-HSCs, ΔST-HSCs, ΔGMPs) with each other to identify genes that were altered in all examined stem and progenitor compartments in AML patients. Using this intersection analysis, we found 11 genes that were consistently dysregulated in AML versus HC in all examined stem and progenitor compartments (Figure 1B).
Transcriptional profiling of phenotypic hematopoietic stem and progenitor compartments of AML patients with monosomy 7 identifies overexpression of IL1RAP. (A) Schematic showing the cell types used for pairwise comparison of gene expression. Phenotypically defined hematopoietic stem and progenitor compartments with potential LSC activity were sorted and compared between healthy and AML individuals. The symbol “Δ” refers to the gene expression differences between groups in each phenotypically defined compartment: LT-HSCs, ST-HSCs, and GMPs. (B) Hierarchical clustering of the 50 most significantly dysregulated genes in −7 AML in phenotypically defined LT-HSCs, ST-HSCs, and GMPs of AML patients compared with HC. Heat maps of log2-transformed gene expression levels are shown (top). Position of IL1RAP is indicated. Venn diagram (bottom) shows the number of differentially expressed genes that are shared between, or restricted to, specific compartments between AML samples and HC. The numbers represent the total of up- or down-regulated genes in each pairwise comparison. Genes in the triple intersection (common overlap) are listed. Red and green arrows indicate overexpression and down-regulation in the AML samples, respectively. (C) Validation of IL1RAP mRNA expression in LT-HSCs, ST-HSCs, and GMPs. Yellow bars represent expression levels in the gene expression array (n = 4 for LT-HSCs, n = 5 for ST-HSCs, and n = 6 for GMPs). Blue bars represent mRNA levels determined by quantitative RT-PCR in amplified RNA (n = 2). Red bars represent the mRNA levels measured by quantitative RT-PCR from unamplified cDNA (n = 1 for LT-HSCs and n = 2 for ST-HSCs and GMPs). mRNA levels were normalized to GAPDH. Fold change compared with HC is shown.
Transcriptional profiling of phenotypic hematopoietic stem and progenitor compartments of AML patients with monosomy 7 identifies overexpression of IL1RAP. (A) Schematic showing the cell types used for pairwise comparison of gene expression. Phenotypically defined hematopoietic stem and progenitor compartments with potential LSC activity were sorted and compared between healthy and AML individuals. The symbol “Δ” refers to the gene expression differences between groups in each phenotypically defined compartment: LT-HSCs, ST-HSCs, and GMPs. (B) Hierarchical clustering of the 50 most significantly dysregulated genes in −7 AML in phenotypically defined LT-HSCs, ST-HSCs, and GMPs of AML patients compared with HC. Heat maps of log2-transformed gene expression levels are shown (top). Position of IL1RAP is indicated. Venn diagram (bottom) shows the number of differentially expressed genes that are shared between, or restricted to, specific compartments between AML samples and HC. The numbers represent the total of up- or down-regulated genes in each pairwise comparison. Genes in the triple intersection (common overlap) are listed. Red and green arrows indicate overexpression and down-regulation in the AML samples, respectively. (C) Validation of IL1RAP mRNA expression in LT-HSCs, ST-HSCs, and GMPs. Yellow bars represent expression levels in the gene expression array (n = 4 for LT-HSCs, n = 5 for ST-HSCs, and n = 6 for GMPs). Blue bars represent mRNA levels determined by quantitative RT-PCR in amplified RNA (n = 2). Red bars represent the mRNA levels measured by quantitative RT-PCR from unamplified cDNA (n = 1 for LT-HSCs and n = 2 for ST-HSCs and GMPs). mRNA levels were normalized to GAPDH. Fold change compared with HC is shown.
Of note, AML-LT-HSCs, AML-ST-HSCs, and AML-GMPs clearly separated in a principal component analysis (supplemental Figure 3A). Furthermore, when we included LT-HSCs, ST-HSCs, and GMPs of age-matched HC in the principal component analysis, we found that they clustered with the respective AML populations, suggesting that AML-LT-HSCs, AML-ST-HSCs, and AML-GMPs are overall molecularly comparable with their normal counterparts (supplemental Figure 3B). To further characterize the cell populations used for gene expression comparison, we performed FISH in sorted AML-LT-HSCs and AML-ST-HSCs. We found that −7/7q− was present in the majority of cells (68% in LT-HSCs, SD 34%; and 92% in ST-HSCs, SD 10%), indicating that these earliest definable cellular compartments were indeed part of the abnormal clone (supplemental Figure 4).
Among the 11 intersecting genes, the IL1RAP was consistently one of the most statistically significant differentially expressed genes in LT-HSCs, ST-HSCs, and GMPs of AML patients with monosomy 7. When we increased the stringency of the cut-off criteria, IL1RAP and IL-8 remained in the intersection as the 2 most significantly up-regulated genes (supplemental Figure 5). Notably, when we compared unfractionated blast cells of patients with −7/7q− AML with healthy CD34+ cells, there was no difference in IL1RAP mRNA expression (supplemental Figure 6), suggesting that IL1RAP plays a role specifically in immature stem and progenitor cells. We confirmed IL1RAP and RASGRP3 overexpression, as well as NFIA down-regulation in sorted LT-HSCs, ST-HSCs, and GMPs by quantitative RT-PCR on the same mRNA samples examined by microarray analysis, as well as samples of additional patients (n = 2; Figure 1C; supplemental Figure 7). We decided to focus on IL1RAP for further studies, as IL1RAP has previously been implicated in the regulation of hematopoietic cells, and its expression on the cell surface may permit direct therapeutic targeting as well as use as a diagnostic marker.27-29
IL1RAP protein is aberrantly expressed on stem and progenitor cells of AML patients with −7/7q−
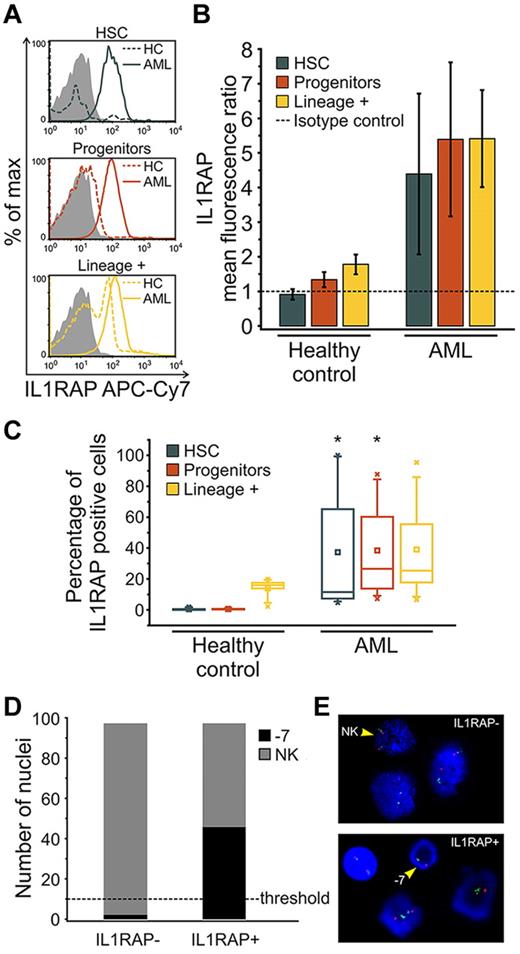
To validate the finding of IL1RAP overexpression at the protein level, we stained HSCs and progenitors from AML patients with −7/7q− with an IL1RAP-directed antibody and measured IL1RAP expression by flow cytometry. We found that HSCs and progenitors of AML patients expressed IL1RAP at the protein level, whereas HSCs and progenitors from HCs did not show detectable IL1RAP surface protein expression (Figure 2A-B). We determined the percentage of IL1RAP-expressing cells within the subpopulations of HSCs, progenitors, as well as lineage-positive cells as a more mature cell population. Although none of the HC HSCs or progenitors contained significant numbers of IL1RAP+ cells, HSCs and progenitors of AML patients displayed a highly significant percentage of IL1RAP-expressing cells (37.3%, P = .006, and 38.5%, P = .002, respectively; Figure 2C). In contrast, the lineage-positive cell fraction of AML patients contained only moderately more IL1RAP-positive cells than HCs, and this difference was not significant (P = .149; Figure 2C).
IL1RAP protein is aberrantly expressed on distinct stem and progenitor cell compartments of AML patients with −7/7q−. Detection of IL1RAP expression at the protein level by flow cytometry in bone marrow-derived cells from HCs (n = 5) and AML patients with −7/7q− (n = 8). (A) Representative histograms show the distribution of IL1RAP protein (cell surface) expression (fluorescence intensity) within phenotypically defined cell compartments of HC (dotted line) and AML (solid line) samples measured using IL1RAP antibody. Gray histograms correspond to the isotype control. (B) Ratios of IL1RAP geometric mean fluorescence intensity relative to the isotype control (arbitrary unit = 1, indicated by the dotted line) for HSCs in blue, progenitors in red, and lineage-positive cells in yellow. Error bars represent SD. (C) Box-and-whisker plots represent the percentage of IL1RAP-positive cells in each cellular compartment, phenotypic HSCs in blue, progenitors in red, and lineage-positive cells in yellow, in HC and AML bone marrow. An isotype control was used to define positive expression in each experiment. The central box represents the values from the 25th to 75th percentile. The middle square represents the mean; and horizontal line, median. A line extends from the minimum to the maximum value. Black star indicates significant difference with HC counterparts. (D) FISH of sorted IL1RAP-positive and IL1RAP-negative cells of an AML patient bearing monosomy 7 hybridized with the Vysis LSI D7S486 (7q31) SpectrumOrange/CEP 7 SpectrumGreen Probe. Bar graph represents the number of nuclei with normal karyotype (NK) and monosomy 7 (−7) for each group (n = 100 nuclei analyzed per group). P < .001 (χ2). The scoring threshold is indicated. (E) Representative FISH image. The arrows indicate a NK FISH pattern with 2 individual green (centromere chromosome 7) and 2 orange (7q31) signals per nuclear section (top), and monosomy 7(−7) with 1 green and 1 orange signal (bottom), respectively.
IL1RAP protein is aberrantly expressed on distinct stem and progenitor cell compartments of AML patients with −7/7q−. Detection of IL1RAP expression at the protein level by flow cytometry in bone marrow-derived cells from HCs (n = 5) and AML patients with −7/7q− (n = 8). (A) Representative histograms show the distribution of IL1RAP protein (cell surface) expression (fluorescence intensity) within phenotypically defined cell compartments of HC (dotted line) and AML (solid line) samples measured using IL1RAP antibody. Gray histograms correspond to the isotype control. (B) Ratios of IL1RAP geometric mean fluorescence intensity relative to the isotype control (arbitrary unit = 1, indicated by the dotted line) for HSCs in blue, progenitors in red, and lineage-positive cells in yellow. Error bars represent SD. (C) Box-and-whisker plots represent the percentage of IL1RAP-positive cells in each cellular compartment, phenotypic HSCs in blue, progenitors in red, and lineage-positive cells in yellow, in HC and AML bone marrow. An isotype control was used to define positive expression in each experiment. The central box represents the values from the 25th to 75th percentile. The middle square represents the mean; and horizontal line, median. A line extends from the minimum to the maximum value. Black star indicates significant difference with HC counterparts. (D) FISH of sorted IL1RAP-positive and IL1RAP-negative cells of an AML patient bearing monosomy 7 hybridized with the Vysis LSI D7S486 (7q31) SpectrumOrange/CEP 7 SpectrumGreen Probe. Bar graph represents the number of nuclei with normal karyotype (NK) and monosomy 7 (−7) for each group (n = 100 nuclei analyzed per group). P < .001 (χ2). The scoring threshold is indicated. (E) Representative FISH image. The arrows indicate a NK FISH pattern with 2 individual green (centromere chromosome 7) and 2 orange (7q31) signals per nuclear section (top), and monosomy 7(−7) with 1 green and 1 orange signal (bottom), respectively.
To investigate whether aberrant IL1RAP expression is indeed a hallmark of immature cells that are part of the AML clone, we sorted IL1RAP+ and IL1RAP− cell fractions of a patient with monosomy 7 and performed FISH analysis. Strikingly, 47% of the IL1RAP-positive cells harbored monosomy 7, whereas IL1RAP-negative cells did not show the cytogenetic aberration at all above thresholds (Figure 2D-E). This observation was confirmed in another patient with 7q− deletion (data not shown), indicating that the occurrence of the clonotypic −7/7q− aberration is indeed restricted to IL1RAP-expressing cells and that IL1RAP expression is a consistent feature of the cells, that are part of the −7/7q− clone.
IL1RAP is overexpressed on stem cells of patients with normal karyotype AML and with high-risk MDS
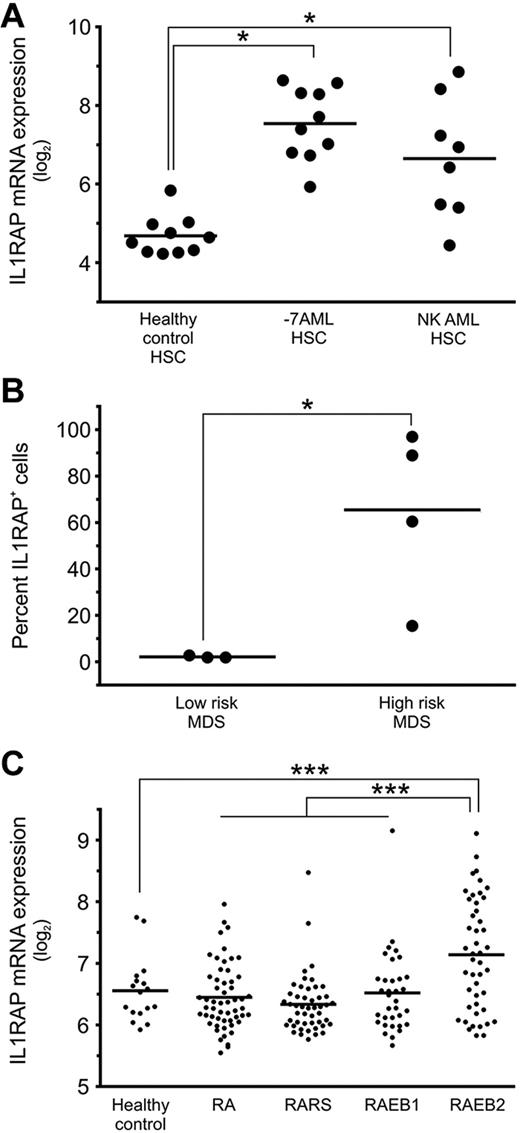
To evaluate whether IL1RAP overexpression is a unique feature of AML patients with −7/7q−, we examined gene expression in other subtypes of AML. We isolated LT-HSCs, ST-HSCs, and GMPs from AML patients with normal karyotype and determined IL1RAP expression. We found mRNA levels of IL1RAP to be 3.8-fold up-regulated in HSCs compared with HCs (Figure 3A). These data indicate that overexpression of IL1RAP is not unique to −7/7q− AML but also occurs in other subsets of AML, suggesting that overexpression of IL1RAP might be part of a more common pathogenetic mechanism effective in immature stem and progenitor cell populations in AML.
IL1RAP is overexpressed on stem cells of patients with AML with monosomy 7, AML with normal karyotype, and high-risk MDS. (A) IL1RAP gene expression in phenotypically defined HSCs (Lin−CD34+CD38−) in HCs (n = 10), AML with monosomy 7 (n = 10), and AML with normal karyotype (n = 8). *Differences compared with the HC group are statistically significant (P < .05 in both cases). (B) IL1RAP protein expression was determined in Lin−CD34+CD38− cells in MDS patients (n = 7: 3 with low-risk and 4 with high-risk MDS). An isotype control was used to define IL1RAP positivity in each experiment. Percentages of IL1RAP+ cells are shown. *P < .05. (C) IL1RAP mRNA expression in CD34+ cells of different types of MDS (n = 183 total) with refractory anemia (RA), refractory anemia with ringed sideroblasts (RARS), and refractory anemia with excess blast types 1 (RAEB1) and 2 (RAEB2), and CD34+ cells of HCs (n = 17). ***P < .001.
IL1RAP is overexpressed on stem cells of patients with AML with monosomy 7, AML with normal karyotype, and high-risk MDS. (A) IL1RAP gene expression in phenotypically defined HSCs (Lin−CD34+CD38−) in HCs (n = 10), AML with monosomy 7 (n = 10), and AML with normal karyotype (n = 8). *Differences compared with the HC group are statistically significant (P < .05 in both cases). (B) IL1RAP protein expression was determined in Lin−CD34+CD38− cells in MDS patients (n = 7: 3 with low-risk and 4 with high-risk MDS). An isotype control was used to define IL1RAP positivity in each experiment. Percentages of IL1RAP+ cells are shown. *P < .05. (C) IL1RAP mRNA expression in CD34+ cells of different types of MDS (n = 183 total) with refractory anemia (RA), refractory anemia with ringed sideroblasts (RARS), and refractory anemia with excess blast types 1 (RAEB1) and 2 (RAEB2), and CD34+ cells of HCs (n = 17). ***P < .001.
MDSs are heterogeneous hematologic disorders that can progress to AML. Transformation to overt AML is particularly frequent in high-risk MDS as defined by the new WHO classification.30,31 To test whether IL1RAP plays a role in MDS, we sorted and analyzed Lin−CD34+CD38− HSCs from patients with MDS. We found that high-risk MDS patients displayed aberrant IL1RAP expression on their stem cells (average, 64%; range, 17%-97%), whereas stem cells of low-risk MDS patients did not show a detectable fraction of IL1RAP-positive stem cells (Figure 3B). When we analyzed gene expression data from enriched CD34+ cells of a larger cohort of MDS patients,23 we found that the subset of patients with refractory anemia with excess blast type 2 had significantly higher IL1RAP expression levels compared with patients with refractory anemia, refractory anemia with ringed sideroblasts, and refractory anemia with excess blast type 1 (P = 4.23 × 10−5), and compared with CD34+ cells from HCs (P = 7.4 × 10−4; Figure 3C). These observations suggest that aberrant IL1RAP expression on stem cells discriminates high-risk from low-risk MDS and may thus be associated with inferior outcome.
IL1RAP overexpression is independently associated with poor clinical outcome
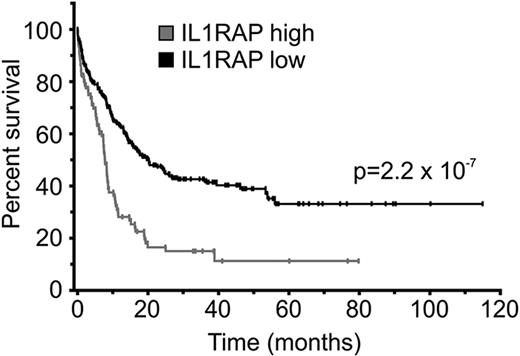
We noted that phenotypic HSCs in AML with normal karyotype showed substantial variability in the expression of IL1RAP (Figure 3A). Some patients displayed IL1RAP levels as high as −7/7q− patients, whereas others had IL1RAP levels not distinguishable from HCs. We sought to examine whether IL1RAP expression levels were associated with known clinical or molecular parameters. For this purpose, we analyzed 3 large published datasets of patients with AML with normal karyotype, for which gene expression data of leukemic bulk (mononuclear) cells and time-to-event data were available (GSE10358, GSE12417 [U133A], GSE12417 [U133plus2]).32,33 As the top quartile of patients with normal karyotype had IL1RAP expression levels very similar to the ones we had observed in patients with −7/7q− (Figure 3A), we decided to use the 75th percentile to dichotomize patients with normal karyotype into “IL1RAP high” and “IL1RAP low” expressers. We compared the overall survival of AML patients with low versus high IL1RAP, and observed that, in each of the 3 different datasets, high levels of IL1RAP expression were associated with inferior overall survival (supplemental Figure 8A-C). Overall survival (irrespective of IL1RAP status) in all examined datasets was very similar, with superimposable survival curves (supplemental Figure 8D), suggesting that the patient populations in these datasets and their clinical outcomes were comparable and could be combined for further analyses. Consistent with the analyses of the individual datasets, the evaluation of the combined set of AML patients with normal karyotype from the GSE10358 and GSE12417 (total n = 317) confirmed that high IL1RAP levels were associated with inferior overall survival (P = 2.2 × 10−7 [log-rank]; hazard ratio [HR] = 2.59; 95% CI, 1.81-3.72); median survival (7.82 months for IL1RAP high, 20 months for IL1RAP low; 5-year survival rate: 11.3% for IL1RAP high, 33.1% for IL1RAP low; Figure 4). To assess whether the impact of IL1RAP expression on overall survival within AML with normal karyotype was independent of other prognostic factors, we performed multivariate analysis using a Cox regression model. In this analysis, high IL1RAP status remained an independent prognostic factor (P = .002; HR = 3.17; 95% CI, 1.52-6.63). Notably, in this analysis, IL1RAP was an even stronger prognostic factor than FLT3 mutation status (P = .006; HR = 3.44; 95% CI, 1.29-4.59), which is a known independent covariate and used in the clinic for risk stratification of patients with normal karyotype.
IL1RAP overexpression is associated with poor clinical outcome in AML with normal karyotype. Combined survival analysis of 3 cohorts of AML patients with normal karyotype (n = 317) dichotomized for IL1RAP gene expression levels at the 75th percentile. Overall survival of patients with low and high IL1RAP expression is shown in black and gray, respectively. Statistical significance is indicated.
IL1RAP overexpression is associated with poor clinical outcome in AML with normal karyotype. Combined survival analysis of 3 cohorts of AML patients with normal karyotype (n = 317) dichotomized for IL1RAP gene expression levels at the 75th percentile. Overall survival of patients with low and high IL1RAP expression is shown in black and gray, respectively. Statistical significance is indicated.
Knockdown of IL1RAP decreases clonogenicity and increases cell death of AML cells
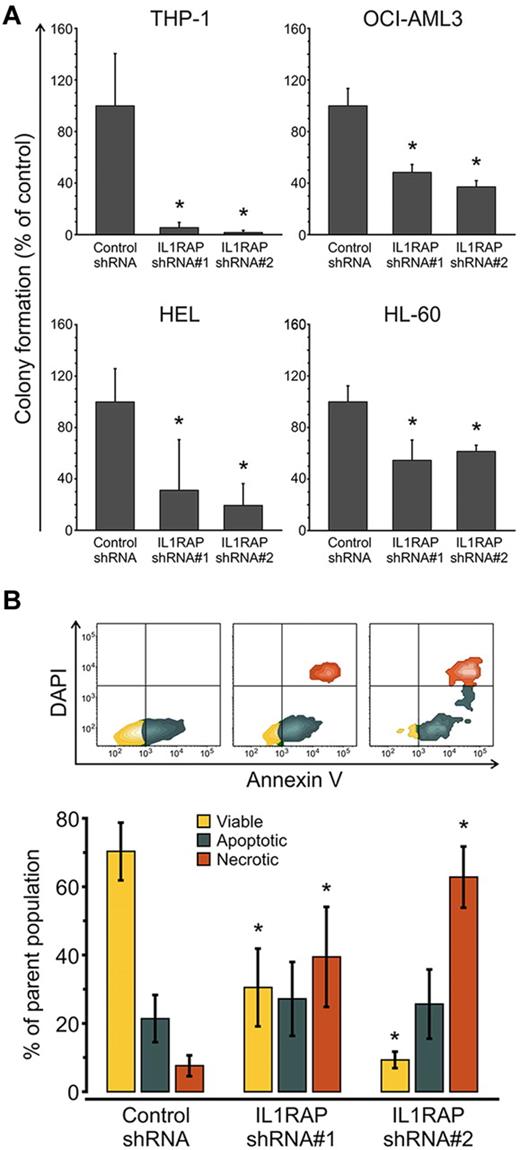
To determine whether IL1RAP is functionally important for the malignant growth of AML cells, we initially performed knockdown experiments in THP1 cells, which express high levels of IL1RAP. We used lentivirally expressed shRNAs directed against IL1RAP and coexpressing GFP as a marker. We tested and identified 2 shRNA constructs, which caused a decrease of IL1RAP expression by 49% and 88%, respectively (supplemental Figure 9). We transduced THP1 cells with these lentiviral shRNAs or with a nonsilencing control, and subjected sorted GFP-positive, annexin V–negative, DAPI-negative cells to clonogenic assays. Knockdown of IL1RAP led to a significant (P < .05) 94% and 98% inhibition of leukemic colony formation of THP1 cells compared with the nonsilencing control (Figure 5A). Moreover, although the cell cycle state was not significantly affected (supplemental Figure 11), IL1RAP knockdown caused a significant increase of cellular death as determined by annexin V/DAPI staining (Figure 5B). Cell death mainly occurred between 24 and 48 hours after infection (supplemental Figure 12). We examined further AML cell lines and found that shRNA-mediated inhibition of IL1RAP reduced clonogenicity by 51.5% (shRNA#1) and 62% (shRNA#2) in OCI-AML3 cells (P < .05), by 69% (shRNA#1) and 81% (shRNA#2) in HEL cells (P < .05), and by 45% (shRNA#1) and 38.5% (shRNA#2) in HL-60 cells (P < .05; Figure 5A; supplemental Figure 9). These results demonstrate that IL1RAP is functionally relevant in AML and that reduction of IL1RAP leads to a decrease of the clonogenic capacity across different AML cell lines. These findings suggest that targeting IL1RAP could be used as a therapeutic strategy to inhibit AML cells.
IL1RAP knockdown decreases clonogenic potential and leads to increased cell death of AML cells. (A) Colony formation assays in semisolid methylcellulose media of AML cells (THP-1, OCI-AML3, HEL, and HL-60) infected with control and 2 IL1RAP-directed shRNAs. Data are mean ± SD. *P < .05. (B) Analysis of apoptosis/necrosis with annexin V/DAPI in THP-1 cells infected with control and IL1RAP shRNAs. Top panel: FACS contour plots of 1 representative experiment. Bottom panel: Mean ± SD of 3 independent experiments. *P < .05.
IL1RAP knockdown decreases clonogenic potential and leads to increased cell death of AML cells. (A) Colony formation assays in semisolid methylcellulose media of AML cells (THP-1, OCI-AML3, HEL, and HL-60) infected with control and 2 IL1RAP-directed shRNAs. Data are mean ± SD. *P < .05. (B) Analysis of apoptosis/necrosis with annexin V/DAPI in THP-1 cells infected with control and IL1RAP shRNAs. Top panel: FACS contour plots of 1 representative experiment. Bottom panel: Mean ± SD of 3 independent experiments. *P < .05.
Discussion
Transcription factors are frequently disrupted in AML,34,35 and we and others have previously shown dysregulated transcription in AML, including in stem and progenitor cells.18,26,32,36-38 However, experimental determination of dysregulated gene expression in AML is challenging because of patient-to-patient heterogeneity as well as cellular heterogeneity within the leukemic cells of individual patients. In addition, LSCs have recently been reported to reside in different phenotypic stem and progenitor compartments in AML and at relatively low frequencies.3,5 In this study, we identified IL1RAP as a novel candidate for stem cell-targeted therapy and diagnosis in AML. We applied an experimental approach, which has features that set it apart from previously used strategies. First, in contrast to several other studies, we included rigorous lineage depletion, including myeloid markers in our strategy, in an attempt to focus on the most immature definable stem cell populations and to most effectively reduce experimental variation caused by cellular heterogeneity. We show that this strategy resulted in a more stringent selection of cellular subpopulations and excluded a significant additional fraction of more mature cells in stem and progenitor populations compared with HSCs sorted without lineage depletion. Second, to decrease experimental noise because of interpatient variability, we initially focused on a genetically defined subset of AML, patients with loss/deletions of chromosome 7 as the sole aberration. Third, we performed parallel transcriptional analysis of 3 different stem and progenitor cell types (LT-HSCs, ST-HSCs, and GMP) and compared them with their respective normal counterparts to circumvent the uncertainty about the precise cellular localization and identity of LSCs and pre-LSCs, and to identify commonly dysregulated transcription, independent of the various differentiation stages at which diverse pre-LSC and LSC types may reside. Using this approach, we were able to identify 11 genes commonly dysregulated in LT-HSCs, ST-HSCs, and GMP in AML with −7/7q−, which included IL1RAP. Of note, IL1RAP could not be found differentially expressed in unfractionated leukemia cells. Some of the differentially expressed genes we found in individual compartments, in particular in AML-GMP, showed a partial overlap with the results of previous analyses using less rigorous sorting (eg, CD97, CD123, PDK1, FYB, TFRC, and HMGN5),4,6,10 thereby validating our findings. Interestingly, we found IL1RAP overexpression on stem cells of some patients with another subtype of leukemia, AML with normal karyotype, as well as in patients with high-risk MDSs, but not in patients with low-risk MDS. It is tempting to speculate that IL1RAP expression on MDS-HSCs may precede transformation to overt AML. High IL1RAP levels were independently associated with poor clinical outcome in AML, supporting the biologic significance of IL1RAP in the clinical setting. Inhibition of IL1RAP led to a significant reduction of growth and colony-forming capacity of AML cells ex vivo, indicating that IL1RAP is functionally relevant in AML and may be a promising diagnostic and therapeutic target. The finding that IL1RAP was preferentially overexpressed in immature HSCs and the observation that all AML cells carrying the clonotypic −7/7q− aberration overexpressed IL1RAP suggest that IL1RAP overexpression may be an early event in AML pathogenesis.
IL1RAP was recently reported to be overexpressed in bcr-abl–positive CD34+ cells of patients with chronic myelogeneous leukemia, and its targeting by cytotoxic T cells inhibited leukemia cell proliferation in vitro.29 This suggests that IL1RAP overexpression is implicated in the molecular pathogenesis of different myeloid malignancies, including chronic myelogeneous leukemia and subsets of AML and MDS. IL1RAP mediates the response to IL-1, IL-33, and IL-36 and has been shown to regulate the inflammatory response, as well as activation of T lymphocytes and mast cells.28,39-42 Previous studies have shown increased IL1β production in patients with AML.43,44 IL1β is produced as an autocrine factor in AML and confers apoptosis resistance to AML blast cells.45 Interestingly, IL1R antagonist (IL1Ra) was previously shown to have an inhibitory effect on the growth of AML blasts,46 and it is available as a drug (anakinra) with promising safety data.42 Our data suggest that aberrant expression of IL1RAP on stem and progenitor cells in AML and high-risk MDS may mediate similar effects and cytokine responsiveness in immature cell populations in AML. The use of IL1Ra could be of particular benefit for AML patients with aberrant IL1RAP expression on stem and progenitor cells. Of note, IL1RAP, as opposed to IL1R1, is only expressed on a subset of blood cell types; thus, its targeting in AML may be more specific and have less severe side effects. As recent data demonstrate that IL1RAP can also associate with and enhance signaling of other receptors,47,48 therapeutic targeting of IL1RAP may offer the opportunity to simultaneously inhibit signaling through multiple pathways in AML.
Several markers have been proposed as leukemia stem cell markers, such as CD123,6 CD44,7 CLL-1,8 TIM3,9,49 CD25,50 CD32,50 CD96,11 and CD47.4 Some of these surface proteins have been reported to enrich for LSCs and to be useful for their therapeutic elimination in preclinical models. Because AML is a heterogeneous disease, different genes might need to be targeted in different subtypes to eradicate the disease. Our data show that diverse sets of molecules are aberrantly expressed in different immature stem and progenitor subsets of individual patients, with only very limited overlap, thus reflecting the cellular heterogeneity of the disease. We found that IL1RAP was one of the few genes aberrantly overexpressed across different stem and progenitor subsets, including putative pre-LSC populations, making it a particularly promising therapeutic and diagnostic target, conceivably in combination with 1 or more of the markers previously listed in this paragraph. Further studies will be required to assess the precise role of IL1RAP overexpression and potentially disturbed downstream signaling in the molecular pathogenesis of AML stem cells. Our results provide a rationale for further evaluating the therapeutic effects of targeting IL1RAP and its specificity for AML stem cells in vitro as well as in preclinical in vivo studies. The examination of IL1RAP surface expression by FACS in larger cohorts is warranted to determine its potential clinical usefulness as a prognostic marker.
In addition to IL1RAP, our study provides an inventory of further targets aberrantly expressed in immature stem and progenitor cells in AML with −7/7q−. These include, for instance, RAS guanyl-releasing protein 3 and IL-8, both of which have previously been implicated in oncogenic signaling51-53 and may offer further opportunities for the development of stem cell-targeted therapies. Functional evaluation of these candidates is warranted in future studies. Our experimental approach may also be used for the characterization of transcriptional dysregulation in stem and progenitor cells in other subtypes of AML.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the ECOG for providing specimens from the ECOG tissue bank, Guillermo Simkin and Swathi-Rao Narayanagari from the Einstein Stem Cell FACS and Xenotransplantation Facility (NYSTEM, C024172) for expert technical assistance, and the Albert Einstein Genomics Core Facilities.
This work was supported by the National Institutes of Health grant U24 CA114737-05S3 (E.P.), Leukemia & Lymphoma Research United Kingdom (J.B. and A.P.), a NYSTEM research grant (CO24350, U.S.), and an American Cancer Society–J. T.Tai & Company Inc postdoctoral fellowship (B.W.). U.S. is the recipient of a Howard Temin Award of the National Cancer Institute (R00 CA131503) and a Medical Research Award of the Gabrielle's Angel Foundation for Cancer Research. U.S. is the Diane and Arthur B. Belfer Faculty Scholar in Cancer Research of the Albert Einstein College of Medicine.
National Institutes of Health
Authorship
Contribution: L.B. designed and performed research, analyzed data, made figures, and wrote the manuscript; B.W. and C.M. performed research and analyzed data; B.B. analyzed data, performed statistical analyses, and made figures; L.Z., T.I.T., R.F.S., and S.B.-N. performed research; S.P. contributed vital reagents and analyzed and interpreted data; A.P. and J.B. analyzed data; E.P. contributed vital reagents, collected data, and analyzed and interpreted data; R.P.K. performed research and collected and analyzed data; L.C., H.F.F., P.L.G., and M.S.T. collected and analyzed data; C.S. and C.S.M. performed research and analyzed and interpreted data; A.V. designed and performed research and analyzed and interpreted data; and U.S. designed the research, analyzed and interpreted data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ulrich Steidl, Albert Einstein College of Medicine, Chanin Bldg, Rm 606, 1300 Morris Park Ave, Bronx, NY 10461; e-mail: ulrich.steidl@einstein.yu.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal