Abstract
We recently generated 2 phenotypically similar Hoxa9+Meis1 overexpressing acute myeloid leukemias that differ by their in vivo biologic behavior. The first leukemia, named FLA2, shows a high frequency of leukemia stem cells (LSCs; 1 in 1.4 cells), whereas the second, FLB1, is more typical with a frequency of LSCs in the range of 1 per several hundred cells. To gain insights into possible mechanisms that determine LSC self-renewal, we profiled and compared the abundance of nuclear and cytoplasmic proteins and phosphoproteins from these leukemias using quantitative proteomics. These analyses revealed differences in proteins associated with stem cell fate, including a hyperactive p38 MAP kinase in FLB1 and a differentially localized Polycomb group protein Ezh2, which is mostly nuclear in FLA2 and predominantly cytoplasmic in FLB1. Together, these newly documented proteomes and phosphoproteomes represent a unique resource with more than 440 differentially expressed proteins and 11 543 unique phosphopeptides, of which 80% are novel and 7% preferentially phosphorylated in the stem cell–enriched leukemia.
Introduction
Pioneer studies by Dick et al exposed that a significant proportion of human leukemias are hierarchically organized.1 Although the majority of cells in these leukemias show limited proliferative capacity, their production is continuously ensured by a much less frequent population termed leukemia stem cells (LSCs), which display unlimited self-renewal potential. In contrast to normal (ie, nonleukemic) hematopoietic stem cells (HSCs), which can be purified by flow cytometry to near-functional homogeneity, the prospective isolation of LSC has proven more difficult to achieve. Interspecimen variation in cell surface marker expression attributed to LSCs is, at least in part, responsible for this difficulty. For this reason, functional in vivo studies remain the “gold standard” assay for quantitative assessment of LSC numbers in any given specimen.2,3
Original studies, which took advantage of NOD-scid recipient mice for LSC evaluation in human acute myeloid leukemia (AML), reported considerable variations in LSC frequencies, ranging from 1 in 104 to 1 per 107 cells.2 Additional depletion of the recipient's NK cells using NOD/ShiLtSz-scid/IL2Rγnull or NOD/ShiJic-scid/IL-2Rγnull mice considerably improved LSC detection.4 Such studies highlighted the important contribution of host factors in LSC frequency assessment. These factors could be minimized through the exploitation of a syngenic model system (eg, mouse leukemias transplanted into syngenic or congenic recipients). For example, LSC frequencies of ∼ 1% were observed in E2a-Pbx1 + Hoxa9 or MLL-AF9–induced mouse AML, whereas frequencies of 0.01% were found in MOZ-TIF AML, respectively.5–7 Together, these studies confirmed the relatively low LSC frequency in AML.
Thus, 2 outstanding issues (one related to low LSC abundance and the other to reliable cell surface markers for LSC isolation) still limit our ability to obtain a sufficiently high number of pure LSCs that can be used for comprehensive proteomic or phosphoproteomic studies. To overcome these hurdles, our group recently generated several closely related Hoxa9 + Meis1 overexpressing AMLs derived from phenotypically primitive mouse fetal liver cells. These leukemias express similar levels of the Hoxa9 and Meis1 oncogenes, have a normal karyotype, and do not harbor any coarse chromosomal abnormalities.8 They also display the same cell surface marker phenotype (c-Kit+Sca1−CD150−CD48+CD34+CD71+Mac1+GR1+), cell cycle profile, and dissemination characteristics. Despite this remarkable similarity, 2 of these leukemias exhibit very different LSC frequencies, ranging from 1 in 1.4 (∼ 71%) for FLA2 to 1 in 347 (∼ 0.3%) for FLB1.8 Importantly, the difference in LSC frequencies between FLA2 and FLB1 leukemias has remained stable over successive transplantations. These leukemias are clinically relevant because the Hoxa9 and Meis1 oncogenes are overexpressed in a large proportion of human leukemias, are transcriptional targets of MLL fusion proteins and have been shown to be key regulators of LSC self-renewal.9–11 Unlike other leukemia mouse models, high numbers of an almost pure population of LSCs can be readily obtained from FLA2 without the need for purification using cell surface markers. Therefore, FLA2 and FLB1 leukemias represent a unique model system to dissect the molecular machinery underlying the self-renewal of LSCs. To this end, transcriptome analysis using next-generation RNA sequencing revealed hundreds of differentially expressed genes and numerous structural differences (alternative splicing and promoter usage).8
Using the same FLA2/FLB1 leukemia model, we now report the first ever described comparative proteome and phosphoproteome of 2 highly similar leukemias, one of which is made of almost pure LSCs. Our results show that several novel candidate proteins are differentially expressed or phosphorylated between FLA2 and FLB1, including the Polycomb Group (PcG) proteins Ezh2 and Suz12. These results represent a unique resource for the dissection of signaling pathways that are active in LSCs.
Methods
Generation of FLA2 and FLB1 primary mouse leukemias
FLA2 and FLB1 primary mouse leukemias were generated as described previously8 (see supplemental Methods for details, see the Supplemental Materials link at the top of the article). Experimental procedures were approved by the Université de Montréal animal ethics committee.
Expression analyses using SDS-PAGE and mass spectrometry
Biologic replicates (n = 3, 20 μg each) of FLA2 and FLB1 nuclear and cytosolic extracts were reduced with tris(2-carboxyethyl)phosphine (Pierce Chemical), alkylated with iodoacetamide (Sigma-Aldrich) and separated on a 4%-12% precast NuPAGE gel (Invitrogen). The gel was stained with colloidal Coomassie, and 12 bands of equal size were excised using a custom cutting device. Proteins were reduced, alkylated with iodoacetamide, digested with trypsin, and peptides extracted as described before.12 Samples were analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) on an LTQ-Orbitrap mass spectrometer (Thermo Fischer Scientific) using a 65-minute linear gradient (supplemental Methods).
Phosphopeptide enrichment and mass spectrometry
Nuclear and cytosolic protein extracts (pooled from 5 mice, 1.2 mg each, n = 3 independent experiments) were reduced, alkylated with iodoacetamide, and digested with trypsin (Promega). Phosphopeptides were enriched on custom TiO2-microcolumns (GL Science) as described previously.12,13 Phosphopeptides were analyzed using online 2D-LC-MS/MS on an LTQ-Orbitrap mass spectrometer. Phosphopeptide samples were loaded on a self-packed 45 mm × 300 μm Polysulfoethyl column (Nest Group) and eluted with increasing salt fractions (0, 0.05, 0.075, 0.1, 0.5, and 2.0M ammonium acetate in 2% ACN, 0.2% formic acid, pH 3.0). Eluted salt fractions were then analyzed directly by LC-MS/MS as described in “Expression analyses using SDS-PAGE and mass spectrometry” (supplemental Methods).12
Protein identification and bioinformatic analyses
Results
Subcellular quantitative analysis of proteome and phosphoproteome of FLA2 and FLB1 leukemias
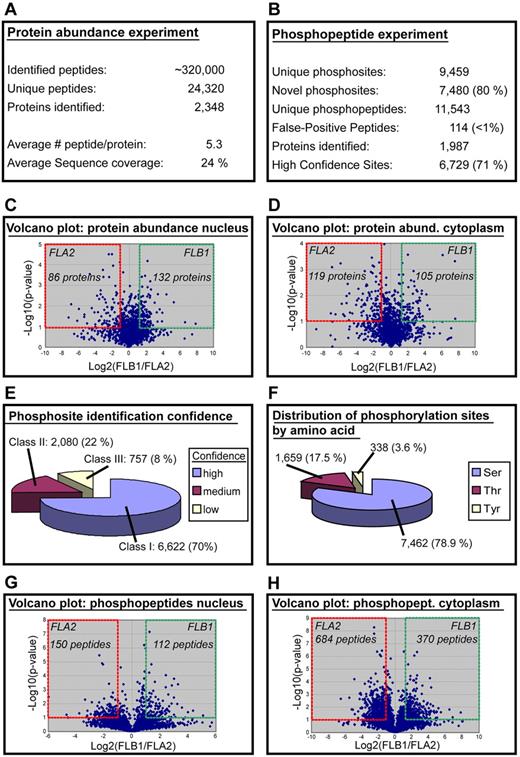
Nuclear and cytosolic protein extracts were generated from freshly harvested FLA2 and FLB1 leukemias acquired from 15 different mice. Three biologic replicates were generated by combining the protein extracts from 5 mice. Purity of the nuclear and cytosolic fractions was assessed for each specimen (supplemental Figure 1A). A 2-pronged approach based on label-free quantitative proteomics was used to profile changes in both protein abundance and phosphorylation stoichiometry in the corresponding nuclear and cytosolic extracts (supplemental Figure 1B). Comparative MS analysis of these extracts on an LTQ-Orbitrap mass spectrometer led to the identification of 24 320 unique peptides from 2348 proteins with an average sequence coverage of 24% and a false discovery rate < 1% using a forward and reverse IPI mouse database. We correlated the changes in abundance of 12 749 unique peptides (2655 proteins) using label-free quantitative proteomics where 95% of peptide clusters showed a relative SD < 53% (1.53-fold) across biologic replicates (supplemental Figure 1C). Although expression levels between FLB1 and FLA2 remained unaffected for most identified proteins, a subset of 442 proteins (∼ 19%) showed more than a 2-fold change in abundance in either nuclear or cytosolic fractions (Figure 1A-D; supplemental Table 1). Among the most differentially abundant proteins in nucleus and cytosol fractions, we observed the up-regulation of Lef1 in FLA2 LSCs, a transcription factor and critical effector of the canonical Wnt/β-catenin signaling pathway involved in HSC self-renewal.15 Ectopic expression of Lef1 in bone marrow cells leads to development of AML when transplanted in mice.16 The increased expression of the tumor suppressor Ing5 in the nucleus of FLB1 cells is also interesting. This protein is part of the chromatin remodeling MOZ/MORF and HBO1 complexes and stimulates their acetyltransferase activity.17 ING5 also interacts with p53 and enhances its transcriptional activity associated with growth arrest and apoptosis.18 High nuclear expression of ING5 was found to correlate with differentiation status of head and neck squamous cell carcinomas.19 Most of the differentially expressed proteins identified have no known function in stem cell self-renewal or cancer. Of the 205 proteins most highly differentially expressed in FLA2, only 5 have previously been associated with HSC activity, namely, Dnmt1, Nup98, Fgfr1, Lef1, and CD34 (supplemental Table 1).16,20–23 To determine whether differences in protein expression were also observed at the mRNA transcript level, we compared the proteomic data with previous RNA-seq data of FLA2 and FLB1 leukemias8 (supplemental Figure 2). An up-regulation of gene and protein expression levels was observed for Car2, Gsn (gelsolin), S100a9, BC117090, Stfa1, and 2010005H15Rik (supplemental Figure 2 bottom left quadrant). Gsn is required for the assembly and disassembly of actin filaments, and negatively regulate p53 mediated apoptosis by preventing its nuclear translocation.24 S100a9 is a Ca2+ binding protein and a regulator of proinflammatory responses and premetastatic niche formation.25 Interestingly, BC117090, Stfa1, and 2010005H15Rik are contiguous genes on chromosome 16, but their functions are either unknown or not fully characterized. However, we noted a lack of correlation in expression levels for a large proportion of genes and proteins from our datasets, suggesting intensive posttranscriptional and posttranslational regulation.
Large-scale proteome and phosphoproteome analyses of FLA2 and FLB1 leukemias. (A) Results of protein expression experiments. (B) Results of phosphoproteome experiments. (C-D) Volcano plot of the protein abundance data: levels of 218 and 224 proteins change significantly in nuclear and cytosolic extracts, respectively (≥ 2-fold change in abundance). (E) Phosphosite confidence level. A total of 6622 phosphosites were identified with a high confidence level (class I), whereas 2080 and 757 sites were assigned with medium and low confidence, respectively. (F) Distribution of phosphorylation sites by amino acids showing a relatively large proportion of phosphorylation at serine residues. (G-H) Volcano plot of the phosphopeptide experiments: 262 and 1054 phosphopeptides change significantly in nuclear and cytosolic extracts, respectively (≥ 2-fold change in abundance).
Large-scale proteome and phosphoproteome analyses of FLA2 and FLB1 leukemias. (A) Results of protein expression experiments. (B) Results of phosphoproteome experiments. (C-D) Volcano plot of the protein abundance data: levels of 218 and 224 proteins change significantly in nuclear and cytosolic extracts, respectively (≥ 2-fold change in abundance). (E) Phosphosite confidence level. A total of 6622 phosphosites were identified with a high confidence level (class I), whereas 2080 and 757 sites were assigned with medium and low confidence, respectively. (F) Distribution of phosphorylation sites by amino acids showing a relatively large proportion of phosphorylation at serine residues. (G-H) Volcano plot of the phosphopeptide experiments: 262 and 1054 phosphopeptides change significantly in nuclear and cytosolic extracts, respectively (≥ 2-fold change in abundance).
We next examined the differences in the status of protein phosphorylation between nuclear and cytosolic extracts derived from FLA2 and FLB1 leukemias (supplemental Figure 1B). This analysis led to the identification of 11 543 unique phosphopeptides from 1987 phosphoproteins (9459 phosphorylation sites, false discovery rate < 1%). Label-free quantitative proteomics enabled the correlation of abundance for 9220 phosphopeptides (1812 proteins), of which 1316 phosphopeptides were differentially abundant in either cytosolic or nuclear extracts of FLA2 and FLB1 leukemic cells (≥ 2-fold change; Figure 1B-H; supplemental Table 2). These phosphopeptides represented 707 unique proteins, indicating that almost 36% of the identified phosphoproteins are differentially regulated. The analysis of FLA2 and FLB1 leukemias yielded an unprecedented number of new phosphorylation sites from an in vivo model of LSCs. Comparison of the identified phosphorylation sites with those from SwissProt (Version 51.0) and PhosphoELM (http://phospho.elm.eu.org/) indicated that 7480 sites (∼ 80%) have not been previously reported. We also evaluated the confidence level in the site localization using a probability score function12,26 and determined that 70% of the identified sites (6622 sites) corresponded to high confidence assignments (Figure 1E). Analysis of the distribution of phosphorylation sites by amino acids indicates that phosphotyrosines account for 3.6% of all identified phosphorylation sites (Figure 1F). Among the most differentially abundant phosphoproteins in nucleus and cytosol, increased phosphorylation of Son at serines 1987/1989/1991 in the cytosolic fraction of FLA2 cells is interesting because this protein interacts with ETO and inhibits the leukemogenic activity of the AML1-ETO fusion protein often found in AMLs.27 However, abnormal cytoplasmic localization of SON has been found in AML1-ETO–positive cell lines and in patient samples, and appears to collaborate with AML1-ETO to generate leukemia. Phosphorylation of the high mobility group protein Hmga1 at S102 and S103 is increased 14-fold in FLA2 cells. HMGA1 is overexpressed in human leukemias and many other types of tumors.28 Other members of this family have been shown to expand HSCs when overexpressed (Hmgb1) and promote neural stem cell self-renewal (Hmga2).29,30 Interestingly, many of the differentially phosphorylated proteins that have been previously associated with HSC activity are chromatin-modifying proteins, such as Myst3, Dnmt3a/b, and Ezh2 (supplemental Table 2).31–33 These results suggest that post-translational mechanisms affect the protein content of FLA2 and FLB1 leukemias and that phosphorylation-mediated signaling in the LSC-enriched leukemia (FLA2) differs from a leukemia with a lower content of self-renewing leukemic blasts (FLB1).
Protein interaction network analysis in LSCs
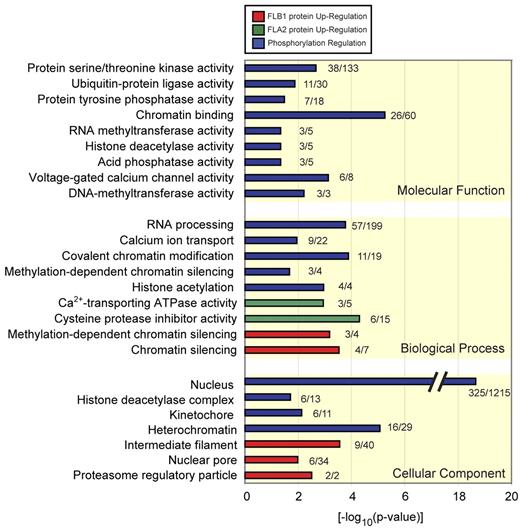
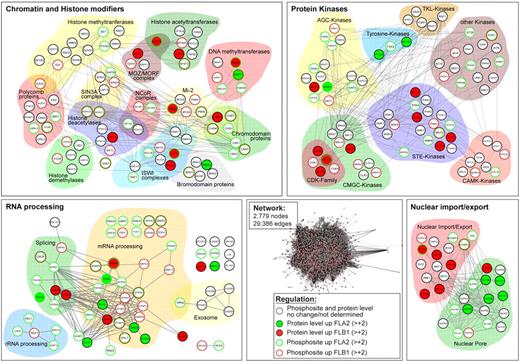
To get more functional insights into the proteins and pathways differentially regulated in LSCs, we conducted a bioinformatic analysis of our proteomics and phosphoproteomics data. First, we used the protein expression and phosphorylation data to develop an interaction network using the STRING program (http://string.embl.de). We combined interactions found in mouse and their human orthologs, resulting in a network of 2779 proteins (nodes) and 29 386 connections (edges). From this complex network, we extracted subnetworks using GO. Groups of proteins with specific GO terms changing in abundance or phosphorylation status were compared with all identified proteins. GO terms were considered significant when they had adjusted P values < .05 in a Fisher exact test, resulting in 165 significant terms. Protein groups were then sorted according to cellular component, biologic processes, and molecular function GO categories (Figure 2; supplemental Table 3). From this analysis, statistically significant enrichment of 4 major groups of differentially regulated protein networks between FLA2 and FLB1 were identified (Figure 3). These groups include: (1) chromatin and histone modifiers (P = .0005), (2) proteins involved in nuclear import/export (P = .01), (3) protein kinases (P = .011), and (4) proteins involved in RNA processing (P = .015).
GO analysis of regulated proteins and phosphopeptides. Significant GO terms (P < .05) of proteins regulated by phosphorylation (blue) or increased protein abundance in FLB1 (red) and FLA2 (green). GO terms are grouped according to biologic process, molecular function, and cellular component, and the number of regulated proteins versus the background of all identified proteins is given. Significant terms include protein serine/threonine kinase activity, chromatin binding, RNA processing, cysteine protease inhibitor activity, and nuclear pore.
GO analysis of regulated proteins and phosphopeptides. Significant GO terms (P < .05) of proteins regulated by phosphorylation (blue) or increased protein abundance in FLB1 (red) and FLA2 (green). GO terms are grouped according to biologic process, molecular function, and cellular component, and the number of regulated proteins versus the background of all identified proteins is given. Significant terms include protein serine/threonine kinase activity, chromatin binding, RNA processing, cysteine protease inhibitor activity, and nuclear pore.
Regulated protein networks in FLA2 versus FLB1 leukemias. Protein abundance and phosphorylation data were analyzed with the STRING database (http://string.embl.de) resulting in a network composed of 2779 nodes and 29 386 edges. A GO analysis was performed for extracting significant (P < .05) subsets of proteins differentially regulated in FLA2 and FLB1 leukemias. Shown are the 4 most significant subnetworks affected: chromatin and histone modifiers, protein kinases, RNA processing, and nuclear import/export. Proteins/nodes are grouped according to their function and/or family. Color code indicates differentially expressed and/or phosphorylated proteins in FLA2 and FLB1 leukemias as described in the caption.
Regulated protein networks in FLA2 versus FLB1 leukemias. Protein abundance and phosphorylation data were analyzed with the STRING database (http://string.embl.de) resulting in a network composed of 2779 nodes and 29 386 edges. A GO analysis was performed for extracting significant (P < .05) subsets of proteins differentially regulated in FLA2 and FLB1 leukemias. Shown are the 4 most significant subnetworks affected: chromatin and histone modifiers, protein kinases, RNA processing, and nuclear import/export. Proteins/nodes are grouped according to their function and/or family. Color code indicates differentially expressed and/or phosphorylated proteins in FLA2 and FLB1 leukemias as described in the caption.
Many chromatin and histone modifiers are known to affect stem cells or are implicated in cancer.34,35 More than 50 different chromatin-modifying proteins were found differentially regulated between FLA2 and FLB1 cells, mostly by phosphorylation (11 differentially expressed proteins vs 46 differentially phosphorylated; Figure 3; supplemental Tables 1 and 2). In addition to PcG proteins, members of the ISWI and Mi-2 complexes as well as DNA methyltransferases and methylated DNA binding proteins were differentially regulated. Histone deacetylases (Hdac1/2, Sirt2/7) and chromodomain proteins of the CHD and HP1 families also displayed multiple differentially phosphorylated residues. Interestingly, the only 2 proteins found to be more abundant in FLA2 LSCs, Dnmt1 and Brd4, are involved in the transmission of transcriptional programs to daughter cells. The former is thought to maintain DNA methylation patterns after DNA replication, whereas the latter is involved in gene bookmarking, an epigenetic mechanism allowing daughter cells to reproduce an inherited pattern of gene expression after mitosis.36,37
Strikingly, more than 27% of the 199 identified proteins involved in RNA processing were differentially expressed (n = 13) or regulated by phosphorylation (n = 44), suggesting that post-transcriptional regulation of mRNAs and ncRNAs by degradation, splicing, or differential transport differs significantly between FLA2 and FLB1 leukemic cells (Figure 3; supplemental Tables 1 and 2). In support of this, analysis by RNA-seq revealed numerous structural differences between the FLA2 and FLB1 transcriptomes, namely, alternative splicing and promoter usage.8
Nuclear pore proteins and factors involved in nuclear import/export were also significantly enriched in our proteomics analysis. Of the 20 most differentially regulated proteins from this group, 11 varied in expression, whereas 10 were phosphorylated in either FLA2 or FLB1. Differential regulation of proteins forming the nuclear pore was considerably more prevalent in FLA2 cells than FLB1, with 9 nucleoporins either more expressed (n = 5) or phosphorylated (n = 4; Figure 3; supplemental Tables 1 and 2). Notably, expression of the nucleoporin Nup98 was increased ∼ 3-fold in FLA2 LSCs. This protein is often implicated in recurrent chromosomal translocation in AMLs.38 Contrary to nuclear pore constituents, import/export proteins, such as Ntf2, Importins, and Rae1l, were more expressed or phosphorylated in FLB1 cells (Figure 3; supplemental Tables 1 and 2). Together, these results suggest that the nuclear transport machinery of proteins and RNA behaves differently within the 2 leukemias.
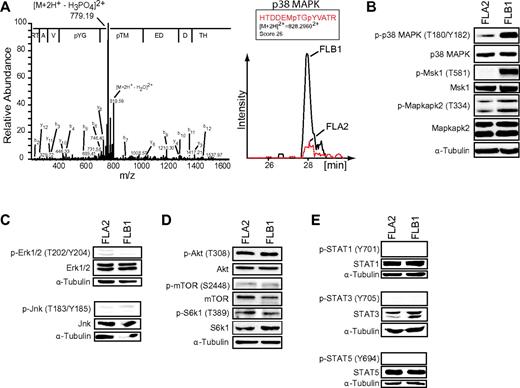
Several serine/threonine kinases displayed differential regulation between FLA2 and FLB1 cells. Of the 81 protein kinases identified, 12 showed differences in protein abundance, whereas 32 were differentially phosphorylated (Figure 3; supplemental Tables 1 and 2). The CMGC and STE families represented almost half of the differentially regulated kinases (Figure 3). Most of the phosphosites detected in our study fall outside of the kinase activation loops, suggesting alternate regulatory roles in modulating kinase function (eg, target specificity, protein interaction, subcellular localization). One of the few kinases for which activity could be correlated by independent Western blot experiments was p38 MAP kinase (Mapk14). Strikingly, we found that p38 MAP kinase was considerably more activated in FLB1 than FLA2 cells (Figure 4A; supplemental Table 2). RNA transcript and total p38 MAPK protein levels were similar in both leukemias (Figure 4B).8 Interestingly, we observed that the levels of p38 activation were inversely correlated with the self-renewal capacity of LSCs (supplemental Figure 4). Furthermore, replicate experiments performed on mice injected with FLB1 cells and treated with the p38 inhibitor SB203580 during the last week of leukemia development failed to show a reduction in LSC frequency (supplemental Figure 5). These results suggest that other effectors besides p38 are implicated in LSC self-renewal.
Increased activation of p38 MAP kinase is inversely correlated with LSC activity. (A) MS/MS spectrum (left panel) of phosphopeptide-enriched FLB1 extracts showing phosphorylation on threonine 180 (T180) and tyrosine 182 (Y182) in the activation loop of p38 MAPK. The amino acid sequence is displayed with the corresponding to y-type ion fragments. Ion chromatogram (right panel) of the p38 MAPK phosphopeptide HTDDEMpTGpYVATR (encompassing the activation site) in cytosolic extracts shows an ∼ 3-fold up-regulation in FLB1. Mass spectrometry analyses were performed in biologic triplicates. (B) Western blot analysis of the phosphorylation status of p38 MAPK, Msk1, and Mapkapk2 in FLA2 and FLB1 specimens. (C) Western blot analysis of the phosphorylation status of Erk1/2 and Jnk kinases in FLA2 and FLB1 specimens. (D) Western blot analysis of the phosphorylation status of Akt, mTOR, and S6k1 kinases in FLA2 and FLB1 specimens. (E) Western blot analysis of the phosphorylation status of STAT1/3/5 in FLA2 and FLB1 specimens. α-tubulin was used as a loading control. All Western blots shown are representative results from 3 independent experiments using 5 different samples for each leukemia per experiment.
Increased activation of p38 MAP kinase is inversely correlated with LSC activity. (A) MS/MS spectrum (left panel) of phosphopeptide-enriched FLB1 extracts showing phosphorylation on threonine 180 (T180) and tyrosine 182 (Y182) in the activation loop of p38 MAPK. The amino acid sequence is displayed with the corresponding to y-type ion fragments. Ion chromatogram (right panel) of the p38 MAPK phosphopeptide HTDDEMpTGpYVATR (encompassing the activation site) in cytosolic extracts shows an ∼ 3-fold up-regulation in FLB1. Mass spectrometry analyses were performed in biologic triplicates. (B) Western blot analysis of the phosphorylation status of p38 MAPK, Msk1, and Mapkapk2 in FLA2 and FLB1 specimens. (C) Western blot analysis of the phosphorylation status of Erk1/2 and Jnk kinases in FLA2 and FLB1 specimens. (D) Western blot analysis of the phosphorylation status of Akt, mTOR, and S6k1 kinases in FLA2 and FLB1 specimens. (E) Western blot analysis of the phosphorylation status of STAT1/3/5 in FLA2 and FLB1 specimens. α-tubulin was used as a loading control. All Western blots shown are representative results from 3 independent experiments using 5 different samples for each leukemia per experiment.
Increased activation of p38 and its downstream targets Msk1 and Mapkapk2 in the lower self-renewing FLB1 cells was confirmed by Western blot analysis (Figure 4B). Importantly, high activated p38 MAPK levels have been shown to impair stem cell self-renewal and induce differentiation of immature cells.39–41 Activation status of other kinases known to have a role in stem cells and cancer, such as Erk1/2, Jnk, Akt1, mTOR, and Jak-STAT pathways, was further assessed. Despite being activated, no major differences between FLA2 and FLB1 cells were observed (Figure 4C-D). Together, this suggests that activation of p38 MAPK and its downstream targets is a major signaling pathway that inversely correlates with the self-renewal potential of LSCs.
Polycomb group protein phosphorylation and localization in FLA2 versus FLB1
Analysis of the FLA2 and FLB1 phosphoproteome datasets revealed multiple phosphorylation sites in chromatin-modifying PcG proteins (Table 1). Phosphopeptides for 2 proteins of the PRC2 complex (Ezh2 and Suz12) and 6 of the PRC1 complex (Cbx8, Bmi1, Phc3, L3mbtl2, L3mbtl3, and Sfmbt1) were identified by mass spectrometry (Figure 3; Table 1). For some of these proteins, only a few peptides were identified and protein expression data could not be obtained for all of them (supplemental Table 1). At the mRNA level, quantitative RT-PCR analysis of all 37 known PcG genes revealed no significant expression differences between FLA2 and FLB1 cells (supplemental Figure 3A-B). Interestingly, phosphorylation of L3mbtl3 on serine 605 was found to be 3.5-fold higher in FLA2 cells (Table 1). Conversely, phosphorylation of the Ezh2 threonine 487 (T487) and Suz12 serines 548 and 585 is more abundant in FLB1 than FLA2 leukemic cells (Table 1; Figure 5A-B; supplemental Figure 3C-D). This suggests that the function of both PRC1 and PRC2 complexes is differentially regulated by phosphorylation in LSCs. The Jarid2 histone demethylase, which associates with the PRC2 complex and modulates its methyltransferase activity,42,43 was also 5.7-fold more phosphorylated in FLB1 than FLA2 (supplemental Table 2).
Polycomb group phosphopeptides identified in FLA2 and FLB1 leukemias
| Name . | Phosphopeptide sequence . | Phosphosites . | Ratio FLA2/FLB1* . | Confidence score . | Novel . |
|---|---|---|---|---|---|
| EZH2 | LPNNSpSRPpSTPTISVLESK | S363, S366 | 2.0 | 0.77, 0.87 | No/no |
| GRLPNNSpSRPpSTPTISVLESK | S363, S366 | 1.3 | 0.87, 0.88 | No/no | |
| LPNNSSRPSpTPTISVLESK | T367 | 1.1 | 0.42 | No | |
| ESSIIAPVPTEDVDpTPPR | T487 | 0.56 | 0.96 | No | |
| SUZ12 | ASMSEFLEpSEDGEVEQQR | S548 | 0.34 | 0.99 | No |
| LYFHSDTCLPLRPQEMEVDpSEDEKDPEWLR | S585 | 0.28 | 0.90 | No | |
| CBX8 | VDDKPSpSPGDSSK | S164 | 0.71 | 0.90 | No |
| BMI1 | KSpSLNGSSATSSG | S314 | 3.5 | 0.5 | Yes |
| PHC3 | KGGpSPGLESR | S281 | 1.4 | 1.00 | No |
| SFMBT1 | DAQTSSVpSDDENKPPpSPK | S764, S772 | NA | 0.95, 1.00 | No/no |
| KSSSpSSPTQSETPTPLPPDpTQTNK | S736, T751 | NA | 1.00, 1.00 | Yes/yes | |
| L3MBTL2 | SPpSPQLPLPIESIK | S687 | 1.4 | 0.84 | No |
| EEHQDISSLDRpSPpSPQLPLPIESIK | S685, S687 | NA | 1.00, 1.00 | No/no | |
| L3MBTL3 | LSGDTpSPPTTPSFPR | S605 | 3.5 | 0.82 | No |
| Name . | Phosphopeptide sequence . | Phosphosites . | Ratio FLA2/FLB1* . | Confidence score . | Novel . |
|---|---|---|---|---|---|
| EZH2 | LPNNSpSRPpSTPTISVLESK | S363, S366 | 2.0 | 0.77, 0.87 | No/no |
| GRLPNNSpSRPpSTPTISVLESK | S363, S366 | 1.3 | 0.87, 0.88 | No/no | |
| LPNNSSRPSpTPTISVLESK | T367 | 1.1 | 0.42 | No | |
| ESSIIAPVPTEDVDpTPPR | T487 | 0.56 | 0.96 | No | |
| SUZ12 | ASMSEFLEpSEDGEVEQQR | S548 | 0.34 | 0.99 | No |
| LYFHSDTCLPLRPQEMEVDpSEDEKDPEWLR | S585 | 0.28 | 0.90 | No | |
| CBX8 | VDDKPSpSPGDSSK | S164 | 0.71 | 0.90 | No |
| BMI1 | KSpSLNGSSATSSG | S314 | 3.5 | 0.5 | Yes |
| PHC3 | KGGpSPGLESR | S281 | 1.4 | 1.00 | No |
| SFMBT1 | DAQTSSVpSDDENKPPpSPK | S764, S772 | NA | 0.95, 1.00 | No/no |
| KSSSpSSPTQSETPTPLPPDpTQTNK | S736, T751 | NA | 1.00, 1.00 | Yes/yes | |
| L3MBTL2 | SPpSPQLPLPIESIK | S687 | 1.4 | 0.84 | No |
| EEHQDISSLDRpSPpSPQLPLPIESIK | S685, S687 | NA | 1.00, 1.00 | No/no | |
| L3MBTL3 | LSGDTpSPPTTPSFPR | S605 | 3.5 | 0.82 | No |
NA indicates not applicable.
Ratio normalized for changes in protein expression.
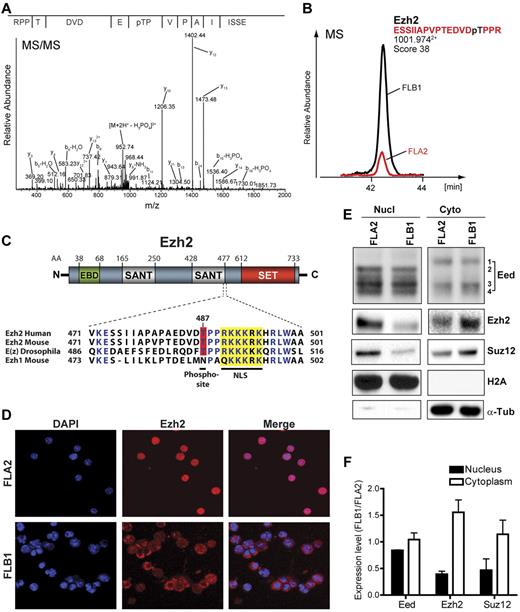
Differential phosphorylation and localization of PRC2 proteins in LSCs. Ezh2 is differentially phosphorylated at threonine 487 (T487) in FLB1 and FLA2 leukemias. (A) MS/MS spectrum of phosphopeptide-enriched FLB1 extracts showing the phosphosite on Ezh2 T487. (B) Ion chromatogram of the phosphopeptide ESSIIAPVPTEDVDpTPPR in nuclear extracts shows an ∼ 3-fold up-regulation in FLB1 cells. Mass spectrometry analyses were performed in triplicates. (C) Ezh2 primary protein structure and amino acid alignment of the region encompassing the identified T487 phosphorylated residue. The putative nuclear localization site (NLS, yellow box) is well conserved through evolution and in the mouse Ezh1 paralog. The phosphosite T487 is also well conserved but not present in Ezh1. Amino acid delimitation of Eed-binding domain (EBD), SANT, and SET domains are shown. (D) Differential localization of Ezh2 in FLB1 and FLA2 cells as shown by confocal microscopy using anti-Ezh2 antibody. Although Ezh2 is mainly nuclear in FLA2, it is predominantly cytoplasmic in FLB1 leukemic cells. (E) Western blot analysis of nuclear and cytosolic extracts from FLA2 and FLB1 leukemias for the core PRC2 complex proteins Eed, Ezh2, and Suz12 (representative results from replicate experiments using 4 different samples for each leukemia per experiment). Ezh2, Suz12, and specific isoforms of Eed are differentially localized in the nucleus and the cytoplasm. (F) Differences in nuclear and cytoplasmic abundance for Eed, Ezh2, and Suz12 in FLA2 and FLB1 cells. Levels were determined by analyzing the density of immunoblot signals using the Multi-Gauge imaging software from FUJIFilm. Graph represents the mean from 4 different samples for each leukemia.
Differential phosphorylation and localization of PRC2 proteins in LSCs. Ezh2 is differentially phosphorylated at threonine 487 (T487) in FLB1 and FLA2 leukemias. (A) MS/MS spectrum of phosphopeptide-enriched FLB1 extracts showing the phosphosite on Ezh2 T487. (B) Ion chromatogram of the phosphopeptide ESSIIAPVPTEDVDpTPPR in nuclear extracts shows an ∼ 3-fold up-regulation in FLB1 cells. Mass spectrometry analyses were performed in triplicates. (C) Ezh2 primary protein structure and amino acid alignment of the region encompassing the identified T487 phosphorylated residue. The putative nuclear localization site (NLS, yellow box) is well conserved through evolution and in the mouse Ezh1 paralog. The phosphosite T487 is also well conserved but not present in Ezh1. Amino acid delimitation of Eed-binding domain (EBD), SANT, and SET domains are shown. (D) Differential localization of Ezh2 in FLB1 and FLA2 cells as shown by confocal microscopy using anti-Ezh2 antibody. Although Ezh2 is mainly nuclear in FLA2, it is predominantly cytoplasmic in FLB1 leukemic cells. (E) Western blot analysis of nuclear and cytosolic extracts from FLA2 and FLB1 leukemias for the core PRC2 complex proteins Eed, Ezh2, and Suz12 (representative results from replicate experiments using 4 different samples for each leukemia per experiment). Ezh2, Suz12, and specific isoforms of Eed are differentially localized in the nucleus and the cytoplasm. (F) Differences in nuclear and cytoplasmic abundance for Eed, Ezh2, and Suz12 in FLA2 and FLB1 cells. Levels were determined by analyzing the density of immunoblot signals using the Multi-Gauge imaging software from FUJIFilm. Graph represents the mean from 4 different samples for each leukemia.
The PRC2 polycomb complex catalyzes the trimethylation of lysine 27 of histone H3 (H3K27me3), a crucial silencing epigenetic mark with roles in cell fate determination and cancer progression.44 Two recent studies showed that Ezh2, the enzyme responsible for H3K27me3, can be phosphorylated at T487 by CDK1.45,46 Whether this posttranslational modification inhibits Ezh2 methyltransferase activity remains unclear as both studies reported conflicting results. We therefore tested whether the increased phosphorylation of Ezh2 T487 observed in FLB1 cells correlated with changes in global H3K27me3 levels. Western blots analysis of H3K27me3 failed to reveal any difference between FLA2 and FLB1 leukemias (supplemental Figure 3E). Global levels of other histone modifications, such as H3K4me3 and H3K9ac, were also similar between both leukemias (supplemental Figure 3E). Interestingly, the phosphopeptide IGEGpTpYGVVYK, common to the first 3 Cdk isoforms, is 5.1-fold more abundant in FLB1 cells despite having similar cell cycle profile as FLA2 cells (supplemental Table 2; and data not shown). The inhibitory phosphorylation status of one or all of these Cdk isoforms thus appears to be inversely correlated with the higher phosphorylation of Ezh2 T487 in FLB1 leukemic cells, suggesting that either another kinase targets this residue or that phosphorylation status varies significantly among these Cdks.
The Ezh2 T487 residue is located close to the second SANT domain. It is highly conserved from Drosophila to humans but is replaced by an asparagine in the Ezh1 paralog (Figure 5C). This T487 residue is located only 3 amino acids upstream of a putative nuclear localization site (NLS), suggesting that phosphorylation of Ezh2 T487 may affect its localization. Strikingly, immunofluorescence staining of freshly isolated FLA2 and FLB1 cells reveals that Ezh2 is predominantly localized in the nucleus of FLA2 LSCs, whereas it is predominantly cytoplasmic in FLB1 cells (Figure 5D). Accordingly, mass spectrometry results revealed an ∼ 2-fold decrease in protein levels of both Ezh2 and its partner Suz12 in the nucleus of FLB1 cells compared with FLA2 (data not shown). These results were confirmed by Western blot analyses (Figure 5E-F). Whether phosphorylation of Ezh2 T487 is directly involved in this differential localization remains to be tested.
As for the third core component of the PRC2 complex, Eed, neither mass spectrometry nor Western blot analyses revealed changes in nuclear or cytoplasmic protein levels (Figure 5E-F; and data not shown). Previous studies revealed the existence of 4 alternative translation initiation products (isoforms 1-4) for this protein.47 Until now, their subcellular localization had never been assessed. Surprisingly, we found specific cytoplasmic localization of Eed isoform 1 in both FLA2 and FLB1 whereas isoform 2 is solely nuclear, suggesting subcellular localization-specific differences in the composition of the core PRC2 complex (Figure 5E). Interestingly, Eed isoform 2 has only been identified in undifferentiated ES cells and in tumors.48 Together, these results indicate that differences in posttranslational modifications and subcellular localization of PRC2 proteins distinguish highly self-renewing leukemic cells (FLA2) from non-reconstituting leukemic cells (FLB1) without affecting global levels of H3K27me3.
Discussion
We report here the first large-scale quantitative proteome and phosphoproteome analyses of mouse AMLs, which include the most comprehensive datasets from a highly enriched primary stem cell population (FLA2: frequency of 1 in 1.4 LSCs). Several other groups have analyzed the proteome and the phosphoproteome of self-renewing ES cells revealing quantitative changes of the protein content on differentiation.49–51 In this study, when comparing LSCs to leukemic blasts with lower self-renewal potential, major differences in protein abundance and phosphorylation were found, indicating distinct regulation of signaling pathways in LSCs (FLA2). Importantly, proteins implicated in chromatin and histone modification, RNA processing, nuclear import/export, and protein kinases constitute the most differentially regulated networks. Among these, activation levels of the p38 MAPK pathway and differential phosphorylation and subcellular localization of Polycomb PRC2 proteins distinguish LSCs from lower self-renewing leukemic cells.
The PcG proteins are critical epigenetic regulators of cell fate and cancer development.44 By inducing a repressive chromatin state and other nonhistone functions, PcG proteins regulate multiple cellular programs essential for stem cell activity, such as cell cycle checkpoints, DNA damage repair, differentiation, apoptosism and senescence. Differential regulation of Polycombs between FLA2 and FLB1 leukemias is therefore likely to have an impact on their self-renewal potential. The effect of phosphorylation on PcG protein activity is still poorly understood. However, studies found that phosphorylation of BMI1 by the MK3 kinase induces its dissociation from chromatin, whereas AKT-mediated phosphorylation of Ezh2 at serine 21 suppresses its H3K27 methyltransferase activity and derepresses silenced genes.52,53 Consistent with a role in cell fate, recent evidence now indicates that p38 MAPK-mediated phosphorylation of Ezh2 threonine 367 (T367) promotes formation of a repressive PRC2 complex that prevents expression of Pax7 required for muscle stem cell activity.40 Together, this suggests that cell signaling pathways can posttranslationally modify PRC2 activity to alter cell fate.
In this study, we found increased phosphorylation of L3mbtl3 S605 in FLA2 LSCs, whereas Ezh2 T487 and Suz12 S548 and S585 were more phosphorylated in the lower self-renewing FLB1 cells. The effect of these posttranslational modifications on LSC activity is still obscure and will have to be further characterized. The Ezh2 T487 is embedded in a putative proline-directed kinase site. Recently, activation of the proline-directed CDK1 kinase was found to phosphorylate Ezh2 T487, thereby promoting derepression of osteogenic genes in mesenchymal stem cell.46 This argues for a role of Ezh2 T487 phosphorylation in stem cell differentiation. However, we observed that increased phosphorylation of Ezh2 T487 in FLB1 cells correlated with an increase in the inhibited form of one or all of the first 3 Cdk isoforms. Thus, it remains possible that Ezh2 T487 may be phosphorylated by another proline-directed kinase in FLB1 leukemic blasts.
Interestingly, an inverse correlation between the activation state of the p38 MAPK kinase pathway and the self-renewal activity of FLA2 LSCs was observed. The proline-directed p38 MAPK kinase plays a major role in the self-renewal potential of stem cells.39–41 High p38 MAPK activation levels restrict cell proliferation, induce differentiation, and limit the life span of HSCs.39 Accordingly, pharmacologic inhibition of p38 MAPK restores the long-term repopulating potential of HSCs in vivo. The p38 MAPK kinase downstream targets mediating these critical effects are much less characterized. However, the recent finding that p38 MAPK-mediated Ezh2 T367 phosphorylation induces muscle stem cells to differentiate40 indicates that posttranslational regulation of PcG proteins is a key step. In this study, Ezh2 T367 phosphorylation levels did not change between FLA2 and FLB1 cells (Table 1). However, it remains possible that p38 MAPK also target Ezh2 at T487. It will be interesting to test whether p38 MAPK can indeed phosphorylate this residue and affect Ezh2's function.
The differential cellular distribution of Ezh2 and Suz12, located in the nucleus of LSCs (FLA2) and mostly in the cytoplasm of lower self-renewing leukemic cells (FLB1) is striking. In addition, we found, for the first time, differential localization of Eed isoforms 1 and 2 in leukemic cells. PRC2 proteins have been found previously in the cytoplasm. Activation of the TCR or PDGF receptors induces the translocation of Ezh2 in the cytoplasm where it regulates actin polymerization.54 Similarly, activation of the TNF receptor 1 (TNFR1) induces translocation of EED in the cytoplasm where it interacts with a TNFR1-FAN-RACK1 complex to activate neutral sphingomyelinase 2, an enzyme that generates stress-induced ceramide.55 Interestingly, blocking of TNFR1 signaling using anti–TNF-α antibodies inhibits p38 MAPK-mediated phosphorylation of Ezh2 at T367 leading to an accumulation of muscle stem cells.40 This suggests a potential link between TNFR1 signaling, p38 activation, Ezh2 phosphorylation, and cytoplasmic localization of PRC2 proteins. Other receptors involved in stem cells and cancer development, such as integrins, have also been shown to induce cytoplasmic translocation of PRC2 proteins when activated,56,57 further supporting a nonchromatin cytoplasmic function for PRC2 proteins. The mechanism of PRC2 protein nuclear/cytoplasmic shuttling is still unknown. However, it is possible that phosphorylation of the Ezh2 T487 residue, next to a putative nuclear localization site, regulates this process. Further studies will have to be conducted to test this.
Global levels of the PRC2-mediated H3K27me3 chromatin mark were similar between FLA2 and FLB1 cells, despite Ezh2 being more present in the cytoplasm of FLB1 cells. Changes in H3K27me3 may affect only a small proportion of the histone population undetectable by immunoblots. Alternatively, the remaining levels of Ezh2 in the nucleus of FLB1 cells may be sufficient to maintain global levels of H3K27me3. The Ezh1 paralog may also compensate for lower Ezh2 levels in the nucleus. In any case, this does not mean that there are no differences between the chromatin landscapes of FLA2 and FLB1 leukemic cells. Local differences in PcG-repressed target genes between FLA2 and FLB1 cells have not been tested but probably differ abundantly, as suggested by the differences in their transcriptome profile.8
In addition to p38 MAPK signaling and PcG proteins, the fact that 3 of the most differentially regulated groups of proteins between FLA2 and FLB1 are involved in RNA processing, nuclear import/export, and chromatin remodeling is particularly interesting. By establishing epigenetic chromatin states and participating in multiple essential processes, such as DNA replication, DNA damage repair, chromosome condensation/segregation, and transmission of transcriptional programs through cell divisions, chromatin modifiers are increasingly recognized as key factors of cell fate.35 They are also widely implicated in cancer development. Regulation of chromatin modifiers function by phosphorylation has not been extensively studied; but given the importance of establishing specific epigenetic states for stem cell activity, post-translational modulation of these proteins offers a link between signaling pathways and chromatin to regulate self-renewal and tumorigenesis.
The potential impact of distinct regulation of RNA processing proteins on the transcriptional program of LSCs is supported by a previous RNA-sequencing study on FLA2 and FLB1 cells showing considerable structural differences in their transcriptome, such as alternative splicing and promoter usage.8 The importance of RNA processing is further substantiated by studies reporting increased alternative splicing in stem cells for many signaling pathway components known to govern self-renewal and differentiation.58 This assuredly induces important changes in the self-renewal properties of stem cells (whether normal of cancerous). Furthermore, modifications in the nuclear pore complex and other proteins involved in nuclear-cytoplasmic transport is expected to affect many cellular processes by regulating the proper localization of RNAs and signaling proteins. The nuclear pore protein NUP98 is well known for its leukemic potential when fused to a variety of genes.38 Moreover, fusion of NUP98 to the homeodomain of HOXA10 can massively expand HSCs ex vivo when expressed in these cells, suggesting an important role for this protein in regulating self-renewal pathways.20 A recent report also indicates that nuclear pore complex composition is important for differentiation of stem cells and embryo development, as Nup133-deficient ES cells fail to differentiate correctly, although the nuclear pore complex assembles correctly.59
In addition, these processes are intimately connected to each other. Recent studies found that splicing factors interact with chromatin remodeling enzymes. Specific chromatin structures and histone modifications also appear to dictate alternative splicing patterns.60 Multiple interactions also occur between RNA processing proteins and the nuclear pore import/export machinery, allowing coordination between transcription and mRNA transport.61 Moreover, nucleoporins have been found to interact directly with chromatin and chromatin modifiers to regulate the transcriptional output of target genes.62,63 Nucleoporins are thought to help define heterochromatin-euchromatin boundaries and a large part of the nuclear architecture and higher-order chromatin structure occurs through nuclear pore complexes serving as docking sites for chromatin. The nuclear lamina, which makes contacts with nuclear pores, chromatin, and chromatin modifiers, is also involved in chromatin looping and spatial organization of the genome.64 Accumulating data suggest that spatial organization of chromatin through the nuclear lamina may be important in establishing transcriptional programs and maintaining specific cell states.65 Interestingly, many nuclear lamina-associated proteins, such as Lmnb1/2, Lbr, Nsun2, Tmpo, and Safb, were differentially regulated between FLA2 and FLB1 (supplemental Tables 1 and 2). Together, this suggests that differential regulation of RNA processing, nuclear import/export, chromatin modifiers, and the nuclear lamina may collaborate together in establishing a permissive or restrictive self-renewal program.
This study constitutes a unique resource for cancer and stem cell biologists to start dissecting candidate proteins and pathways implicated in self-renewal of LSCs. The vast number of differentially regulated proteins identified suggests that multiple pathways are at play, mainly chromatin modifiers, RNA processing, and nuclear import/export. Future work will show if and how they are connected to allow LSCs to self-renew.
This article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Olivier Caron-Lizotte, Kevin Eng, and Eric Bonneil for help with bioinformatic and mass spectrometry analyses and Mélanie Fréchette and Angèle Fournier for help with animal care.
This work was supported by the Cancer Society Research Institute (team grant, G.S.) and the Canadian Institutes of Health Research as well as the Terry Fox Foundation (Research Studentship, M.S.). G.S., S.M., and P.T. hold Canada Research Chairs in Molecular Genetics of Stem Cells, Cellular Signaling and Proteomics, and Bioanalytical Spectrometry, respectively. Institute for Research in Immunology and Cancer is supported in part by the Canadian Center of Excellence in Commercialization and Research, the Canada Foundation for Innovation, and Fonds de la Recherche en Sante du Quebec.
Authorship
Contribution: M.T., M.S., S.M., G.S., and P.T. carried out the experimental design; M.T., M.S., O.H., P.D., J.C., C.P., and N.M. performed the experimental work; M.T. and M.S. analyzed data; and M.S., M.T., G.S., and P.T. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Guy Sauvageau, Institute for Research in Immunology and Cancer, Université de Montréal, PO Box 6128, Station Centre-Ville, Montreal, QC, Canada H3C 3J7; e-mail: guy.sauvageau@umontreal.ca; and Pierre Thibault, Institute for Research in Immunology and Cancer, Université de Montréal, PO Box 6128, Station Centre-Ville, Montreal, QC, Canada H3C 3J7; e-mail: pierre.thibault@umontreal.ca.
References
Author notes
M.T. and M.S. contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal