Key Points
Dnmt3b acts as a haploinsufficient tumor suppressor in Myc-induced lymphomas.
Abstract
The drivers of abnormal DNA methylation in human cancers include widespread aberrant splicing of the DNMT3B gene, producing abnormal transcripts that encode truncated proteins that may act as dominant negative isoforms. To test whether reduced Dnmt3b dosage can alter tumorigenesis, we bred Dnmt3b+/− mice to Eµ-Myc mice, a mouse model susceptible to B-cell lymphomas. Eµ-Myc/Dnmt3b+/− mice showed a dramatic acceleration of lymphomagenesis, greater even than that observed in Eµ-Myc mice that express a truncated DNMT3B isoform found in human tumors, DNMT3B7. This finding indicates that Dnmt3b can act as a haploinsufficient tumor suppressor gene. Although reduction in both Dnmt3b dosage and expression of DNMT3B7 within the Eµ-Myc system had similar effects on tumorigenesis and DNA hypermethylation, different molecular mechanisms appear to underlie these changes. This study offers insight into how de novo DNA methyltransferases function as tumor suppressors and the sensitivity of Myc-induced lymphomas to DNA methylation.
Introduction
Cancer cells are characterized by abnormal DNA methylation, causing aberrant activation of some genes and silencing of others.1 Three DNA methyltransferases (DNMTs) catalyze DNA methylation in eukaryotic cells DNMT1, DNMT3A, and DNMT3B.2 Our laboratory and others discovered aberrant splicing of the DNMT3B gene in human cancer cells that produces transcripts encoding truncated proteins that may act as dominant negative isoforms.3,4 Transgenic mice expressing DNMT3B7, an aberrant transcript expressed most commonly in human cancers, have little cancer predisposition on their own.5 However, when crossed to Eµ-Myc mice, which are susceptible to B-cell lymphomas,6 Eµ-Myc/DNMT3B7 mice showed accelerated mediastinal tumorigenesis, with overall increases in global DNA methylation and alterations in site-specific DNA methylation.5
DNMT3B7 transgenic mice demonstrated developmental defects including subaortic ventriculoseptal defects,5 reminiscent of those in Dnmt3b knockout mice.7,8 Given the similar phenotype between DNMT3B7 transgenic mice and Dnmt3b knockout mice, we hypothesized that DNMT3B7 acts as a dominant negative isoform of full-length Dnmt3b. Recent work has demonstrated that inactive DNMT3B isoforms can alter the enzymatic activity of full-length DNMT3B in vitro.9 Because we observed acceleration of tumorigenesis in Eµ-Myc mice with DNMT3B7 introduction, we tested whether Dnmt3b gene dosage (Dnmt3b+/−) could influence Myc-induced lymphomagenesis.
Study design
Mice and monitoring of tumors
Chromatin immunoprecipitation
Chromatin immunoprecipitation assays were performed using a Myc (Millipore, Billerica, MA) or Dnmt3b antibody (Santa Cruz Biotechnology, Santa Cruz, CA). Real-time polymerase chain reaction (PCR) primers are listed in supplemental Table 1.
Detailed materials and methods are included in “Supplemental Materials.”
Results and Discussion
Given the acceleration of Myc-induced lymphomagenesis in the presence of the dominant-negative DNMT3B7 protein,5 we tested directly whether reducing Dnmt3b levels would influence tumorigenesis in Eµ-Myc mice.6 We found dramatic acceleration in mediastinal lymphomagenesis in Eµ-Myc/Dnmt3b+/− mice (Figure 1A), whereas the incidence of peripheral lymphomas was not significantly different (supplemental Figure 1A). Overall lymphoma incidence was dramatically accelerated in both Eµ-Myc/DNMT3B7 and Eµ-Myc/Dnmt3b+/− mice (supplemental Figure 1B). Similar to the Eµ-Myc/DNMT3B7 mice, mediastinal tumors isolated from Eµ-Myc/Dnmt3b+/− mice were composed of pre-B or mature B-lymphocytes (supplemental Figure 2), and the mice had high leukocyte counts (supplemental Table S2). Cell lines generated from mediastinal tumors recapitulated the enhanced growth (Figure 1B). Cell-cycle profile analysis demonstrated that the growth difference arose from Eµ-Myc cells accumulating in the S-phase, and Eµ-Myc/DNMT3B7 and Eµ-Myc/Dnmt3b+/− cells overcoming this block in progression from S to G2 phase (supplemental Figure 3). There were no significant differences in apoptosis in the 3 cell lines (data not shown). The Dnmt3b cDNA expressed from the remaining allele was wild-type in sequence (data not shown), establishing that Dnmt3b has a haploinsufficient tumor suppressor function in the Eµ-Myc mouse model.
Incidence of mediastinal lymphomas and changes in DNA methylation in Eμ-Myc, Eμ-Myc/DNMT3B7, and Eμ-Myc/Dnmt3b+/− mice. (A) Eµ-Myc/Dnmt3b+/− (dotted line) vs Eµ-Myc/DNMT3B7 (dashed line) and Eµ-Myc mice (solid line). Cumulative incidence curves were compared using the K-sample test (Eµ-Myc vs Eµ-Myc/Dnmt3b+/−: P < .0001; Eµ-Myc vs Eµ-Myc/DNMT3B7: P = .009).5 (B) Growth curves of cell lines established from mediastinal lymphomas of Eµ-Myc (solid line), Eµ-Myc/DNMT3B7 (dashed line), and Eµ-Myc/Dnmt3b+/− mice (dotted line). Curves were compared using 2-way analysis of variance (ANOVA) (P = .012). (C) Total 5-methylcytosine levels were quantified by liquid chromatography-electrospray ionization/tandem mass spectometry,10 n ≥ 4 for each genotype. DNA methylation levels were normalized to wild-type (WT) peripheral blood B cells (gray bar). The 2-tailed Student t test was used to compare global DNA methylation. (D-G) Relative quantification of Dnmt levels, normalized to β-Actin levels. Dnmt1 (D), Dnmt3a (E), Dnmt3b (F), and Dnmt3l (G) levels in WT lymph node (LN) (gray bar) and Eµ-Myc (white bar), Eµ-Myc/DNMT3B7 (hatched bar), and Eµ-Myc/Dnmt3b+/− (striped bar) cell lines. P values were calculated by ANOVA, using WTLN as the control.
Incidence of mediastinal lymphomas and changes in DNA methylation in Eμ-Myc, Eμ-Myc/DNMT3B7, and Eμ-Myc/Dnmt3b+/− mice. (A) Eµ-Myc/Dnmt3b+/− (dotted line) vs Eµ-Myc/DNMT3B7 (dashed line) and Eµ-Myc mice (solid line). Cumulative incidence curves were compared using the K-sample test (Eµ-Myc vs Eµ-Myc/Dnmt3b+/−: P < .0001; Eµ-Myc vs Eµ-Myc/DNMT3B7: P = .009).5 (B) Growth curves of cell lines established from mediastinal lymphomas of Eµ-Myc (solid line), Eµ-Myc/DNMT3B7 (dashed line), and Eµ-Myc/Dnmt3b+/− mice (dotted line). Curves were compared using 2-way analysis of variance (ANOVA) (P = .012). (C) Total 5-methylcytosine levels were quantified by liquid chromatography-electrospray ionization/tandem mass spectometry,10 n ≥ 4 for each genotype. DNA methylation levels were normalized to wild-type (WT) peripheral blood B cells (gray bar). The 2-tailed Student t test was used to compare global DNA methylation. (D-G) Relative quantification of Dnmt levels, normalized to β-Actin levels. Dnmt1 (D), Dnmt3a (E), Dnmt3b (F), and Dnmt3l (G) levels in WT lymph node (LN) (gray bar) and Eµ-Myc (white bar), Eµ-Myc/DNMT3B7 (hatched bar), and Eµ-Myc/Dnmt3b+/− (striped bar) cell lines. P values were calculated by ANOVA, using WTLN as the control.
Our observation is consistent with recent work demonstrating that complete ablation of Dnmt3b in T-lymphocytes promotes proliferation and accelerates lymphomagenesis in the context of MYC overexpression.11 Conditional deletion of Dnmt3a in a mouse model of lung tumorigenesis also promotes tumor growth and progression.12 Heterozygous DNMT3A mutations, often in exons encoding the catalytic domain, have been found in myeloid neoplasms.13-15 Taken together, these data suggest that the de novo DNA methyltransferases DNMT3A and DNMT3B function as haploinsufficient tumor suppressors.
We found a progressive increase in the global 5-methylcytosine levels in Eµ-Myc, Eµ-Myc/DNMT3B7, and Eµ-Myc/Dnmt3b+/− tumors (Figure 1C).5 Because Dnmt3b+/− transgenic mice themselves do not demonstrate any overt DNA methylation defect,7 this was an unexpected observation. A CpG island hypermethylation signature was defined in a mouse model of MYC-induced lymphomas,16 supporting the hypothesis that MYC drives DNA methylation. DNA methylation levels in B1 repetitive elements, major and minor satellites, and retroviruses were similar across all Eµ-Myc genotypes (data not shown). The expression of Dnmt1, Dnmt3a, Dnmt3b, and Dnmt3l did not differ significantly between the Eµ-Myc, Eµ-Myc/DNMT3B7, and Eµ-Myc/Dnmt3b+/− cell lines; however, almost all Dnmts increased in expression with Myc overexpression (Figure 1D-G). In a proliferative germinal center B-cell model, repatterning of DNA methylation correlated with gene expression changes and upregulation of DNMT1 expression.17 Chromosomal abnormalities in Eµ-Myc/Dnmt3b+/− tumors were not significantly different compared with Eµ-Myc tumors (supplemental Table 3).
Because virtually all human tumors and cell lines express aberrant DNMT3B transcripts, we focused on testing whether the molecular mechanism of tumorigenesis in Eµ-Myc/DNMT3B7 and Eµ-Myc/Dnmt3b+/− tumors was similar. We hypothesized that Myc, Dnmt3b, and DNMT3B7 could form a complex when DNMT3B7 was expressed, because Myc binds to Dnmt3a and Dnmt3b,18 and DNMT3B7 binds to full-length Dnmt3b.5 We hypothesized that this complex could bind at Myc-binding E-box(es) upstream of the transcriptional start site (TSS), leading to hypermethylation and gene repression. Because Myc binds preferentially at hypomethylated E-boxes,19 we predicted hypomethylation around the E-box of the Myc target gene tested.
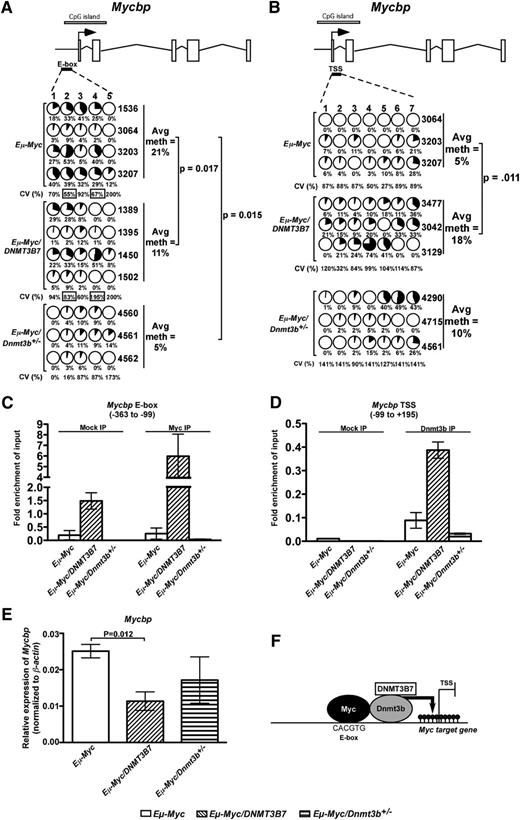
To test this hypothesis, we examined DNA methylation in promoters of genes that are known Myc targets and are repressed in Eµ-Myc/DNMT3B7 tumors (see “Supplemental Methods”). Mycbp, which encodes a protein that binds c-Myc and enhances its ability to activate transcription, was hypomethylated around the E-box in both Eµ-Myc/DNMT3B7 and Eµ-Myc/Dnmt3b+/− tumors relative to Eµ-Myc tumors (Figure 2A). We observed more heterogeneity in DNA methylation levels of particular CpG dinucleotides in Eµ-Myc/DNMT3B7 tumors relative to the Eµ-Myc tumors as we had observed previously,5 in contrast to Eµ-Myc/Dnmt3b+/− tumors that failed to demonstrate this heterogeneity (Figure 2A). The Mycbp TSS was hypermethylated in Eµ-Myc/DNMT3B7 tumors relative to Eµ-Myc tumors, a feature not shared by Eµ-Myc/Dnmt3b+/− tumors (Figure 2B).
DNA methylation changes and binding of Myc and Dnmt3b at Mycbp promoter elements. (A) DNA methylation changes measured by bisulfite sequencing around the E-box upstream of Mycbp TSS. Schematic diagram for Mycbp is shown with exons represented by vertical rectangles, and the location of the CpG island shown with a horizontal shaded rectangle. The black arrow indicates the TSS. The smaller, black horizontal rectangle indicates the location of CpGs analyzed for changes in DNA methylation. Each row represents DNA methylation in a single lymphoma, indicated to the right. Numbers across the top indicate specific CpG dinucleotides in a region of the CpG island. Changes in DNA methylation are indicated by shaded circles, with the shading indicating average amount of DNA methylation at each CpG; numbers below represent percent methylated cytosine. Average percent methylation is indicated to the right, and P values calculated using the 2-tailed Student t test. Coefficient of variance (CV) was used to calculate the heterogeneity of DNA methylation at each CpG within tumors of a single genotype. The average CV is given as a percentage underneath each CpG position. CV values with a black box around them denote P < .05 for Eµ-Myc/DNMT3B7 or Eµ-Myc/Dnmt3b+/− tumors relative to Eµ-Myc tumors. (B) DNA methylation changes as measured by bisulfite sequencing in the Mycbp TSS. Detailed description is as outlined in (A). (C) Fold enrichment of Myc in chromatin from at least 3 independent Eµ-Myc, Eµ-Myc/DNMT3B7, and Eµ-Myc/Dnmt3b+/− cell lines. The plot shows real-time PCR data for the region around the Mycbp E-box (−363 to −99 relative to Mycbp TSS) from Myc-immunoprecipitated or control-immunoprecipitated chromatin normalized to input. (D) Fold enrichment of Dnmt3b in chromatin from at least 3 independent Eµ-Myc, Eµ-Myc/DNMT3B7, and Eµ-Myc/Dnmt3b+/− cell lines. The plot shows real-time PCR data for the region around the Mycbp TSS (−99 to +195 relative to Mycbp TSS) from Dnmt3b-immunoprecipitated or control-immunoprecipitated chromatin normalized to input. (E) Expression of Mycbp in Eµ-Myc, Eµ-Myc/DNMT3B7, and Eµ-Myc/Dnmt3b+/− tumors. Relative expression of Mycbp was measured in all Eµ-Myc tumor types and expressed normalized to β-Actin. (F) Proposed model for the increased acceleration of lymphomagenesis induced by hypermethylation in Eµ-Myc/DNMT3B7 mice. The presence of an unmethylated CpG in the Myc-binding E-box promotes Myc binding, which then assembles a complex that includes Dnmt3b and DNMT3B7. The complex locks into position at the Mycbp TSS and leads to hypermethylation around the TSS.
DNA methylation changes and binding of Myc and Dnmt3b at Mycbp promoter elements. (A) DNA methylation changes measured by bisulfite sequencing around the E-box upstream of Mycbp TSS. Schematic diagram for Mycbp is shown with exons represented by vertical rectangles, and the location of the CpG island shown with a horizontal shaded rectangle. The black arrow indicates the TSS. The smaller, black horizontal rectangle indicates the location of CpGs analyzed for changes in DNA methylation. Each row represents DNA methylation in a single lymphoma, indicated to the right. Numbers across the top indicate specific CpG dinucleotides in a region of the CpG island. Changes in DNA methylation are indicated by shaded circles, with the shading indicating average amount of DNA methylation at each CpG; numbers below represent percent methylated cytosine. Average percent methylation is indicated to the right, and P values calculated using the 2-tailed Student t test. Coefficient of variance (CV) was used to calculate the heterogeneity of DNA methylation at each CpG within tumors of a single genotype. The average CV is given as a percentage underneath each CpG position. CV values with a black box around them denote P < .05 for Eµ-Myc/DNMT3B7 or Eµ-Myc/Dnmt3b+/− tumors relative to Eµ-Myc tumors. (B) DNA methylation changes as measured by bisulfite sequencing in the Mycbp TSS. Detailed description is as outlined in (A). (C) Fold enrichment of Myc in chromatin from at least 3 independent Eµ-Myc, Eµ-Myc/DNMT3B7, and Eµ-Myc/Dnmt3b+/− cell lines. The plot shows real-time PCR data for the region around the Mycbp E-box (−363 to −99 relative to Mycbp TSS) from Myc-immunoprecipitated or control-immunoprecipitated chromatin normalized to input. (D) Fold enrichment of Dnmt3b in chromatin from at least 3 independent Eµ-Myc, Eµ-Myc/DNMT3B7, and Eµ-Myc/Dnmt3b+/− cell lines. The plot shows real-time PCR data for the region around the Mycbp TSS (−99 to +195 relative to Mycbp TSS) from Dnmt3b-immunoprecipitated or control-immunoprecipitated chromatin normalized to input. (E) Expression of Mycbp in Eµ-Myc, Eµ-Myc/DNMT3B7, and Eµ-Myc/Dnmt3b+/− tumors. Relative expression of Mycbp was measured in all Eµ-Myc tumor types and expressed normalized to β-Actin. (F) Proposed model for the increased acceleration of lymphomagenesis induced by hypermethylation in Eµ-Myc/DNMT3B7 mice. The presence of an unmethylated CpG in the Myc-binding E-box promotes Myc binding, which then assembles a complex that includes Dnmt3b and DNMT3B7. The complex locks into position at the Mycbp TSS and leads to hypermethylation around the TSS.
A chromatin immunoprecipitation assay around the E-box demonstrated enrichment of Myc binding in the Eµ-Myc/DNMT3B7 cell lines and absence of Myc binding in both Eµ-Myc and Eµ-Myc/Dnmt3b+/− cell lines (Figure 2C). We also found Dnmt3b enrichment at the Mycbp TSS in Eµ-Myc/DNMT3B7 cell lines, a phenomenon not observed in Eµ-Myc and Eµ-Myc/Dnmt3b+/− cell lines (Figure 2D). Consistent with DNA methylation patterns around the TSS, there was Mycbp repression in Eµ-Myc/DNMT3B7 tumors, but not in Eµ-Myc/Dnmt3b+/− tumors (Figure 2E). We also observed enrichment of Myc binding at the Mycbp TSS and increased enrichment of Dnmt3b binding at the Mycbp E-box (supplemental Figure 4). Similar results were seen for Gadd45a and Maf, which are other Myc targets (supplemental Figures 5A and 6). Gadd45a repression in Eµ-Myc/DNMT3B7 and Eµ-Myc/Dnmt3b+/− tumors (supplemental Figure 5B) provides a mechanism for the hypermethylation observed in these tumors. Taken together, our data indicate that Eµ-Myc/DNMT3B7 tumors have increased Myc binding at the Mycbp E-box, which enhances recruitment of Dnmt3b to form a repressive complex at its promoter, leading to Mycbp repression (Figure 2F), a mechanism distinct from that present in Eµ-Myc/Dnmt3b+/− tumors.
Overall, our observations suggest that Myc-induced lymphomas are exquisitely sensitive to DNA methylation, which may suggest that targeting epigenetic pathways may be particularly effective for MYC-induced hematopoietic malignancies. Epigenetic silencing of RASSF1A, a tumor suppressor gene, has been shown to occur via HOXB3-mediated induction of DNMT3B, which binds MYC and polycomb repressor at the RASSF1A promoter in multiple human cancer cell lines.20 The ability of JQ1, an inhibitor of BRD4-mediated reading of the histone code, to suppress myeloid leukemia cell growth further supports the importance of epigenetic therapy.21 Finally, our findings, combined with the identification of heterozygous DNMT3A mutations in myeloid malignancies, suggest that both de novo DNA methyltransferases can act as haploinsufficient tumor suppressors.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank En Li and Taiping Chen for the gift of the Dnmt3b knockout mice and Scott Lowe for the Eµ-Myc transgenic mice, and Bryan Zahakaylo (Mass Spectometry, Metabolomics, and Proteomics Facility, University of Illinois at Chicago) and members of the L.A.G. and M.M.L.B. laboratories for critical discussion of data, especially Kelly Ostler, Isabelle Lucas, Mrinal Shah, and Jozef Madzo.
This work was supported by the grant National Institutes of Health grant CA129831 (to L.A.G.). A.V. is supported by a National Institutes of Health grant F32.
National Institutes of Health
Authorship
Contribution: A.V. designed and performed experiments, analyzed data, and wrote the manuscript; J.B.L., M.H.Z., M.K., M.S., E.M.D., and P.A.L. performed experiments and analyzed data; J.A., M.M.L.B., and A.R.K. helped design experiments; L.A.G. conceived the study, analyzed data, and edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for A.R.K. is Eppley Institute for Cancer and Allied Diseases, University of Nebraska Medical Center, Omaha, NE.
Correspondence: Lucy A. Godley, The University of Chicago, Department of Medicine Section of Hematology/Oncology, 5841 S. Maryland Ave, MC2115, Chicago, IL 60637-1470; e-mail: lgodley@medicine.bsd.uchicago.edu.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal