Key Points
In adult patients with core binding factor AML, intensified induction is not associated with a better outcome in the context of intensive postremission therapy.
Minimal residual disease, rather than KIT or FLT3 gene mutations, should be used to identify core binding factor AML patients at higher risk of relapse.
Abstract
Not all patients with core binding factor acute myeloid leukemia (CBF-AML) display a good outcome. Modern risk factors include KIT and/or FLT3 gene mutations and minimal residual disease (MRD) levels, but their respective values have never been prospectively assessed. A total of 198 CBF-AML patients were randomized between a reinforced and a standard induction course, followed by 3 high-dose cytarabine consolidation courses. MRD levels were monitored prospectively. Gene mutations were screened at diagnosis. Despite a more rapid MRD decrease after reinforced induction, induction arm did not influence relapse-free survival (RFS) (64% in both arms; P = .91). Higher WBC, KIT, and/or FLT3-ITD/TKD gene mutations, and a less than 3-log MRD reduction after first consolidation, were associated with a higher specific hazard of relapse, but MRD remained the sole prognostic factor in multivariate analysis. At 36 months, cumulative incidence of relapse and RFS were 22% vs 54% (P < .001) and 73% vs 44% (P < .001) in patients who achieved 3-log MRD reduction vs the others. These results suggest that MRD, rather than gene mutations, should be used for future treatment stratifications in CBF-AML patients. This trial was registered at EudraCT as #2006-005163-26 and at www.clinicaltrials.gov as #NCT 00428558.
Introduction
Acute myeloid leukemia (AML) carrying t(8;21) chromosomal translocation or inv(16)/t(16;16) chromosomal rearrangement corresponding to the RUNX1-RUNX1T1 or CBFB-MYH11 fusion genes, respectively, belongs to the favorable-risk AML subset.1,2 Both subtypes are referred as core binding factor (CBF) AML, because RUNX1 (formerly AML1) and CBFB genes respectively code for the α (CBFα) and β (CBFβ) subunits of the CBF complex, a transcription factor involved in normal hematopoiesis. Both genetic events are responsible for loss of function of the CBF complex and differentiation blockade, participating in the development of AML.3 Both subtypes are more frequently observed in younger AML patients and display a high sensitivity to standard chemotherapeutic agents used in AML, leading to high complete remission (CR) rates and a usually good prognosis.4 Nonetheless, about 30% of patients with CBF-AML relapse and not all may be cured. In this context, the place of allogeneic stem cell transplantation (SCT) in first CR remains an open debate.
Numerous prognostic factors have been identified in an attempt to characterize early CBF-AML patients at higher risk of relapse.1,2 These factors include older age, high white blood cell count (WBC), loss of Y chromosome (in the CBFα subtype), trisomy 22 (in the CBFβ subtype), and deletion of the long arm of chromosome 9.5-8 On the other hand, the use of real-time quantitative polymerase chain reaction to assess residual fusion transcript levels after CR induction and/or consolidation has been shown as a potentially strong prognostic factor in these patients,9-14 only prospectively evaluated in a very recent British report in 2012.15 Additionally, frequent mutations of the KIT, FLT3, and N/K-RAS genes have been described in CBF-AML, and mutations in the two receptor tyrosine kinase (RTK) genes KIT and FLT3 have been retrospectively associated with a worse outcome.16-25 To date, comparative evaluation of gene mutations and minimal residual disease (MRD) levels has not been prospectively studied.
To address these issues, we enrolled all younger adult patients with diagnosed CBF-AML in France in a prospective dedicated trial between 2007 and 2010 (ClinicalTrials.gov ID #NCT 00428558). To test chemotherapy intensification, these patients were randomized to receive a reinforced timed-sequential induction, as used by the Acute Leukemia French Association (ALFA), or a more standard 7+3 induction, as used by the Groupe Ouest-Est des Leucémies et Autres Maladies du Sang (GOELAMS). Patients were monitored for postinduction and postconsolidation MRD levels, and KIT, FLT3, and N/K-RAS gene mutations were examined at diagnosis. In addition, MRD levels were used as a treatment-stratifying criterion prompting allogeneic SCT in first CR in poor MRD responders.
Patients and methods
Patients
Between July 2007 and November 2010, 200 patients from 46 French centers, aged 18 to 60 years and with newly diagnosed CBF-AML, were randomized in the CBF-2006 trial (EudraCT #2006-005163-26; ClinicalTrials.gov ID #NCT 00428558). The study, approved by the ethics committee of Nimes University Hospital and by the Institutional Review Board of the French Regulatory Agency, was conducted in accordance with the Declaration of Helsinki. The diagnosis of CBF-AML, defined by the presence of the t(8;21) translocation or the inv(16)/t(16;16) rearrangement by karyotype and/or fluorescence in situ hybridization analysis and/or evidence of RUNX1-RUNX1T1 or CBFB-MYH11 fusion transcripts, was required within a maximal 5-day period prior to trial entry. Patients with AML-M4eo in the FAB classification and WBC > 30 × 109/L could be randomized, provided later confirmation of the CBF anomaly was obtained. Eligibility also included signed informed consent, an ECOG performance status of 2 or less, no uncontrolled severe infection or other malignancy, AST and ALT levels ≤ 2.5 × upper normal limit (UNL), bilirubin ≤ 1.5 × UNL, and serum creatinine ≤ 1.5 × UNL. Patients with CBF-AML secondary to prior chemotherapy and/or radiotherapy were eligible, as were those with central nervous system (CNS) involvement at diagnosis. The randomization sequence was stratified according to age, CBF subtype, and de novo vs secondary AML. Among the 200 randomized patients, CBF-AML was not confirmed in only 2, leading to 198 evaluable patients.
Treatments
Treatment arm A comprised a first sequence with daunorubicin (DNR) at 60 mg/m2/d by 30-minute IV infusion on days 1 to 3 and cytarabine at 500 mg/m2/d by continuous IV infusion from days 1 to 3, systematically followed by a second sequence starting at day 8 with DNR at 35 mg/m2/d by 30-minute IV infusion on days 8 and 9 and cytarabine at 1000 mg/m2/12 h by 2-hour IV infusion on days 8 to 10. Treatment arm B comprised DNR at 60 mg/m2/d by 30-minute IV infusion on days 1 to 3 and cytarabine at 200 mg/m2/d by continuous IV infusion from days 1 to 7. In arm B patients, a peripheral blood and bone marrow (BM) evaluation was performed at day 15. In patients with ≥5% marrow blasts and/or presence of Auer rods at day 15, the second sequence of chemotherapy mentioned above was administered starting on day 16. In patients not reaching CR, a salvage course comprising cytarabine at 3000 mg/m2/12 h by 2-hour IV infusion on days 1 to 4 and amsacrine at 100 mg/m2/d by 30-minute IV infusion on days 5 to 7 was indicated, followed by lenograstim granulocyte colony-stimulating factor starting at day 8 until myeloid recovery. Patients reaching CR then received 3 monthly consolidation cycles with cytarabine at 3000 mg/m2/12 h by 2-hour IV infusion on days 1, 3, and 5, followed by lenograstim granulocyte colony-stimulating factor starting at day 8 until neutrophil recovery. Patients with CBFβ-AML and WBC >100 × 109/L received CNS prophylaxis with 4 triple intrathecal infusions (methotrexate 15 mg, cytarabine 40 mg, methylprednisolone 40 mg). Those with CNS disease at diagnosis received intrathecal infusions twice a week until disappearance of blast cells in the cerebrospinal fluid. According to the trial design, patients not reaching at least a 3-log reduction in MRD ratio before initiation of the second consolidation cycle were candidates for allogeneic SCT in first CR if they had a matched sibling or 10/10 HLA allele fully matched unrelated donor, as were those who needed the salvage course to reach CR. Standard myeloablative or reduced-intensity conditioning regimens were allowed, depending on the patient age and health status. Patients with less than 3-log MRD reduction before second consolidation but no available donor could enter a companion single-agent dasatinib phase 2 trial, not described here, after having received the 3 planned consolidation cycles.
Gene mutations
A systematic screening for KIT exon 8 and 17 mutations, FLT3 internal tandem duplication (ITD), FLT3 tyrosine kinase domain (TKD) mutation, and N/K-RAS exon 2 and 3 mutations was performed in 2 central laboratories (Lille, Reims) as described elsewhere.20 FLT3-ITD was screened by Genescan analysis and FLT3-TKD by polymerase chain reaction and restriction fragments length polymorphism, as described previously.20 Detected mutations were controlled in 2 separate experiments. A total of 194 of 198 patients, including 93 t(8;21) and 101 inv(16)/t(16;16) cases, could be tested for these gene mutations. In “Results,” “FLT3 mutations” collectively designates ITD and TKD mutations, except when specified. Similarly, “RTK mutations” collectively designates FLT3 and KIT gene mutations.
MRD evaluation
MRD levels were serially monitored for RUNX1-RUNX1T1 or CBFB-MYH11 transcripts by real-time quantitative polymerase chain reaction in 4 central laboratories (Angers, Lille, Paris Saint-Louis, Toulouse), as described previously.26 Calibration curves were performed using Ipsogen plasmids (Ipsogen SA, Marseille, France) and ABL1 was amplified concomitantly as internal reference. Results were expressed as a [fusion gene/ABL1] × 100 transcript ratio. MRD level evaluation was scheduled in BM samples before initiation of the first, second, and third consolidation courses (MRD1, MRD2, and MRD3 time points, respectively), and at the end of treatment. BM MRD1, MRD2, and MRD3 levels could be respectively analyzed in 176, 176, and 160 patients in first CR after having received the planned chemotherapy and no SCT. In addition, 133 patients were tested at the end of treatment after the third consolidation course. As mentioned above, the MRD2 time point was retained for SCT decision.
Statistical methods
For the randomized comparison, the primary end point was RFS. Assuming a 2-year RFS of 50% in patients treated in arm B, it required 96 patients per arm and a total of 78 events to detect an increase in 2-year RFS estimate from 50% to 70% in arm A patients (hazard ratio [HR], 0.71), with a type I error rate of 5% and a statistical power of 80%, and assuming an exponential RFS. To take into account potential induction deaths, a total of 100 patients were planned in each arm. Secondary end points were (1) CR rate, MRD response, cumulative incidence of relapse (CIR), and overall survival (OS) in both randomization arms; and (2) prognostic evaluation of clinical features, gene mutation status, and MRD response. Outcome data were updated as of June 2012, for a median follow-up of 32 months. Failure time data were analyzed and compared after censoring at transplant time patients who received allogeneic SCT in first CR. RFS and OS were estimated by the Kaplan-Meier method,27 then compared by the log-rank test.28 CIR was estimated taking into account death in first CR for competing risk and then compared by cause-specific hazard Cox models. Specific hazards of relapse (SHRs) and HRs are given with 95% confidence interval (CI). Comparisons were stratified on treatment arm and CBF-AML subtype. Multivariate analysis was performed using the Cox regression model.29 Proportional hazards assumptions were graphically checked. CR rates and MRD response across randomized groups were compared using Fisher’s exact test. All statistical tests were performed with the Stata/IC 12.1 software (StataCorp, College Station, TX).
Results
Patient characteristics at diagnosis
Patient characteristics are shown in Tables 1 and 2, according to randomization arm (Table 1) and CBF subtype (Table 2). Patient characteristics were correctly balanced between the two treatment arms. Patients with inv(16)/t(16;16) AML had higher WBC than those with t(8;21) AML. Exon 17 KIT gene mutations were more frequent in the t(8;21) subset, whereas FLT3-TKD and N/K-RAS mutations were more frequent in the inv(16)/t(16;16) subset. Overall, the incidence of RTK mutations reached 33%. Higher WBC and BM blast percentage were associated with KIT (P = .037 and <.001, respectively) and FLT3 (P = .020 and .028, respectively) mutations. Patients with inv(16)/t(16;16) AML and N/K-RAS mutation had lower BM blast percentage (P < .001). At diagnosis, the median fusion transcript ratio was higher in RUNX1-RUNX1T1 patients than in CBFB-MYH11 patients (P < .001). This ratio was more widely distributed in the RUNX1-RUNX1T1 subset. Patients with N/K-RAS mutation had a significantly lower median ratio at diagnosis (P = .0013).
Patient characteristics according to randomization arm
| . | Arm A . | Arm B . | All . |
|---|---|---|---|
| Patients, n | 100 | 98 | 198 |
| CBF subset (t[8;21]/inv[16] or t[16;16]), n | 48/52 | 48/50 | 100/98 |
| Median age, y (range) | 42 (18-60) | 42 (18-59) | 42 (18-60) |
| Gender (male/female), n | 54/46 | 52/46 | 106/92 |
| Median WBC, 109/L (range) | 15.6 (0.7-232) | 14.7 (0.51-254) | 14.9 (0.7-254) |
| Median BM blasts, % (range) | 54 (9-94) | 53 (17-98) | 54 (9-98) |
| Median fusion transcript ratio, % (range) | 141 (23-947) | 171 (32-1090) | 150 (23-1090) |
| Secondary AML | 7 | 6 | 13 |
| CNS involvement | 3 | 1 | 4 |
| Additional cytogenetic abnormalities, n* | |||
| Loss of Y | 15 | 18 | 33 |
| Trisomy 8 | 7 | 10 | 17 |
| Trisomy 22 | 10 | 8 | 18 |
| del(9q) | 7 | 8 | 15 |
| del(7q)/-7 | 6 | 3 | 9 |
| Gene mutations, n† | |||
| KIT exon 8 | 10 | 8 | 18 |
| KIT exon 17 | 11 | 12 | 23 |
| KIT, all | 20 | 20 | 40 |
| FLT3-TKD | 7 | 13 | 20 |
| FLT3-ITD | 4 | 7 | 11 |
| FLT3, all | 11 | 19 | 30 |
| RTK, all | 29 | 37 | 66 |
| N-RAS | 21 | 20 | 41 |
| K-RAS | 6 | 7 | 13 |
| RAS, all | 27 | 27 | 54 |
| . | Arm A . | Arm B . | All . |
|---|---|---|---|
| Patients, n | 100 | 98 | 198 |
| CBF subset (t[8;21]/inv[16] or t[16;16]), n | 48/52 | 48/50 | 100/98 |
| Median age, y (range) | 42 (18-60) | 42 (18-59) | 42 (18-60) |
| Gender (male/female), n | 54/46 | 52/46 | 106/92 |
| Median WBC, 109/L (range) | 15.6 (0.7-232) | 14.7 (0.51-254) | 14.9 (0.7-254) |
| Median BM blasts, % (range) | 54 (9-94) | 53 (17-98) | 54 (9-98) |
| Median fusion transcript ratio, % (range) | 141 (23-947) | 171 (32-1090) | 150 (23-1090) |
| Secondary AML | 7 | 6 | 13 |
| CNS involvement | 3 | 1 | 4 |
| Additional cytogenetic abnormalities, n* | |||
| Loss of Y | 15 | 18 | 33 |
| Trisomy 8 | 7 | 10 | 17 |
| Trisomy 22 | 10 | 8 | 18 |
| del(9q) | 7 | 8 | 15 |
| del(7q)/-7 | 6 | 3 | 9 |
| Gene mutations, n† | |||
| KIT exon 8 | 10 | 8 | 18 |
| KIT exon 17 | 11 | 12 | 23 |
| KIT, all | 20 | 20 | 40 |
| FLT3-TKD | 7 | 13 | 20 |
| FLT3-ITD | 4 | 7 | 11 |
| FLT3, all | 11 | 19 | 30 |
| RTK, all | 29 | 37 | 66 |
| N-RAS | 21 | 20 | 41 |
| K-RAS | 6 | 7 | 13 |
| RAS, all | 27 | 27 | 54 |
RTK, receptor tyrosine kinase (KIT and/or FLT3).
A total of 196 the 198 patients, including 96 t(8;21) and 101 inv(16)/t(16;16) AML patients, had an available karyotype.
A total of 194 of the 198 patients, including 93 t(8;21) and 101 inv(16)/t(16;16) AML patients, were tested for KIT, FLT3 and RAS mutations.
Patient characteristics according to CBF subtype
| . | t(8;21) AML . | inv(16)/t(16;16) . | P value . |
|---|---|---|---|
| Patients, n | 96 | 102 | |
| Treatment arm (A/B), n | 48/48 | 52/50 | .99 |
| Median age, y (range) | 42 (18-60) | 42 (18-60) | .37 |
| Gender (male/female), n | 54/42 | 52/50 | .48 |
| Median WBC, 109/L (range) | 11.0 (0.7-94) | 22.0 (1-254) | <.001 |
| Median BM blasts, % (range) | 52 (9-98) | 57 (21-92) | .10 |
| Median fusion transcript ratio, % (range) | 303 (34-1090) | 88 (23-549) | <.001 |
| Secondary AML | 10 | 3 | .044 |
| CNS involvement | 3 | 1 | .36 |
| Additional cytogenetic abnormalities, n* | |||
| Loss of Y | 33 | 0 | <.001 |
| Trisomy 8 | 2 | 15 | .002 |
| Trisomy 22 | 0 | 18 | <.001 |
| del(9q) | 14 | 1 | <.001 |
| del(7q)/-7 | 4 | 5 | .99 |
| Gene mutations, n† | |||
| KIT exon 8 | 6 | 12 | .22 |
| KIT exon 17 | 17 | 6 | .013 |
| KIT, all | 22 | 18 | .37 |
| FLT3-TKD | 4 | 16 | .009 |
| FLT3-ITD | 6 | 5 | .76 |
| FLT3, all | 10 | 20 | .11 |
| RTK, all | 30 | 36 | .65 |
| N-RAS | 13 | 28 | .022 |
| K-RAS | 2 | 11 | .02 |
| RAS, all | 15 | 39 | .001 |
| . | t(8;21) AML . | inv(16)/t(16;16) . | P value . |
|---|---|---|---|
| Patients, n | 96 | 102 | |
| Treatment arm (A/B), n | 48/48 | 52/50 | .99 |
| Median age, y (range) | 42 (18-60) | 42 (18-60) | .37 |
| Gender (male/female), n | 54/42 | 52/50 | .48 |
| Median WBC, 109/L (range) | 11.0 (0.7-94) | 22.0 (1-254) | <.001 |
| Median BM blasts, % (range) | 52 (9-98) | 57 (21-92) | .10 |
| Median fusion transcript ratio, % (range) | 303 (34-1090) | 88 (23-549) | <.001 |
| Secondary AML | 10 | 3 | .044 |
| CNS involvement | 3 | 1 | .36 |
| Additional cytogenetic abnormalities, n* | |||
| Loss of Y | 33 | 0 | <.001 |
| Trisomy 8 | 2 | 15 | .002 |
| Trisomy 22 | 0 | 18 | <.001 |
| del(9q) | 14 | 1 | <.001 |
| del(7q)/-7 | 4 | 5 | .99 |
| Gene mutations, n† | |||
| KIT exon 8 | 6 | 12 | .22 |
| KIT exon 17 | 17 | 6 | .013 |
| KIT, all | 22 | 18 | .37 |
| FLT3-TKD | 4 | 16 | .009 |
| FLT3-ITD | 6 | 5 | .76 |
| FLT3, all | 10 | 20 | .11 |
| RTK, all | 30 | 36 | .65 |
| N-RAS | 13 | 28 | .022 |
| K-RAS | 2 | 11 | .02 |
| RAS, all | 15 | 39 | .001 |
RTK, receptor tyrosine kinase (KIT and/or FLT3).
A total of 196 the 198 patients, including 96 t(8;21) and 101 inv(16)/t(16;16) AML patients, had an available karyotype.
A total of 194 of the 198 patients, including 93 t(8;21) and 101 inv(16)/t(16;16) AML patients, were tested for KIT, FLT3 and RAS mutations.
Outcome
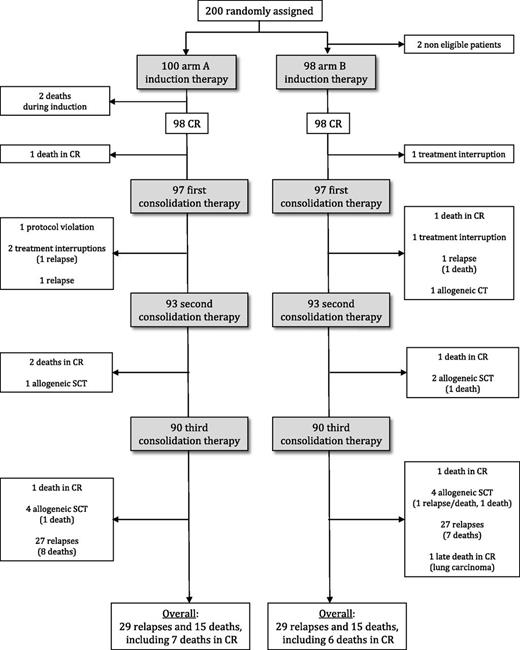
Trial flowchart is shown in Figure 1. All but 2 patients, who died early from infection, entered CR (CR rate, 99%). In arm B, 24 patients received the optional day-16 chemotherapy sequence. Only one arm B patient needed the planned salvage course to reach CR. A total of 12 of the 196 CR patients received allogeneic SCT in first CR (5 arm A, 7 arm B), 11 patients died in first CR (5 arm A, 6 arm B; 3 after allogeneic SCT), 58 patients had hematologic relapse (29 arm A, 29 arm B; 1 after allogeneic SCT), and 30 patients died (15 arm A, 15 arm B), including the 2 induction deaths. Among the 57 nontransplanted relapsing patients, 49 achieved a second CR (86%; 19/26 t[8;21] AML patients and in 30/31 inv[16]/t[16;16] AML patients, P = .018) after salvage therapy and 38 and 4 respectively received allogeneic and autologous SCT in second CR.
Patient outcome is shown in Figure 2. After censoring the 12 patients who received allogeneic SCT in first CR, RFS was estimated at 64% (95% CI, 56-71) at 36 months, with no difference between the treatment arms (Figure 2A; primary end point of the randomized comparison) or the CBF-AML subsets (Figure 2B). At 36 months, CIR and OS from CR were estimated at 32% (95% CI, 25-39) and 85% (95% CI, 78-90), respectively, still with no difference between randomization arms (Figure 2C,E) or CBF-AML subsets (Figure 2D,F).
Outcome by CBF-AML subtype and treatment arm. (A-B) At 36 months, RFS was estimated at 64% (95% CI, 53-73) in arm A patients as compared with 64% (95% CI, 52-73) in arm B patients (Figure 2A; HR, 0.97 [95% CI, 0.59-1.57]; P = .89 by the log-rank test). At 36 months, RFS was estimated at 68% (95% CI, 57-76) in t(8;21) AML patients as compared with 61% (95% CI, 49-70) in inv(16)/t(16;16) AML patients (Figure 2B; HR, 1.17 [95% CI, 0.72-1.91]; P = .53 by the log-rank test). (C-D) At 36 months, CIR was estimated at 32% (95% CI, 23-42) in arm A patients as compared with 32% (95% CI, 23-43) in arm B patients (Figure 2C; SHR, 0.97 [95% CI, 0.58-1.64]; P = .92 by cause-specific hazard Cox model). At 36 months, CIR was estimated at 29% (95% CI, 21-40) in t(8;21) AML patients as compared with 34% (95% CI, 25-45) in inv(16)/t(16;16) AML patients (Figure 2D; SHR, 1.10 [95% CI, 0.65-1.86]; P = .71 by cause-specific hazard Cox model). (E-F) Overall survival from CR. At 36 months, OS from CR was estimated at 87% (95% CI, 77-93) in arm A patients as compared with 83% (95% CI, 71-90) in arm B patients (Figure 2E; HR, 1.03 [95% CI, 0.46-2.29]; P = .95 by the log-rank test). At 36 months, OS from CR was estimated at 84% (95% CI, 74-91) in t(8;21) AML patients as compared with 86% (95% CI, 75-93) in inv(16)/t(16;16) AML patients (Figure 2F; HR, 0.77 [95% CI, 0.34-1.72]; P = .52 by the log-rank test).
Outcome by CBF-AML subtype and treatment arm. (A-B) At 36 months, RFS was estimated at 64% (95% CI, 53-73) in arm A patients as compared with 64% (95% CI, 52-73) in arm B patients (Figure 2A; HR, 0.97 [95% CI, 0.59-1.57]; P = .89 by the log-rank test). At 36 months, RFS was estimated at 68% (95% CI, 57-76) in t(8;21) AML patients as compared with 61% (95% CI, 49-70) in inv(16)/t(16;16) AML patients (Figure 2B; HR, 1.17 [95% CI, 0.72-1.91]; P = .53 by the log-rank test). (C-D) At 36 months, CIR was estimated at 32% (95% CI, 23-42) in arm A patients as compared with 32% (95% CI, 23-43) in arm B patients (Figure 2C; SHR, 0.97 [95% CI, 0.58-1.64]; P = .92 by cause-specific hazard Cox model). At 36 months, CIR was estimated at 29% (95% CI, 21-40) in t(8;21) AML patients as compared with 34% (95% CI, 25-45) in inv(16)/t(16;16) AML patients (Figure 2D; SHR, 1.10 [95% CI, 0.65-1.86]; P = .71 by cause-specific hazard Cox model). (E-F) Overall survival from CR. At 36 months, OS from CR was estimated at 87% (95% CI, 77-93) in arm A patients as compared with 83% (95% CI, 71-90) in arm B patients (Figure 2E; HR, 1.03 [95% CI, 0.46-2.29]; P = .95 by the log-rank test). At 36 months, OS from CR was estimated at 84% (95% CI, 74-91) in t(8;21) AML patients as compared with 86% (95% CI, 75-93) in inv(16)/t(16;16) AML patients (Figure 2F; HR, 0.77 [95% CI, 0.34-1.72]; P = .52 by the log-rank test).
Univariate prognostic analyses
Univariate prognostic analyses for the SHR are indicated in Table 3. Three factors were demonstrated to have a significant impact or a marked trend toward a higher SHR by cause-specific hazard Cox models: (1) higher WBC (SHR, 2.26 [95% CI, 1.29-3.95]; P = .004), especially in t(8;21) AML patients; (2) KIT gene mutations (SHR, 1.78 [95% CI, 0.98-3.24]; P = .057; Figure 3A); and (3) FLT3 gene mutations (SHR, 1.70 [95% CI, 0.88-3.27]; P = .12). When pooling KIT and FLT3 gene mutations in a single covariate, RTK gene mutation appeared to be significantly associated with a higher hazard of relapse (SHR, 1.89 [95% CI, 1.11-3.22]; P = .019) and a shorter RFS (HR= 1.80 [95% CI, 1.09-2.96]; P = .019) (Figure 3B,C). A similar analysis was done for the OS from CR end point. Age, especially in inv(16)/t(16;16) AML patients, and BM blast percentage, especially in t(8;21) AML patients, were the 2 identified prognostic factors, whereas gene mutations did not predict shorter OS.
Univariate analysis for specific hazard of relapse
| . | All patients . | t(8;21) . | inv(16)/t(16;16) . | |||
|---|---|---|---|---|---|---|
| . | SHR . | 95% CI . | SHR . | 95% CI . | SHR . | 95% CI . |
| CBF subset | 1.10 | 0.65-1.86 | ||||
| Treatment arm | 0.97 | 0.58-1.64 | 0.82 | 0.38-1.77 | 1.13 | 0.56-2.28 |
| Age* | 1.00 | 0.98-1.02 | 0.99 | 0.96-1.02 | 1.00 | 0.97-1.04 |
| Gender | 1.44 | 0.85-2.43 | 1.15 | 0.53-2.49 | 1.74 | 0.85-3.60 |
| Log(WBC)* | 2.26 | 1.29-3.95 | 3.43 | 1.27-9.26 | 1.86 | 0.96-3.62 |
| BM blasts %* | 1.01 | 0.99-1.02 | 1.01 | 0.99-1.03 | 1.005 | 0.98-1.025 |
| Diagnosis fusion transcript ratio* | 1.00 | 0.99-1.002 | 1.00 | 0.99-1.002 | 1.00 | 0.99-1.004 |
| Additional cytogenetic abnormalities | ||||||
| Loss of Y | 1.02 | 0.45-2.31 | 1.02 | 0.45-2.31 | NA | NA |
| Trisomy 8 | 0.50 | 0.15-1.63 | NA | NA | 0.36 | 0.08-1.49 |
| Trisomy 22 | 0.82 | 0.31-2.15 | NA | NA | 0.82 | 0.31-2.15 |
| del(9q) | 1.49 | 0.56-3.95 | 1.15 | 0.39-3.33 | NA | NA |
| del(7q)/-7 | 1.09 | 0.34-3.51 | 0.87 | 0.12-6.47 | 1.26 | 0.30-5.29 |
| Gene mutations | ||||||
| KIT exon 8 | 1.89 | 0.88-4.09 | 1.90 | 0.53-6.82 | 1.89 | 0.72-4.97 |
| KIT exon 17 | 1.73 | 0.81-3.72 | 2.04 | 0.81-5.14 | 1.24 | 0.30-5.24 |
| KIT, all | 1.78 | 0.98-3.24 | 1.84 | 0.80-4.27 | 1.72 | 0.74-4.02 |
| FLT3-TKD | 1.81 | 0.86-3.80 | 2.32 | 0.55-9.90 | 1.67 | 0.71-3.93 |
| FLT3-ITD | 1.13 | 0.35-3.71 | 1.44 | 0.32-6.43 | 0.81 | 0.11-6.00 |
| FLT3, all | 1.70 | 0.88-3.27 | 1.88 | 0.63-5.67 | 1.60 | 0.71-3.63 |
| RTK, all | 1.89 | 1.11-3.22 | 1.90 | 0.87-4.15 | 1.88 | 0.91-3.89 |
| N-RAS | 1.30 | 0.70-2.41 | 1.15 | 0.40-3.35 | 1.39 | 0.65-2.97 |
| K-RAS | 0.51 | 0.12-2.10 | 1.70 | 0.23-12.64 | 0.29 | 0.04-2.15 |
| RAS, all | 1.07 | 0.59-1.95 | 1.26 | 0.47-3.34 | 0.98 | 0.47-2.07 |
| . | All patients . | t(8;21) . | inv(16)/t(16;16) . | |||
|---|---|---|---|---|---|---|
| . | SHR . | 95% CI . | SHR . | 95% CI . | SHR . | 95% CI . |
| CBF subset | 1.10 | 0.65-1.86 | ||||
| Treatment arm | 0.97 | 0.58-1.64 | 0.82 | 0.38-1.77 | 1.13 | 0.56-2.28 |
| Age* | 1.00 | 0.98-1.02 | 0.99 | 0.96-1.02 | 1.00 | 0.97-1.04 |
| Gender | 1.44 | 0.85-2.43 | 1.15 | 0.53-2.49 | 1.74 | 0.85-3.60 |
| Log(WBC)* | 2.26 | 1.29-3.95 | 3.43 | 1.27-9.26 | 1.86 | 0.96-3.62 |
| BM blasts %* | 1.01 | 0.99-1.02 | 1.01 | 0.99-1.03 | 1.005 | 0.98-1.025 |
| Diagnosis fusion transcript ratio* | 1.00 | 0.99-1.002 | 1.00 | 0.99-1.002 | 1.00 | 0.99-1.004 |
| Additional cytogenetic abnormalities | ||||||
| Loss of Y | 1.02 | 0.45-2.31 | 1.02 | 0.45-2.31 | NA | NA |
| Trisomy 8 | 0.50 | 0.15-1.63 | NA | NA | 0.36 | 0.08-1.49 |
| Trisomy 22 | 0.82 | 0.31-2.15 | NA | NA | 0.82 | 0.31-2.15 |
| del(9q) | 1.49 | 0.56-3.95 | 1.15 | 0.39-3.33 | NA | NA |
| del(7q)/-7 | 1.09 | 0.34-3.51 | 0.87 | 0.12-6.47 | 1.26 | 0.30-5.29 |
| Gene mutations | ||||||
| KIT exon 8 | 1.89 | 0.88-4.09 | 1.90 | 0.53-6.82 | 1.89 | 0.72-4.97 |
| KIT exon 17 | 1.73 | 0.81-3.72 | 2.04 | 0.81-5.14 | 1.24 | 0.30-5.24 |
| KIT, all | 1.78 | 0.98-3.24 | 1.84 | 0.80-4.27 | 1.72 | 0.74-4.02 |
| FLT3-TKD | 1.81 | 0.86-3.80 | 2.32 | 0.55-9.90 | 1.67 | 0.71-3.93 |
| FLT3-ITD | 1.13 | 0.35-3.71 | 1.44 | 0.32-6.43 | 0.81 | 0.11-6.00 |
| FLT3, all | 1.70 | 0.88-3.27 | 1.88 | 0.63-5.67 | 1.60 | 0.71-3.63 |
| RTK, all | 1.89 | 1.11-3.22 | 1.90 | 0.87-4.15 | 1.88 | 0.91-3.89 |
| N-RAS | 1.30 | 0.70-2.41 | 1.15 | 0.40-3.35 | 1.39 | 0.65-2.97 |
| K-RAS | 0.51 | 0.12-2.10 | 1.70 | 0.23-12.64 | 0.29 | 0.04-2.15 |
| RAS, all | 1.07 | 0.59-1.95 | 1.26 | 0.47-3.34 | 0.98 | 0.47-2.07 |
Comparisons were based on cause-specific hazard Cox models, stratified on treatment arm and CBF subset when applicable, and the 12 patients who received allogeneic SCT in first hematologic CR were censored at SCT time.
NA, not applicable; RTK: receptor tyrosine kinase (KIT and/or FLT3).
Tested as continuous variable.
Outcome by KIT and RTK gene mutations. (A) At 36 months, CIR was estimated at 44% (95% CI, 24-70) and 34% (95% CI, 18-59) in patients with exon 8 and exon 17 KIT-mutated gene, respectively, as compared with 30% (95% CI, 23-39) in those with wild-type KIT gene (SHR= 1.78 [95% CI, 0.98-3.24] for KIT-mutated AML patients; P = .057 by cause-specific hazard Cox model). (B-C) RTK (KIT and/or FLT3-ITD/TKD) gene mutations. (B) At 36 months, CIR was estimated at 42% (95% CI, 30-56) in patients with RTK-mutated genes, as compared with 27% (95% CI, 20-36) in those with wild-type RTK genes (SHR= 1.89 [95% CI, 1.11-3.22] for RTK-mutated AML patients; P = .019 by cause-specific hazard Cox model). (C) At 36 months, RFS was estimated at 53% (95% CI, 39-65) in patients with RTK-mutated genes, as compared with 69% (95% CI, 59-77) in those with wild-type RTK genes (HR= 1.80 [95% CI, 1.09-2.96] for RTK-mutated AML patients; P = .019 by the log-rank test).
Outcome by KIT and RTK gene mutations. (A) At 36 months, CIR was estimated at 44% (95% CI, 24-70) and 34% (95% CI, 18-59) in patients with exon 8 and exon 17 KIT-mutated gene, respectively, as compared with 30% (95% CI, 23-39) in those with wild-type KIT gene (SHR= 1.78 [95% CI, 0.98-3.24] for KIT-mutated AML patients; P = .057 by cause-specific hazard Cox model). (B-C) RTK (KIT and/or FLT3-ITD/TKD) gene mutations. (B) At 36 months, CIR was estimated at 42% (95% CI, 30-56) in patients with RTK-mutated genes, as compared with 27% (95% CI, 20-36) in those with wild-type RTK genes (SHR= 1.89 [95% CI, 1.11-3.22] for RTK-mutated AML patients; P = .019 by cause-specific hazard Cox model). (C) At 36 months, RFS was estimated at 53% (95% CI, 39-65) in patients with RTK-mutated genes, as compared with 69% (95% CI, 59-77) in those with wild-type RTK genes (HR= 1.80 [95% CI, 1.09-2.96] for RTK-mutated AML patients; P = .019 by the log-rank test).
MRD response
MRD levels.
Reductions in the median fusion transcript ratio level are depicted in Figure 4. Median levels were statistically comparable in both CBF subsets at the 3 MRD1, MRD2, and MRD3 time points (Figure 4A). However, they decreased more rapidly in arm A than in arm B patients (Figure 4B), although the difference between both arms became nonsignificant after the last third consolidation course (66 CBFα and 67 CBFβ; 68 arm A and 65 arm B; median ratio, 0.003 vs 0.010% in arm A and arm B, respectively; P = .17).
Reduction in fusion transcript ratio by CBF subset and treatment arm. A total of 176 (90 CBFα and 86 CBFβ; 89 arm A and 87 arm B), 176 (91 CBFα and 85 CBFβ; 88 arm A and 88 arm B), and 160 (79 CBFα and 81 CBFβ; 80 arm A and 80 arm B) patients in first CR after having received the planned chemotherapy and no SCT were tested for MRD response at MRD1, MRD2, and MRD3 time point, respectively. Overall, 86 of 176 (50%), 124 of 176 (70.5%), and 130 of 160 (81%) achieved a 3-log MRD reduction at MRD1, MRD2, and MRD3 time point, respectively. (A) Median ratio was 0.20% vs 0.145%, 0.055% vs 0.05%, and 0.020% vs 0.025% at the MRD1, MRD2, and MRD3 time point in the RUNX1-RUNX1T1 vs CBFB-MYH11 subset, respectively (P = .71, 0.19, and 0.17, respectively, Mann-Whitney U test). The rate of patients who reached the 0.1% level at MRD2 was 60% (57% and 62% in the RUNX1-RUNX1T1 and CBFB-MYH11 subset, respectively; P = .54, Fisher’s exact test). (B) Median ratio was 0.10% vs 0.26%, 0.023% vs 0.10%, and 0.013% vs 0.030% at the MRD1, MRD2, and MRD3 time point in arm A and arm B patients, respectively (P = .006, 0.0002, and 0.013, respectively, Mann-Whitney U test).
Reduction in fusion transcript ratio by CBF subset and treatment arm. A total of 176 (90 CBFα and 86 CBFβ; 89 arm A and 87 arm B), 176 (91 CBFα and 85 CBFβ; 88 arm A and 88 arm B), and 160 (79 CBFα and 81 CBFβ; 80 arm A and 80 arm B) patients in first CR after having received the planned chemotherapy and no SCT were tested for MRD response at MRD1, MRD2, and MRD3 time point, respectively. Overall, 86 of 176 (50%), 124 of 176 (70.5%), and 130 of 160 (81%) achieved a 3-log MRD reduction at MRD1, MRD2, and MRD3 time point, respectively. (A) Median ratio was 0.20% vs 0.145%, 0.055% vs 0.05%, and 0.020% vs 0.025% at the MRD1, MRD2, and MRD3 time point in the RUNX1-RUNX1T1 vs CBFB-MYH11 subset, respectively (P = .71, 0.19, and 0.17, respectively, Mann-Whitney U test). The rate of patients who reached the 0.1% level at MRD2 was 60% (57% and 62% in the RUNX1-RUNX1T1 and CBFB-MYH11 subset, respectively; P = .54, Fisher’s exact test). (B) Median ratio was 0.10% vs 0.26%, 0.023% vs 0.10%, and 0.013% vs 0.030% at the MRD1, MRD2, and MRD3 time point in arm A and arm B patients, respectively (P = .006, 0.0002, and 0.013, respectively, Mann-Whitney U test).
MRD log reduction.
In this study, the treatment-stratifying MRD end point was a ≥3-log reduction between diagnosis and MRD2. This targeted reduction level was reached in 71 of 91 t(8;21) AML patients and in 53 of 85 inv(16)/t(16;16) AML patients (78% vs 62%, P = .03), for an overall MRD2 response rate of 70%. It was reached in 67 of 88 arm A patients and in 57 of 88 arm B patients (76% vs 65%, P = .14). The 3-log MRD2 reduction was less frequently achieved in patients with KIT mutations (54% vs 75%, P = .02), whereas no differences were observed according to the FLT3 (70% vs 71%, P = .99) or N/K-RAS (66% vs 72%, P = .45) status. As illustrated in Figure 5, the 3-log MRD2 reduction was significantly associated with a lower hazard of relapse (SHR = 0.27 [95% CI, 0.15-0.49], P < .001), longer RFS (HR = 0.34 [95% CI, 0.19-0.59], P < .001), and with a trend for a longer OS from CR (HR = 0.43 [95% CI, 0.17-1.08], P = .066). Similar results were observed when using an absolute MRD2 level ≤0.1% rather than MRD2 reduction (HR = 0.38, 0.44, and 0.55; P < .001, P = .0017, and P = .16 for SHR, RFS, and OS from CR, respectively). Median CR duration was 17.5 months in the 52 patients who did not achieve MRD2 reduction ≥3 logs, whereas it was not reached in other patients. Among these 52 patients, only 2 relapsed within 3 months after MRD2 evaluation.
Outcome by MRD2 response. (A) At 36 months, CIR was estimated at 22% (95% CI, 16-32) in patients who achieved a 3-log MRD2 reduction vs 54% (95% CI, 39-69) in those who did not (SHR= 0.27 [95% CI, 0.15-0.49]; P < .001 by cause-specific hazard Cox model). (B) At 36 months, RFS was estimated at 73% (95% CI, 64-81) in patients who achieved a 3-log MRD2 reduction vs 44% (95% CI, 29-58) in those who did not (HR= 0.34 [95% CI, 0.19-0.59]; P < .001, log-rank test). (C) At 36 months, OS from CR was estimated at 90% (95% CI, 83-94) in patients who achieved a 3-log MRD2 reduction vs 71% (95% CI, 51-85) in those who did not (HR= 0.43 [95% CI, 0.17-1.08]; P = .066, log-rank test).
Outcome by MRD2 response. (A) At 36 months, CIR was estimated at 22% (95% CI, 16-32) in patients who achieved a 3-log MRD2 reduction vs 54% (95% CI, 39-69) in those who did not (SHR= 0.27 [95% CI, 0.15-0.49]; P < .001 by cause-specific hazard Cox model). (B) At 36 months, RFS was estimated at 73% (95% CI, 64-81) in patients who achieved a 3-log MRD2 reduction vs 44% (95% CI, 29-58) in those who did not (HR= 0.34 [95% CI, 0.19-0.59]; P < .001, log-rank test). (C) At 36 months, OS from CR was estimated at 90% (95% CI, 83-94) in patients who achieved a 3-log MRD2 reduction vs 71% (95% CI, 51-85) in those who did not (HR= 0.43 [95% CI, 0.17-1.08]; P = .066, log-rank test).
Multivariate prognostic analysis
According to the univariate results, the covariates entering multivariate analysis were WBC, RTK gene mutations, and MRD2 response. Table 4 shows that MRD2 response remained the sole factor significantly influencing SHR in this multivariate setting in the whole patient cohort as well as in the t(8;21) and inv(16)/t(16;16) AML subsets. Table 5 shows multivariate analysis results for OS from CR. Table 6 gives 36-month CIR and OS from CR estimations according to the different RTK mutation/MRD2 patient subsets. Similar results were observed when replacing MRD2 reduction by absolute MRD2 level ≤0.1% (data not shown).
Multivariate analysis for specific hazard of relapse
| . | All patients . | t(8;21) . | inv(16)/t(16;16) . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| . | SHR . | 95% CI . | P value . | SHR . | 95% CI . | P value . | SHR . | 95% CI . | P value . |
| Log(WBC)* | 1.96 | 0.99-3.85 | .051 | 2.10 | 0.73-6.04 | .17 | 1.83 | 0.77-4.34 | .17 |
| RTK gene mutation | 1.62 | 0.89-2.94 | .11 | 1.51 | 0.65-3.54 | .34 | 1.63 | 0.69-3.88 | .27 |
| MRD2 reduction ≥ 3 logs | 0.31 | 0.17-0.57 | <.001 | 0.24 | 0.10-0.57 | .001 | 0.40 | 0.18-0.91 | .029 |
| . | All patients . | t(8;21) . | inv(16)/t(16;16) . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| . | SHR . | 95% CI . | P value . | SHR . | 95% CI . | P value . | SHR . | 95% CI . | P value . |
| Log(WBC)* | 1.96 | 0.99-3.85 | .051 | 2.10 | 0.73-6.04 | .17 | 1.83 | 0.77-4.34 | .17 |
| RTK gene mutation | 1.62 | 0.89-2.94 | .11 | 1.51 | 0.65-3.54 | .34 | 1.63 | 0.69-3.88 | .27 |
| MRD2 reduction ≥ 3 logs | 0.31 | 0.17-0.57 | <.001 | 0.24 | 0.10-0.57 | .001 | 0.40 | 0.18-0.91 | .029 |
Comparisons were based on cause-specific hazard Cox models, stratified on treatment arm and CBF subset when applicable, and the 12 patients who received allogeneic SCT in first hematological CR were censored at SCT time.
RTK, receptor tyrosine kinase (KIT and/or FLT3).
Tested as continuous variable.
Multivariate analysis for OS from CR
| . | All patients . | t(8;21) . | inv(16)/t(16;16) . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| HR . | 95% CI . | P value . | HR . | 95% CI . | P value . | HR . | 95% CI . | P value . | |
| Log(WBC)* | 1.06 | 0.39-2.89 | .91 | 1.57 | 0.35-7.13 | 0.56 | 0.84 | 0.23-3.17 | .80 |
| RTK gene mutation | 1.73 | 0.70-4.30 | .24 | 1.34 | 0.41-4.44 | 0.63 | 2.21 | 0.50-9.78 | .30 |
| MRD2 reduction ≥ 3 logs | 0.51 | 0.20-1.31 | .16 | 0.31 | 0.09-1.03 | 0.056 | 0.92 | 0.21-4.13 | .91 |
| . | All patients . | t(8;21) . | inv(16)/t(16;16) . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| HR . | 95% CI . | P value . | HR . | 95% CI . | P value . | HR . | 95% CI . | P value . | |
| Log(WBC)* | 1.06 | 0.39-2.89 | .91 | 1.57 | 0.35-7.13 | 0.56 | 0.84 | 0.23-3.17 | .80 |
| RTK gene mutation | 1.73 | 0.70-4.30 | .24 | 1.34 | 0.41-4.44 | 0.63 | 2.21 | 0.50-9.78 | .30 |
| MRD2 reduction ≥ 3 logs | 0.51 | 0.20-1.31 | .16 | 0.31 | 0.09-1.03 | 0.056 | 0.92 | 0.21-4.13 | .91 |
Comparisons were based on cause-specific hazard Cox models, stratified on treatment arm and CBF subset when applicable, and the 12 patients who received allogeneic SCT in first hematological CR were censored at SCT time.
RTK, receptor tyrosine kinase (KIT and/or FLT3).
Tested as continuous variable.
CIR and OS from CR estimations at 36 months in the different RTK mutation and MRD patient subsets
| Patient subsets . | All patients . | t(8;21) . | inv(16)/t(16;16) . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Patients (n) . | CIR (95% CI) . | OS from CR (95% CI) . | Patients (n) . | CIR (95% CI) . | OS from CR (95% CI) . | Patients (n) . | CIR (95% CI) . | OS from CR (95% CI) . | |
| RTK unmutated with 3-log MRD2 reduction | 84 | 18% (11-29) | 89% (79-94) | 50 | 19% (10-34) | 90% (77-96) | 34 | 17% (7-38) | 87% (65-96) |
| RTK mutated with 3-log MRD2 reduction | 38 | 33% (19-52) | 92% (76-97) | 19 | 31% (14-59) | 89% (63-97) | 19 | 35% (17-62) | 95% (68-99) |
| RTK unmutated without 3-log MRD2 reduction | 28 | 45% (28-67) | 83% (55-94) | 10 | 55% (28-86) | 73% (28-93) | 18 | 40% (20-68) | 90% (47-98) |
| RTK mutated without 3-log MRD2 reduction | 23 | 61% (40-83) | 56% (18-83) | 10 | 70% (37-96) | 53% (12-82) | 13 | 55% (29-83) | 60% (8-90) |
| Patient subsets . | All patients . | t(8;21) . | inv(16)/t(16;16) . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Patients (n) . | CIR (95% CI) . | OS from CR (95% CI) . | Patients (n) . | CIR (95% CI) . | OS from CR (95% CI) . | Patients (n) . | CIR (95% CI) . | OS from CR (95% CI) . | |
| RTK unmutated with 3-log MRD2 reduction | 84 | 18% (11-29) | 89% (79-94) | 50 | 19% (10-34) | 90% (77-96) | 34 | 17% (7-38) | 87% (65-96) |
| RTK mutated with 3-log MRD2 reduction | 38 | 33% (19-52) | 92% (76-97) | 19 | 31% (14-59) | 89% (63-97) | 19 | 35% (17-62) | 95% (68-99) |
| RTK unmutated without 3-log MRD2 reduction | 28 | 45% (28-67) | 83% (55-94) | 10 | 55% (28-86) | 73% (28-93) | 18 | 40% (20-68) | 90% (47-98) |
| RTK mutated without 3-log MRD2 reduction | 23 | 61% (40-83) | 56% (18-83) | 10 | 70% (37-96) | 53% (12-82) | 13 | 55% (29-83) | 60% (8-90) |
Comparisons were based on cause-specific hazard Cox models, stratified on treatment arm and CBF subset when applicable, and the 12 patients who received allogeneic SCT in first hematological CR were censored at SCT time.
RTK, receptor tyrosine kinase (KIT and/or FLT3).
Discussion
This large prospective study demonstrates a similar efficacy for intensified timed-sequential (arm A) or conventional (arm B) induction in younger patients with CBF-AML. Despite lower MRD levels after initial treatment, arm A patients experienced a similar relapse incidence as arm B patients. This might be explained by the intensity of postremission therapy, which eventually allowed reaching similar MRD response in both arms, even if it was delayed in arm B patients. This observation raises the issue of the possible redundancy of high-dose cytarabine consolidation courses, at least in responding patients.
Hierarchical evaluation of leukemia-related risk factors, such as genomic anomalies, and response-related risk factors, such as MRD, is becoming a general issue in the management of patients with acute leukemia. This was recently addressed in children with acute lymphoblastic leukemia.30 This study is the first to prospectively and simultaneously evaluate gene mutations and MRD response in adults with favorable CBF-AML. Results show that KIT and FLT3 mutations, which have been suggested to be strong prognostic factors in retrospective studies, allow for a relatively poor discrimination of high-risk vs low-risk patients when prospectively evaluated, although statistically significant differences were retrieved in univariate analysis. MRD response appeared as the major and unique predictor of relapse in multivariate analysis, validating its use for treatment stratification, as we did in this study with allogeneic SCT. A similar study should be performed in the other subset of good-prognosis patients (ie, those with cytogenetically normal AML carrying NPM1 gene mutation and no FLT3-ITD) who could also be studied for MRD and other DNMT3A and IDH1/2 gene mutations.
When dealing with nontargeted treatment adaptation, there is still no reason to manage t(8;21) and inv(16)/t(16;16) AML patients differently. Based on the present results, both the 3-log MRD reduction and the absolute 0.1% MRD level can be used to differentiate high-risk from low-risk patients in both CBF subsets. Individual genomic characterization may, however, be useful to indicate or not indicate targeted interventions, such as the use of RTK inhibitors, for instance dasatinib in patients with activating KIT and/or FLT3 gene mutations.
Despite a relapse incidence higher than 30%, OS from CR remained as high as 85%, underlying the relatively good salvage rate in CBF-AML patients, especially in those with inv(16)/t(16;16) CBF anomaly. This might partly explain why investigators transplanted only 12 of the 52 poor MRD2 responders in first CR, while it was planned by the protocol. It should also be stressed than heterogeneity of relapse treatment, including the use of gemtuzumab ozogamicin in some patients, limits the interpretation of survival predictors in these patients. To prevent relapse and prolong RFS should probably remain the primary objective in these “favorable AML” patients.
In conclusion, this study strongly supports the interest of implementing prospective MRD monitoring in the design of CBF-AML trials. Rather than RTK gene mutations, MRD levels should be used for treatment stratification. This recommendation could nonetheless evolve if a better characterization of CBF-AML risk heterogeneity could be demonstrated using refined high-resolution genomic analysis.31,32
There is an Inside Blood commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors are grateful to Karine Celli-Lebras, Corinne Duguet, and Virginie Bacca for her help with data management, Julie Lejeune for data handling and statistical help, and Lamya Haddaoui and Catherine Lacombe (Tumour Bank for the GOELAMS group, Hôpital Cochin, Paris) and Christophe Roumier and Olivier Nibourel (Tumour Bank for the ALFA group, CHU Lille) for handling, conditioning, and storing patient samples. The work of all clinical research assistants of the GOELAMS and ALFA groups is also acknowledged here.
The work was supported by the French National Cancer Institute and French Ministry of Health (PHRC ID, 2006-0213).
Authorship
Contribution: E.J. was PI coordinator of the study. H.D., M.C.B., C. Preudhomme, and S.C. controlled the database. H.D. and S.C. performed the statistical analysis. E.J., N.B., C.R., E.R., J.D., A.P., C.-E.B., C.B., C. Pautas, N.V., B.L., X.T., P.G., N.I., and H.D. enrolled patients in the study. I.L. and C.T. centrally reviewed cytogenetic data. O.B., A.R., P.C., J.-M.C., E.D., and C. Preudhomme centrally performed and reviewed gene mutation and/or MRD analyses. E.J., H.D., M.C.B., N.I., and S.C. wrote the manuscript, which was approved by all the authors.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Hervé Dombret, Hôpital Saint-Louis, 1, Ave Claude Vellefaux, 75010 Paris, France; e-mail: herve.dombret@sls.aphp.fr.

![Figure 2. Outcome by CBF-AML subtype and treatment arm. (A-B) At 36 months, RFS was estimated at 64% (95% CI, 53-73) in arm A patients as compared with 64% (95% CI, 52-73) in arm B patients (Figure 2A; HR, 0.97 [95% CI, 0.59-1.57]; P = .89 by the log-rank test). At 36 months, RFS was estimated at 68% (95% CI, 57-76) in t(8;21) AML patients as compared with 61% (95% CI, 49-70) in inv(16)/t(16;16) AML patients (Figure 2B; HR, 1.17 [95% CI, 0.72-1.91]; P = .53 by the log-rank test). (C-D) At 36 months, CIR was estimated at 32% (95% CI, 23-42) in arm A patients as compared with 32% (95% CI, 23-43) in arm B patients (Figure 2C; SHR, 0.97 [95% CI, 0.58-1.64]; P = .92 by cause-specific hazard Cox model). At 36 months, CIR was estimated at 29% (95% CI, 21-40) in t(8;21) AML patients as compared with 34% (95% CI, 25-45) in inv(16)/t(16;16) AML patients (Figure 2D; SHR, 1.10 [95% CI, 0.65-1.86]; P = .71 by cause-specific hazard Cox model). (E-F) Overall survival from CR. At 36 months, OS from CR was estimated at 87% (95% CI, 77-93) in arm A patients as compared with 83% (95% CI, 71-90) in arm B patients (Figure 2E; HR, 1.03 [95% CI, 0.46-2.29]; P = .95 by the log-rank test). At 36 months, OS from CR was estimated at 84% (95% CI, 74-91) in t(8;21) AML patients as compared with 86% (95% CI, 75-93) in inv(16)/t(16;16) AML patients (Figure 2F; HR, 0.77 [95% CI, 0.34-1.72]; P = .52 by the log-rank test).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/121/12/10.1182_blood-2012-10-462879/4/m_2213f2.jpeg?Expires=1765894655&Signature=zCK~V9jEUus2Q4rVLHLV-cvGYSgTX4c2PgnjJ0JxGzFmitMfEUjE6jVQdin6TVgkbCtOntDh5YKu6E3gfm-ZOQ8FtM1XwjhHSxe9IXOFw3mXrC-edUjM7bBjkSUZbWbl-eIFl6L1GBxO2T3R0BvlXbDEd3gUfzje3OTqn295T8lsKXhCy9LMCk2cjxMuAA5NfS-D7uEbA~Ew5XcYIO3L2VrczVyupAz~9qdZ2QS5NaUGgszNbeICkq7sgwCRXME6v0mUeGYMzxVRoeZdkCmfwVikEPbkUQqF8c2rtH10sWCOTHqtNFNjETTU-wGbfy7vYnTLETiQl~7K-gbSd24nUw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 3. Outcome by KIT and RTK gene mutations. (A) At 36 months, CIR was estimated at 44% (95% CI, 24-70) and 34% (95% CI, 18-59) in patients with exon 8 and exon 17 KIT-mutated gene, respectively, as compared with 30% (95% CI, 23-39) in those with wild-type KIT gene (SHR= 1.78 [95% CI, 0.98-3.24] for KIT-mutated AML patients; P = .057 by cause-specific hazard Cox model). (B-C) RTK (KIT and/or FLT3-ITD/TKD) gene mutations. (B) At 36 months, CIR was estimated at 42% (95% CI, 30-56) in patients with RTK-mutated genes, as compared with 27% (95% CI, 20-36) in those with wild-type RTK genes (SHR= 1.89 [95% CI, 1.11-3.22] for RTK-mutated AML patients; P = .019 by cause-specific hazard Cox model). (C) At 36 months, RFS was estimated at 53% (95% CI, 39-65) in patients with RTK-mutated genes, as compared with 69% (95% CI, 59-77) in those with wild-type RTK genes (HR= 1.80 [95% CI, 1.09-2.96] for RTK-mutated AML patients; P = .019 by the log-rank test).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/121/12/10.1182_blood-2012-10-462879/4/m_2213f3.jpeg?Expires=1765894655&Signature=ZAhpZSJ~5V5AFTHpTmXtx~wkKnE3Mdjqlg6yTcXPkg7o0cieJQx4GSDj~M7neXwSl70ossgx4Soo74-DZm~vM1HX0UxjnRV92tomdJq9mISWXQ1YoqaTaqcGPcQKp9B883lPXw8hUq6TBtxS6hV9iNKXiVGLayoeVmqkFC6V2b0UOjVcTZvBElvBdHJnrQ6Jvk6bE4xz75YuxtMAJkV7g2KFt2q-is0cse3vTp55hes1vvHAfBgL0g5lqOKP8Am1DlVB3vCqxH0Ia7s~nhjJtroBjpgh3R2qWgODeiwPDyWx9cBoPwAhC-ypBD5sDumaeR9HVdbxQ3L9C-vKZ5xWxg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Figure 5. Outcome by MRD2 response. (A) At 36 months, CIR was estimated at 22% (95% CI, 16-32) in patients who achieved a 3-log MRD2 reduction vs 54% (95% CI, 39-69) in those who did not (SHR= 0.27 [95% CI, 0.15-0.49]; P < .001 by cause-specific hazard Cox model). (B) At 36 months, RFS was estimated at 73% (95% CI, 64-81) in patients who achieved a 3-log MRD2 reduction vs 44% (95% CI, 29-58) in those who did not (HR= 0.34 [95% CI, 0.19-0.59]; P < .001, log-rank test). (C) At 36 months, OS from CR was estimated at 90% (95% CI, 83-94) in patients who achieved a 3-log MRD2 reduction vs 71% (95% CI, 51-85) in those who did not (HR= 0.43 [95% CI, 0.17-1.08]; P = .066, log-rank test).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/121/12/10.1182_blood-2012-10-462879/4/m_2213f5.jpeg?Expires=1765894655&Signature=IC5iG5W4tfOAqlWvX3OmPAQjv7SByW4dtqMY1IMuNIFohSHDvQDFGhNKHhJ2ahmB0X5gtE7ZghWUKvjpDb8AiHvic94rtJjvybD2sGT8GeRLFO~MDJPI7XPXSOhwq8SOSxkPfY611-i79X5vDsE5qh~PFeyFVlt85KHrLmlvfnmGidKEpmZ3LBxDLW-vNXbs0g2RLGRgSot-WJXX2VQlmzcq4ulltWYeAfJ11IIF55f68YcIR50nExdZTwZxF5U9P~YTsl6VbRj64mclbKV9g7kSkMYpV74fnmaPoV1wxQBiwJ0aPuIaibFM3-VDS1Et~o7WueK7ZRcaXVF2EtlJ~Q__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal