Key Points
Lenalidomide treatment of primary CLL/nurse-like cell cocultures resulted in significantly decreased viability of CLL cells.
Lenalidomide increased IL-10 levels, activation of STAT1, expression of ICAM-1, and migration-related genes, and reduced CLL cell motility.
Abstract
Chronic lymphocytic leukemia (CLL) cells depend on microenvironmental stimuli for their survival, provided for example by monocyte-derived nurse-like cells (NLCs). The immunomodulatory drug lenalidomide shows therapeutic effects in subgroups of CLL patients, and is believed to act via the microenvironment. To investigate the effects of lenalidomide on the survival support of NLCs, cocultures of monocytes and CLL cells were treated for 14 days with lenalidomide, which resulted in significantly decreased viability of CLL cells. Among the changes induced by this drug, we observed reduced expression of HLA-DR in NLCs as well as increased secretion of interleukin-10 (IL-10), indicating an altered inflammatory milieu in the cocultures. The increase in IL-10 levels lead to an induction of STAT1 phosphorylation in CLL cells and to enhanced cell-surface expression of intercellular adhesion molecule 1 and altered expression of cytoskeletal and migration-related genes. Chemotaxis assays with lenalidomide-treated CLL cells revealed an impaired migration capability. Our data show that lenalidomide reduces the survival support of NLCs for CLL cells in vitro, suggesting that this drug affects the myeloid microenvironment in CLL in vivo. Furthermore, lenalidomide acts on the migratory potential of CLL cells, which may affect circulation and homing of CLL cells in vivo.
Introduction
Chronic lymphocytic leukemia (CLL) is the most common leukemia in adults in the Western world, with an incidence of 3.9 per 100 000 people, and is still an incurable disease.1 Survival and proliferation of CLL cells is regulated by their microenvironment, because the cells die within days if cultured ex vivo but can be rescued by coculture with various nonmalignant accessory cells.2 Of interest is the support of CLL cells by myeloid cells, because peripheral blood-derived monocytes differentiate in the presence of CLL cells to so-called nurse-like cells (NLCs), which are round or fibroblast-shaped adherent cells that promote survival of CLL cells in vitro and were detected in lymph nodes of CLL patients.3 Spontaneous apoptosis of CLL cells is prevented by NLCs by the release of soluble factors and by cell-cell contact.4,5 As observed by expression profiling, NLCs share characteristics with tumor-associated macrophages,6 which display high cell surface expression of HLA-DR, CD163, and CD206.7 Recently, our group highlighted the importance of undifferentiated, peripheral blood-derived monocytes in the CLL microenvironment.8,9
Lenalidomide is an immunomodulatory drug that is currently undergoing clinical trial as a treatment of different B-cell malignancies, including CLL.10-13 Here, it is believed to act mainly via the microenvironment because lenalidomide shows no direct cytotoxicity for CLL cells.14 Analysis of blood serum after lenalidomide treatment of CLL patients revealed changes in the levels of several inflammatory cytokines.12,15 Further, an increased expression of several activation markers was observed.14,16 By in vitro studies and in the murine TCL1 mouse model for CLL, lenalidomide treatment was shown to reverse a defect in immunologic synapse formation between T cells and CLL cells, which was induced by the malignant cells.17,18 Moreover, immunomodulatory drugs like lenalidomide were shown to change the expression of several cytoskeletal molecules like cdc42 and Rac family members by modulating ρ-GTPases.19
Because myeloid cells provide survival support for CLL cells in vitro, we sought to investigate the effects of lenalidomide in long-term cocultures of CLL cells and monocytes. Even though lenalidomide had no effect on the viability of CLL cells alone, it reduced survival rates of the malignant cells in the cocultures. This study revealed alterations induced by lenalidomide in CLL cells and cocultured monocytes/NLCs, which can explain at least part of the mode of action of this immunomodulatory drug in CLL.
Methods
Lenalidomide
Lenalidomide was generously provided by Celgene (San Diego, CA).
Cell culture and viability
Blood samples from patients that matched standard diagnosis criteria for CLL were obtained from the University Hospital of Ulm or Heidelberg, Germany (Table 1). All patients provided written informed consent, validated by the respective ethics committee, in accordance with the declaration of Helsinki. Monocytes and B cells were obtained from buffy coats of healthy donors (HD) from the blood bank of the University Hospital of Heidelberg. Primary blood cells were prepared by Ficoll density centrifugation as formerly described.9 CD19+ B cells or CD14+ monocytes were enriched by magnetic-activated cell sorting (MACS) according to the manufacturer’s protocol (Miltenyi Biotech, Bergisch Gladbach, Germany) resulting in purities of >98% for CLL or normal B cells and >90% for monocytes.
Patient data, including age, gender, clinical stage at time of experiment (Binet), IGHV mutational status, FISH results, ZAP70 expression, and experiments performed with this sample
| Number . | Age (y) . | Gender . | Binet . | IGHV . | FISH . | ZAP70 . | Experiments performed . |
|---|---|---|---|---|---|---|---|
| 34 | 46 | m | A | UM | 12q+ | Positive | Survival; CCL2 |
| 39 | 59 | m | C | UM | 13q− | nd | Survival; CCL2 |
| 101 | 59 | f | A | M | 13q−; 14q− | Negative | Survival; CCL2; IL-10; HLA-DR |
| 102 | 49 | m | nd | nd | nd | nd | CD54 |
| 111 | 66 | m | A | UM | 11q−; 12p+; t(14q) | Positive | Survival; CCL2; IL-10; HLA-DR |
| 113 | 76 | m | nd | M | 13q− | Negative | CD54 |
| 114 | 67 | m | A | UM | 13q− | nd | CD54 |
| 115 | 62 | f | A | M | 13q− | nd | Survival; CCL2; IL-10; HLA-DR |
| 122 | 51 | m | B | M V3-53 | 13q− | Negative | Survival; CCL2; IL-10 |
| 123 | 76 | m | C | UM V1-69 | 11q−; 13q−; 14q− | Negative | Survival; CCL2; IL-10 |
| 124 | 49 | m | A | M | Normal | Negative | Survival; CCL2; IL-10 |
| 127 | 53 | m | A | M | 13q− | Negative | CD54 |
| 138 | 44 | m | B | UM | 13q− | Negative | CD54 |
| 139 | 60 | f | A | M | 13q−; 14q− | Negative | CD54 |
| 149 | 65 | f | A | M | Normal | Negative | Survival; CCL2; IL-10 |
| 150 | 61 | m | A | M | 13q- | Negative | CD54 |
| 151 | 66 | f | A | M | 13q− | Negative | Survival; CCL2 |
| 155 | 42 | m | A | M | Normal | Negative | Survival; CCL2 |
| 157 | 77 | m | A | M | 13q− | Negative | Survival; CCL2; IL-10 |
| 178 | 54 | m | nd | nd | Tris12, 13q−; 14q−; 17p− | nd | qRT-PCR |
| 180 | 49 | m | nd | nd | 13q bidel | nd | Survival; p-STAT1 |
| 194 | 63 | f | B | M | 13q− | nd | qRT-PCR |
| 195 | 62 | f | A | UM | Tris12; 17p− | Positive | Migration |
| 196 | 71 | m | A | M | 13q− | Negative | Survival; migration; p-STAT1 |
| 197 | 72 | f | nd | nd | Tris12 | nd | Survival; p-STAT1 |
| 199 | 79 | f | nd | nd | 17p−; 14q− | nd | Migration |
| 212 | 42 | m | A | M | Normal | Negative | qRT-PCR; p-STAT1 |
| 222 | 62 | m | C | M | 13q− | Negative | qRT-PCR |
| 229 | 61 | m | B | M | 13q− | nd | p-STAT1 |
| 238 | 60 | f | A | M | 13q−; 14q− | Negative | p-STAT1 |
| 245 | 62 | m | B | M | Normal | Negative | qRT-PCR; p-STAT1 |
| 260 | 43 | m | nd | M | Normal | nd | p-STAT1 |
| 263 | 60 | f | A | M | 13q−; 14q− | Negative | p-STAT1 |
| 264 | 53 | m | A | UM | nd | nd | p-STAT1 |
| 296 | 53 | m | B | M | 13q− | Negative | Migration |
| 299 | 55 | m | C | M | 13q− | Negative | p-STAT1 |
| 300 | 64 | f | C | M | 13q− | nd | Migration |
| Number . | Age (y) . | Gender . | Binet . | IGHV . | FISH . | ZAP70 . | Experiments performed . |
|---|---|---|---|---|---|---|---|
| 34 | 46 | m | A | UM | 12q+ | Positive | Survival; CCL2 |
| 39 | 59 | m | C | UM | 13q− | nd | Survival; CCL2 |
| 101 | 59 | f | A | M | 13q−; 14q− | Negative | Survival; CCL2; IL-10; HLA-DR |
| 102 | 49 | m | nd | nd | nd | nd | CD54 |
| 111 | 66 | m | A | UM | 11q−; 12p+; t(14q) | Positive | Survival; CCL2; IL-10; HLA-DR |
| 113 | 76 | m | nd | M | 13q− | Negative | CD54 |
| 114 | 67 | m | A | UM | 13q− | nd | CD54 |
| 115 | 62 | f | A | M | 13q− | nd | Survival; CCL2; IL-10; HLA-DR |
| 122 | 51 | m | B | M V3-53 | 13q− | Negative | Survival; CCL2; IL-10 |
| 123 | 76 | m | C | UM V1-69 | 11q−; 13q−; 14q− | Negative | Survival; CCL2; IL-10 |
| 124 | 49 | m | A | M | Normal | Negative | Survival; CCL2; IL-10 |
| 127 | 53 | m | A | M | 13q− | Negative | CD54 |
| 138 | 44 | m | B | UM | 13q− | Negative | CD54 |
| 139 | 60 | f | A | M | 13q−; 14q− | Negative | CD54 |
| 149 | 65 | f | A | M | Normal | Negative | Survival; CCL2; IL-10 |
| 150 | 61 | m | A | M | 13q- | Negative | CD54 |
| 151 | 66 | f | A | M | 13q− | Negative | Survival; CCL2 |
| 155 | 42 | m | A | M | Normal | Negative | Survival; CCL2 |
| 157 | 77 | m | A | M | 13q− | Negative | Survival; CCL2; IL-10 |
| 178 | 54 | m | nd | nd | Tris12, 13q−; 14q−; 17p− | nd | qRT-PCR |
| 180 | 49 | m | nd | nd | 13q bidel | nd | Survival; p-STAT1 |
| 194 | 63 | f | B | M | 13q− | nd | qRT-PCR |
| 195 | 62 | f | A | UM | Tris12; 17p− | Positive | Migration |
| 196 | 71 | m | A | M | 13q− | Negative | Survival; migration; p-STAT1 |
| 197 | 72 | f | nd | nd | Tris12 | nd | Survival; p-STAT1 |
| 199 | 79 | f | nd | nd | 17p−; 14q− | nd | Migration |
| 212 | 42 | m | A | M | Normal | Negative | qRT-PCR; p-STAT1 |
| 222 | 62 | m | C | M | 13q− | Negative | qRT-PCR |
| 229 | 61 | m | B | M | 13q− | nd | p-STAT1 |
| 238 | 60 | f | A | M | 13q−; 14q− | Negative | p-STAT1 |
| 245 | 62 | m | B | M | Normal | Negative | qRT-PCR; p-STAT1 |
| 260 | 43 | m | nd | M | Normal | nd | p-STAT1 |
| 263 | 60 | f | A | M | 13q−; 14q− | Negative | p-STAT1 |
| 264 | 53 | m | A | UM | nd | nd | p-STAT1 |
| 296 | 53 | m | B | M | 13q− | Negative | Migration |
| 299 | 55 | m | C | M | 13q− | Negative | p-STAT1 |
| 300 | 64 | f | C | M | 13q− | nd | Migration |
f, female; FISH, fluorescence in situ hybridization; m, male; M, mutated IGHV; nd, not determined; qRT-PCR, quantitative reverse-transcription PCR; UM, unmutated.
Cells were cultured in Dulbecco’s modified Eagle medium containing 4.5 g/L glucose and 4 mM L-glutamine (GibcoBRL/Invitrogen, Karlsruhe, Germany), 10% fetal calf serum (Biochrom AG, Berlin, Germany), and 1% penicillin/streptomycin (GibcoBRL/Invitrogen) at 37°C, 95% humidity, and 10% CO2. Primary cocultures were established using CD19-enriched CLL cells and CD14-enriched HD monocytes as indicated. Lenalidomide was added at a starting concentration of 10 µM at setup and added daily at a concentration of 5 µM. The cell culture medium was not changed during culture time.
Apoptotic cell death was analyzed by flow cytometry using annexin V phycoerythrin (PE) and 7-amino-actinomycin (7-AAD) as described before.9 Double-negative cells were counted as viable cells. All flow cytometry analyses were carried out using a FACSCanto II flow cytometer equipped with FACSDiva software (BD Biosciences, Heidelberg, Germany).
Expression analysis by flow cytometry
The abundance of cell surface proteins was quantified with the following antibodies and respective isotype controls: CD5-fluorescein isothiocyanate (FITC, clone L17F12), CD19-allophycocyanin (APC, clone HIB19), CD14-FITC (clone M5E2), HLA-DR-PE (clone G46-6), and respective controls from BD Biosciences; CD54-FITC (clone 84H10) from Immunotech (Marseille, France). Staining of cells was performed in 50 µL FACS buffer (1 × phosphate-buffered saline, 0.1% bovine serum albumin, 0.02% sodium azide) according to the manufacturer’s instructions. Relative median fluorescence intensities (MFI) were calculated as ratios of the MFI of a specific antibody over the MFI of the isotype control antibody.
Quantification of IL-10
Quantification of interleukin-10 (IL-10) was performed using Flex Set Bead Arrays from BD Biosciences by flow cytometry following the manufacturer’s protocol. Briefly, the cell culture supernatant and recombinant protein in known concentrations for the standard curve were mixed with IL-10–specific capture beads. Thereafter, beads were incubated with PE-labeled antibodies specific for IL-10 and fluorescence was quantified by flow cytometry.
Immunoblots
Cells were incubated with 10 µM lenalidomide, 50 ng/mL recombinant human (rh-)IL-10 (Miltenyi Biotec), or 10 µg/mL neutralizing IL-10–specific or isotype control antibodies (eBioscience, Frankfurt, Germany; clone JES3-9D7) and harvested by centrifugation at indicated time points. After washing with 1% NaVO4 in 1 × phosphate-buffered saline, cells were lysed in PathScan buffer (Cell Signaling Technology, Danvers, MA) containing benzonase (Merck, Darmstadt, Germany) and complete EDTA-free tablets (Roche Diagnostics, Mannheim, Germany). Subsequently, cellular debris was removed by centrifugation. Commercial NuPAGE Kit from Invitrogen (Karlsruhe, Germany) was used for SDS-PAGE (4%–12% gradient). For protein detection, membranes were hybridized with anti-human signal transducer and activator of transcription 1 (STAT1; 1:1000) or phospho-STAT1(Tyr701) (1:1000; clone 58D6; both Cell Signaling Technology) overnight at 4°C, followed by horseradish peroxidase–conjugated anti-rabbit immunoglobulin G (1:5000; Cell Signaling Technology) and visualization using ECL Blotting Detection Reagents (GE Healthcare, Chalfont St. Giles, UK). For loading control, anti-human α-tubulin (1:2000; clone DM1A; Sigma-Aldrich, Munich, Germany) or anti–glyceraldehyde-3-phosphate dehydrogenase (1:5000; clone 6C51; Merck Millipore) followed by HRP-conjugated anti-mouse immunoglobulin G (1:5000; Cell Signaling Technology) was used. Protein bands were quantified using Image J software and normalized to α-tubulin or glyceraldehyde-3-phosphate dehydrogenase values.
Quantitative real-time PCR
For RNA extraction, 2 × 107 CLL cells were lysed in TRIzol reagent. After adding chloroform and separation of phases by centrifugation, the aqueous phase was recovered, mixed 1:2 with 70% ethanol and applied to QIAGEN RNeasy Mikro columns (QIAGEN, Hilden, Germany) for cleanup. After washing, DNase digestion was performed on column. Finally, RNA was eluted in RNase-free H2O and quantified using a Nanodrop ND-100 spectrometer (Wilmington, DE).
Reverse transcription of mRNA was performed using Superscript II and anchored oligo-d(T)20 primer as formerly described.8 Real-time polymerase chain reaction (PCR) was conducted using an ABIPrism7900RT sequence detection system (Applied Biosystems, Foster City, CA) and SYBR green ROX mix as DNA intercalating dye. Calculation of efficiency and ratio to housekeeping genes was performed as described.9 Primer sequences for real-time PCR are listed in supplemental Table 1.
Chemotaxis assay
Chemotaxis assays were performed using transwells for 24-well plates with 5 µm pore size (Corning, NY) applying 3 × 106 CLL cells per well. After 1.5 hours, migrated cells were washed off the lower side of the membrane and counted using a ViCell counter XR 2.03 (Beckmann Coulter, Krefeld, Germany). Complete medium containing 200 ng/mL SDF1-α (Merck Millipore) or CCL19 (Miltenyi Biotech) was used as chemoattractant.
Statistical analyses
Statistical analyses were performed using SigmaPlot 12.0 Software (Systat, Chicago, IL). Data were tested for normal distribution and P values were calculated by Student t test (*P < .05, **P < .01, ***P < .001). Standard error of the mean (SEM) is depicted as error bars.
Results
Lenalidomide reduces survival support of NLCs in CLL cell cocultures
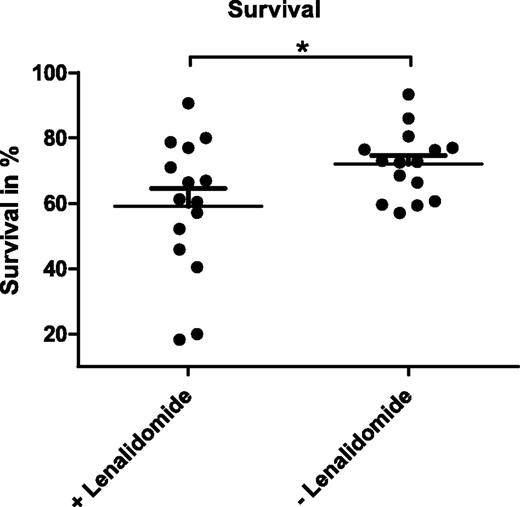
As an immunomodulatory drug, lenalidomide was described to induce apoptosis in CLL cells indirectly via the microenvironment.14 In line with that, we observed no reduction in viability of CD19-enriched CLL cells treated with lenalidomide in vitro (data not shown). To investigate the effects of this drug on myeloid cells within the tumor-supportive microenvironment, we established cocultures of CD19-enriched CLL cells obtained from 15 CLL patients and CD14-enriched monocytes from HD in the presence or absence of 10 µM lenalidomide. Over a culture period of 14 days, NLCs differentiated both in treated and untreated cocultures in comparable kinetics and numbers. After 14 days of coculture, all cells were harvested and the percentage of viable CLL cells was analyzed by annexin V-PE/7-AAD staining and flow cytometry. These analyses revealed that the mean survival rate of CLL cells was significantly reduced from 72.0% (SEM ±2.7) to 59.1% (SEM ±5.4) by the presence of lenalidomide (Figure 1; P = .0187) with a strong patient-to-patient variability of response. We therefore hypothesized that NLC-mediated survival support of CLL cells was impaired by lenalidomide treatment.
Survival of CLL cells in NLC cocultures in the presence and absence of lenalidomide. A total of 5 × 105 CLL cells enriched for CD19+ cells from 15 patients were cocultured with 1 × 105 monocytes isolated by CD14-specific MACS using PBMC of healthy donors in the presence or absence of 10 µM lenalidomide. Cells were cultured in 200 µL complete medium in 48-well plates for 14 days with a daily supplement of lenalidomide. Mean survival rates measured after annexin V-PE/7-AAD staining by flow cytometry of 3 technical replicates per sample are depicted. In addition, the mean value and SEM of all biological replicates within 1 group are indicated. Paired Student t test was performed for significance analysis (*P = .0187).
Survival of CLL cells in NLC cocultures in the presence and absence of lenalidomide. A total of 5 × 105 CLL cells enriched for CD19+ cells from 15 patients were cocultured with 1 × 105 monocytes isolated by CD14-specific MACS using PBMC of healthy donors in the presence or absence of 10 µM lenalidomide. Cells were cultured in 200 µL complete medium in 48-well plates for 14 days with a daily supplement of lenalidomide. Mean survival rates measured after annexin V-PE/7-AAD staining by flow cytometry of 3 technical replicates per sample are depicted. In addition, the mean value and SEM of all biological replicates within 1 group are indicated. Paired Student t test was performed for significance analysis (*P = .0187).
Lenalidomide alters immunophenotype and cytokine secretion of NLCs
Immune cells communicate and influence each other via the secretion of soluble factors. Therefore, we sought to quantify the abundance of cytokines that are known to be secreted by monocytes or macrophages in CLL/NLC cocultures. We selected CCL2 because it was identified as a deregulated protein in the CLL microenvironment,9 and IL-10, a cytokine that was described to be altered in the serum of CLL patients after lenalidomide treatment.12 Supernatants of primary CLL/NLC cocultures established from 12 patients in the presence or absence of 10 µM lenalidomide were collected after 14 days of culture. Quantification of CCL2 by enzyme-linked immunosorbent assay revealed significantly reduced levels of this chemokine in lenalidomide-treated cultures, showing mean levels of 15.8 ng/mL (SEM ±1.4) in control cultures and 13.4 ng/mL (SEM ±1.1) in treated cultures (P = .0006; supplemental Figure 1).
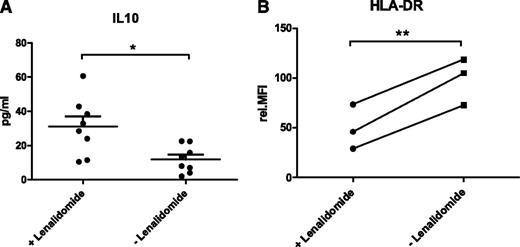
The abundance of IL-10 was quantified in the culture supernatant of 8 different CLL/NLC cocultures by flow cytometry using bead arrays. In contrast to CCL2, the concentration of IL-10 was significantly increased from 11.9 pg/mL (SEM ±2.8) to 31.2 pg/mL (SEM ±5.9) in the presence of lenalidomide (Figure 2A; P = .01).
Lenalidomide alters NLCs. (A) Secretion of IL-10 in CLL/NLC cocultures in the presence and absence of lenalidomide. Cell culture supernatants of cocultures setup as described in Figure 1 were used for the quantification of IL-10 (n = 8) by flow cytometry using bead arrays. Mean concentrations of 3 technical replicates per sample are depicted. In addition, the mean value and SEM of all biological replicates within one group are indicated. Paired Student t test was performed for significance analysis (*P = .01). (B) Expression of HLA-DR in NLCs differentiated in the presence and absence of lenalidomide. A total of 1 × 106 CD19-enriched CLL cells were cocultured with 1 × 106 CD14-enriched monocytes from HD in the presence or absence of 10 µM lenalidomide. Cells were cultured in 400 µL complete medium in 24-well plates for 14 days with daily addition of lenalidomide. NLCs were harvested by trypsin digestion and expression of HLA-DR was quantified by flow cytometry. Mean values of 3 technical replicates are depicted for each of the 3 CLL samples. Values of lenalidomide-treated and untreated samples of each patient are connected by lines. Paired Student t test was performed for significance analysis (**P = .009).
Lenalidomide alters NLCs. (A) Secretion of IL-10 in CLL/NLC cocultures in the presence and absence of lenalidomide. Cell culture supernatants of cocultures setup as described in Figure 1 were used for the quantification of IL-10 (n = 8) by flow cytometry using bead arrays. Mean concentrations of 3 technical replicates per sample are depicted. In addition, the mean value and SEM of all biological replicates within one group are indicated. Paired Student t test was performed for significance analysis (*P = .01). (B) Expression of HLA-DR in NLCs differentiated in the presence and absence of lenalidomide. A total of 1 × 106 CD19-enriched CLL cells were cocultured with 1 × 106 CD14-enriched monocytes from HD in the presence or absence of 10 µM lenalidomide. Cells were cultured in 400 µL complete medium in 24-well plates for 14 days with daily addition of lenalidomide. NLCs were harvested by trypsin digestion and expression of HLA-DR was quantified by flow cytometry. Mean values of 3 technical replicates are depicted for each of the 3 CLL samples. Values of lenalidomide-treated and untreated samples of each patient are connected by lines. Paired Student t test was performed for significance analysis (**P = .009).
We speculated that lenalidomide might influence the tumor-promoting characteristics of NLCs by reversing their immune-suppressive characteristics. Therefore, we quantified the abundance of the macrophage marker proteins CD206, CD163, and HLA-DR on the cell surface of NLCs after 14 days of coculture by flow cytometry. For CD163 and CD206, no difference in protein expression was detected between lenalidomide-treated and untreated NLCs (data not shown). However, the expression of HLA-DR was significantly reduced from a relative MFI of 98.6 (SEM ±13.5) to 49.5 (SEM ±12.8) in NLCs treated with lenalidomide (Figure 2B; P = .009).
Lenalidomide induces phosphorylation of STAT1 in CLL cells mediated by IL-10
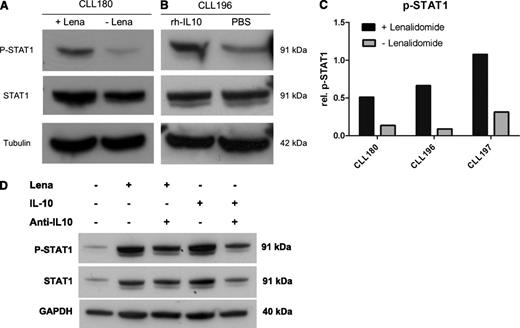
Because lenalidomide enhanced the secretion of IL-10 in CLL/NLC cocultures, we sought to investigate downstream effects of this cytokine in CLL cells. We tested for the activation of the transcription factor STAT1 because this was described as part of a proapoptotic response to IL-10 in CLL cells.20 Unsorted peripheral blood mononuclear cells (PBMC) of CLL patients were cultured for 3 days in the presence or absence of 10 µM lenalidomide and activation of STAT1 was monitored by immunoblotting using STAT1- and phospho-STAT1(Tyr701)–specific antibodies. Detection of α-tubulin protein served as loading control. In all cases tested (n = 3), we observed significantly higher levels of phosphorylated STAT1 in cells that were treated with lenalidomide, whereas the levels of total STAT1 protein remained stable (Figure 3A and 3C; P = .037). Respective immunoblot analyses using CLL cells that were CD19 sorted after culture verified that STAT1 was indeed phosphorylated in the malignant B cells and not in T cells or myeloid cells that were present in unsorted cell preparations in minor amounts (data not shown).
Activation of STAT1 in CLL cells upon lenalidomide or IL-10 treatment. (A) A total of 3 × 107 PBMC from CLL patients were cultured in the presence or absence of 10 µM lenalidomide in 4 mL complete medium in 6-well plates for 3 days. Cell lysates were analyzed by immunoblotting using anti-STAT1, anti-phospho STAT1(Tyr701), and anti-tubulin antibodies. One representative result out of 3 independently performed experiments is shown. (B) A total of 3 × 107 PBMC from CLL patients were cultured in the presence or absence of 50 ng/mL recombinant human (rh-)IL-10 in 4 mL complete medium in 6-well plates for 2 days. Immunoblot analysis was performed as described in panel A. One representative result out of 3 independently performed experiments is shown. (C) Corresponding to Figure 3A: Quantification of protein bands by ImageJ software revealed significantly stronger p-STAT1 signals in lenalidomide-treated samples (P = .037). (D) A total of 1 × 107 CLL PBMC were cultured for 3 days in the presence of 10 µM lenalidomide or 50 ng/mL IL-10 in combination with either 10 µg/mL anti–IL-10 or isotype control antibodies. All supplements were added to the medium at days 0 and 2. After 3 days of culture, cells were harvested and immunoblot analysis was performed as described in panel A.
Activation of STAT1 in CLL cells upon lenalidomide or IL-10 treatment. (A) A total of 3 × 107 PBMC from CLL patients were cultured in the presence or absence of 10 µM lenalidomide in 4 mL complete medium in 6-well plates for 3 days. Cell lysates were analyzed by immunoblotting using anti-STAT1, anti-phospho STAT1(Tyr701), and anti-tubulin antibodies. One representative result out of 3 independently performed experiments is shown. (B) A total of 3 × 107 PBMC from CLL patients were cultured in the presence or absence of 50 ng/mL recombinant human (rh-)IL-10 in 4 mL complete medium in 6-well plates for 2 days. Immunoblot analysis was performed as described in panel A. One representative result out of 3 independently performed experiments is shown. (C) Corresponding to Figure 3A: Quantification of protein bands by ImageJ software revealed significantly stronger p-STAT1 signals in lenalidomide-treated samples (P = .037). (D) A total of 1 × 107 CLL PBMC were cultured for 3 days in the presence of 10 µM lenalidomide or 50 ng/mL IL-10 in combination with either 10 µg/mL anti–IL-10 or isotype control antibodies. All supplements were added to the medium at days 0 and 2. After 3 days of culture, cells were harvested and immunoblot analysis was performed as described in panel A.
We hypothesized that IL-10 was responsible for the phosphorylation of STAT1. To validate this, we cultured PBMC from CLL patients for three days in the presence or absence of 50 ng/mL human recombinant IL-10 and performed immunoblots as described above. As expected, a comparable phosphorylation of STAT1 was observed in CLL cells in the presence of IL-10 (Figure 3B). In addition, we used neutralizing antibodies for IL-10 both in lenalidomide and IL-10–treated cultures and observed reduced STAT1 phosphorylation in the presence of the antibodies confirming the relevance of IL-10 in STAT1 phosphorylation in CLL cells (Figure 3D).
Subsequent immunoblot experiments using CD19-enriched CLL cells and unsorted PBMC from the same patients revealed that activation of STAT1 is more robust and in general stronger in the presence of accessory cells (see supplemental Results and supplemental Figures 2 and 3). In summary, these data suggest that IL-10, secreted mainly by myeloid cells, is responsible for STAT1 phosphorylation upon lenalidomide treatment.
Lenalidomide increases expression of ICAM-1 in CLL cells
STAT1 is known to regulate the expression of the adhesion receptor ICAM-1 (CD54),21 which together with its ligand LFA-1 (CD11a/CD18) is a critical player in lymphocyte migration and an important protein within immunological synapse formation.22 We therefore monitored the expression of these two cell surface proteins in cocultures of CLL cells and monocytes in the presence or absence of 10 µM lenalidomide by flow cytometry. Whereas LFA-1 levels, analyzed by CD11a-specific antibodies, were constantly low in CLL cells and did not change upon lenalidomide treatment, the expression of ICAM-1 increased after 2 days of coculture with monocytes from a mean relative MFI value of 2.3 (SEM ±0.44) in freshly isolated CLL cells to 3.6 (SEM ±0.39) after coculture (Figure 4A). Interestingly, lenalidomide treatment of the coculture induced a stronger increase of ICAM-1 expression with a mean relative MFI value of 5.8 (SEM ±0.90). Using cell preparations of 8 CLL patients, we observed a median increase of ICAM-1 expression of 1.6-fold (standard deviation ±0.29) by lenalidomide treatment (Figure 4B; P = .008). Quantification of ICAM-1 levels on monocytes revealed low expression on freshly isolated cells (relative MFI, 2.9) and an induction after coculture with CLL cells (relative MFI, 13.4). There was, however, no difference in the expression level of ICAM-1 in monocytes upon lenalidomide treatment (relative MFI, 14.1).
Expression of ICAM-1 in CLL and normal B cells upon lenalidomide treatment. (A) A total of 2 × 105 CD19-enriched CLL or normal B cells were cocultured with 2 × 105 monocytes isolated by CD14-specific MACS from PBMC of healthy individuals in 250 µL complete medium in 48-well plates for 2 days in the presence or absence of 10 µM lenalidomide. ICAM-1 (CD54) expression in CLL cells and B cells was quantified by flow cytometry before and after culture. Scatter plots including mean values and SEM of relative MFI obtained in 8 CLL samples and 3 HD B-cell samples by independently performed experiments are shown. Paired Student t test revealed a statistically significant difference between treated and untreated CLL cells (**P = .008). (B) Ratios of relative MFI values for ICAM-1 of treated versus untreated cells were calculated for each analyzed sample and are depicted as dot blots including mean values for CLL and HD B cells. Student t test revealed a statistically significant difference between the two cell types (**P = .005). rel., relative.
Expression of ICAM-1 in CLL and normal B cells upon lenalidomide treatment. (A) A total of 2 × 105 CD19-enriched CLL or normal B cells were cocultured with 2 × 105 monocytes isolated by CD14-specific MACS from PBMC of healthy individuals in 250 µL complete medium in 48-well plates for 2 days in the presence or absence of 10 µM lenalidomide. ICAM-1 (CD54) expression in CLL cells and B cells was quantified by flow cytometry before and after culture. Scatter plots including mean values and SEM of relative MFI obtained in 8 CLL samples and 3 HD B-cell samples by independently performed experiments are shown. Paired Student t test revealed a statistically significant difference between treated and untreated CLL cells (**P = .008). (B) Ratios of relative MFI values for ICAM-1 of treated versus untreated cells were calculated for each analyzed sample and are depicted as dot blots including mean values for CLL and HD B cells. Student t test revealed a statistically significant difference between the two cell types (**P = .005). rel., relative.
Respective coculture experiments were further performed with B cells of healthy individuals. The expression level of ICAM-1 on freshly isolated B cells (relative MFI, 2.8; SEM ±0.74) was comparable to the value of CLL cells (Figure 4A). However, the induced expression after coculture with monocytes was similar in lenalidomide-treated (relative MFI, 4.3; SEM ±0.84) and untreated (relative MFI, 4.2; SEM ±0.48) conditions (Figure 4B).
Lenalidomide alters the expression of migration-related genes in CLL cells
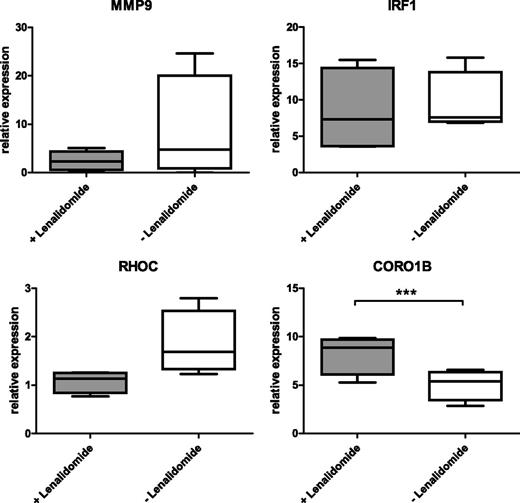
To further monitor downstream effects of lenalidomide treatment in CLL cells, we quantified the expression of known STAT1 (MMP9, IRF1) and putative lenalidomide target genes (RHOC, CORO1B) by quantitative real-time PCR. Therefore, PBMC from 4 CLL patients were cultured for 3 days in the presence or absence of 10 µM lenalidomide. Thereafter, cells were harvested and enriched for CD19+ cells. The obtained data show a mean downregulation of 2.7-fold (SEM ±0.75; P = .25) for MMP9 and of 1.7-fold (SEM ±0.28; P = .08) for RHOC in cells treated with lenalidomide (Figure 5). Whereas the expression of IRF1 showed a heterogeneous pattern, CORO1B was significantly upregulated in CLL cells treated with lenalidomide compared with untreated cells with a mean value of 1.7-fold (SEM ±0.07) and a P value of .001 (Figure 5).
Differential expression of migration-related genes upon lenalidomide treatment. A total of 1 × 107 PBMC from CLL patients were cultured in 400 µL complete medium in 24-well plates in the presence or absence of 10 µM lenalidomide for 3 days. Thereafter, CLL cells were enriched by CD19-specific MACS. Gene expression of MMP9, IRF1, RHOC, and CORO1B was analyzed by quantitative reverse-transcription PCR. Results of 4 independently analyzed patient samples were normalized to the expression of 2 housekeeping genes (PGK1, DCTN2) and are depicted as box plots. Whiskers depict 10th and 90th percentile of data. Paired Student t test was performed. CORO1B was significantly upregulated after lenalidomide treatment (***P = .001).
Differential expression of migration-related genes upon lenalidomide treatment. A total of 1 × 107 PBMC from CLL patients were cultured in 400 µL complete medium in 24-well plates in the presence or absence of 10 µM lenalidomide for 3 days. Thereafter, CLL cells were enriched by CD19-specific MACS. Gene expression of MMP9, IRF1, RHOC, and CORO1B was analyzed by quantitative reverse-transcription PCR. Results of 4 independently analyzed patient samples were normalized to the expression of 2 housekeeping genes (PGK1, DCTN2) and are depicted as box plots. Whiskers depict 10th and 90th percentile of data. Paired Student t test was performed. CORO1B was significantly upregulated after lenalidomide treatment (***P = .001).
Lenalidomide inhibits migration of CLL cells
Because we observed alterations in the expression of migration-related genes in CLL cells upon lenalidomide treatment, we sought to investigate whether lenalidomide affects the migratory potential of CLL cells. Therefore, CLL cells cultured for 3 days in the presence or absence of 10 µM lenalidomide were tested in migration assays using 200 ng/mL SDF1-α as chemoattractant. Quantification of migrated cells after 1.5 hours of incubation showed that lenalidomide treatment impaired migration of CLL cells, with an average fold change in the number of migrated cells of 0.5 (SEM ±0.08; n = 3; Figure 6). Cell viability was monitored in parallel and showed similar survival rates of approximately 60% in all samples (data not shown). FACS analyses of the SDF1-α receptor CXCR4 revealed no altered cell surface expression upon lenalidomide treatment (data not shown). Further, respective migration assays using 200 ng/mL CCL19 as chemoattractant revealed a comparable reduction in CLL cell migration upon lenalidomide treatment (data not shown), suggesting that the overall motility is impaired in CLL cells most likely by the observed alterations in the cytoskeleton.
Impaired SDF1-α-mediated migration of CLL cells upon lenalidomide treatment. A total of 1 × 107 PBMC from CLL patients were cultured in 400 µL complete medium in 24-well plates in the presence or absence of 10 µM lenalidomide for 3 days. The, 3 × 106 of these pretreated cells were seeded in the upper compartment of a transwell with 5 µm pore size. Complete medium containing 200 ng/mL SDF1-α was added to the lower compartment as chemoattractant. After 1.5 hours of incubation at 37°C, migrated cells were quantified using a ViCell counter. Mean values and SEM of 3 technical replicates of 1 representative example out of 3 independently performed experiments are shown.
Impaired SDF1-α-mediated migration of CLL cells upon lenalidomide treatment. A total of 1 × 107 PBMC from CLL patients were cultured in 400 µL complete medium in 24-well plates in the presence or absence of 10 µM lenalidomide for 3 days. The, 3 × 106 of these pretreated cells were seeded in the upper compartment of a transwell with 5 µm pore size. Complete medium containing 200 ng/mL SDF1-α was added to the lower compartment as chemoattractant. After 1.5 hours of incubation at 37°C, migrated cells were quantified using a ViCell counter. Mean values and SEM of 3 technical replicates of 1 representative example out of 3 independently performed experiments are shown.
Discussion
Similar to other cancers, CLL is associated with immune dysfunction, preventing antitumor immunity. Therefore, immunomodulatory drugs like lenalidomide that are known to promote cellular and innate immune activation are promising therapeutic options for this disease.23 Although the exact mechanism of action of lenalidomide is not well understood, it has been shown to reverse a T-cell immune synapse defect in CLL.17,18 In addition, lenalidomide treatment enhanced the levels of several inflammatory cytokines and induced expression of costimulatory molecules CD40, CD80, and CD86 in CLL cells.14,15 Furthermore, lenalidomide was shown to promote CD40L (CD154) expression in CLL cells resulting in an activation phenotype, and may therefore reverse the humoral immune defect observed in this disease.16
Previous studies revealed an important role for monocytes and NLCs in maintaining viability of CLL cells.4,8 Therefore, we aimed to investigate the effect of lenalidomide on the survival support of these cells. Because NLCs differentiate from CD14+ cells of healthy individuals in a similar way as from CLL-derived monocytes,24 we established cocultures of defined numbers of HD monocytes and CD19+ CLL cells. Treatment of these cocultures over 14 days with lenalidomide significantly reduced viability of CLL cells. The magnitude of response to lenalidomide varied from patient to patient, including samples that showed no reduction in CLL cell viability. This is in accordance with clinical data, which report a 47% response rate and 9% complete remission in relapsed patients treated with lenalidomide.25 Response rates in the NLC cocultures did not correlate with prognostic molecular features such as immunoglobulin heavy chain (IGHV) mutational status or genomic abnormalities of the patients.26,27 Most likely, the broad spectrum of response rates in vitro reflects the differences in the dependence of CLL cells on survival factors provided by NLCs. Of major interest for future studies will be the exploration whether response to lenalidomide in vitro correlates with treatment success in vivo. If this holds true, patients who will receive lenalidomide treatment could be selected based on the results of preceding in vitro drug testing.
Because lenalidomide neither affected viability of isolated CLL cells, nor of monocytes or NLCs in culture, we hypothesized that it impairs the survival support of NLCs. Because this support is mediated via intercellular contact as well as secreted cytokines,4,28 we investigated cell-surface protein expression and secretion of cytokines in CLL/NLC cocultures. Interestingly, lenalidomide treatment reduced secretion levels of CCL2 in all samples tested. We previously showed that CCL2 is highly upregulated in monocytes after contact with CLL cells.9 Because this chemokine is known to support survival of monocytes and to induce differentiation to tumor-promoting M2-type macrophages,29 it appears to be an important player in the CLL microenvironment. Because CCL2 is not expressed by CLL cells,9 lenalidomide treatment most likely affects the secretion of CCL2 in NLCs. This was confirmed by quantification of CCL2 expression in CD14-sorted cells after lenalidomide treatment by quantitative reverse-transcription PCR (data not shown).
In addition, lenalidomide treatment resulted in significantly increased levels of the immunosuppressive cytokine IL-10 that was shown to promote tumorigenesis in several cancers. However, there is also evidence that IL-10 has antitumor activity, because it was recently shown that IL-10–deficient mice are more prone to chemical carcinogenesis.30 These studies further showed that IL-10 inhibits the production of inflammatory cytokines and the development of myeloid-derived suppressor cells. More importantly, IL-10 was shown to induce apoptosis of CLL cells via activation of STAT1.20 Interestingly, lenalidomide treatment of CLL patients resulted in increased IL-10 serum levels,12 suggesting that the observed effect in CLL/NLC cocultures reflects the situation in vivo.
Because lenalidomide was shown to reverse immune deficiency of T cells,17 we sought to investigate whether it also effects immunocompetence of myeloid cells. Surprisingly, the expression of the major histocompatibility complex II protein HLA-DR was reduced on the surface of NLCs by lenalidomide treatment in our coculture setup, suggesting that the immune function of NLCs is not increased by this drug. This might be explained by the increased IL-10 level in the coculture, which is known to downregulate HLA-DR expression.31 Whether this observation reflects the effect of lenalidomide in vivo needs to be investigated with patient samples during and after treatment with this drug.
Because IL-10 was described to activate STAT1, we investigated the phosphorylation status of STAT1 in CLL cells and observed a clear induction of phosphorylation on Tyr701 after treatment with lenalidomide. This activation of STAT1 is most likely induced by IL-10, because recombinant IL-10 as well as Toll-like receptor 4 stimulation by lipopolysaccharide, which is known to induce IL-10 secretion in monocytes,32 resulted in an increase in STAT1 phosphorylation. Further, blocking of IL-10 with neutralizing antibodies reduced p-STAT1 levels.
The expression of the adhesion receptor ICAM-1 (CD54) is known to be regulated by STAT1.21 Consistent with that, we observed an upregulation of ICAM-1 in CLL cells upon lenalidomide treatment of cocultures. Interestingly, no increase of ICAM-1 expression was observed in healthy B cells in an equivalent setup. ICAM-1 is an important part of the immunologic synapse, the interface between antigen-presenting cells and lymphocytes.22 Ramsay et al17 previously showed that T cells from patients with CLL exhibit defective immunologic synapse formation with antigen-presenting cells and that lenalidomide was able to enhance both formation and function of the synapse between CLL and T cells. These findings were further validated in vivo by using a CLL mouse model.21 Higher ICAM-1 expression after lenalidomide treatment might therefore be involved in strengthening the immunologic synapse.
Activated STAT1 was shown to transcriptionally inhibit MMP9 expression in CLL cells.20 Accordingly, we observed downregulation of MMP9 after lenalidomide treatment in all cases tested. MMP9, which is frequently upregulated in CLL cells, is an important player in cell migration and was shown to directly promote survival of CLL cells.33,34 The downregulation of MMP9 by lenalidomide could therefore directly contribute to the impaired survival of CLL cells after lenalidomide treatment and/or influence the migratory potential of CLL cells.
Two putative lenalidomide targets, namely RHOC and CORO1B, are both associated with cell migration and were described to be upregulated in T cells after lenalidomide treatment.35-37 RHOC is an important player in cell invasion and migration and is together with RHOA responsible for cytoskeletal rearrangements.38 In our coculture setup, lenalidomide induced downregulation of RHOC in CLL cells in all cases tested, suggesting an impaired migration capability of the malignant cells. As described in T cells, CORO1B was strongly upregulated after lenalidomide treatment in CLL cells. CORO1B is involved in actin filament organization and was shown to negatively regulate smooth muscle cell migration.39
Finally, the presented transwell assays clearly showed that lenalidomide reduces the migration capability of CLL cells. Because this effect was observed using both SDF1-α and CCL19 as chemoattractants, and because CXCR4, the receptor for SDF1-α, was not downregulated upon treatment, we speculate that the motility of CLL cells is impaired by the drug. The observed expression changes in migration-related molecules and cytoskeletal structures (eg, RHOC and CORO1B) are most likely involved in this effect. In lenalidomide-treated patients, this might impact migration and homing of CLL cells to lymph nodes or other hematopoietic tissues. In line with that, a recently published study demonstrated that CLL cells from RhoH-deficient mice show reduced migration toward SDF1-α (CXCL12) and CXCL13 and therefore impaired bone marrow homing and engraftment.40 Of interest, this study further showed that in human CLL cells, lenalidomide treatment in vitro and in vivo resulted in reduced RHOH expression.
In summary, our data provide evidence that lenalidomide has an impact on the myeloid microenvironment in CLL by altering cell surface receptor expression and cytokine secretion of monocytes and NLCs. Thereby, the chronic inflammation characteristic of the CLL microenvironment is diminished, exemplified by the reduction of proinflammatory CCL2 and the induction of anti-inflammatory IL-10. These changes might impact on CLL cell viability directly and result in an altered expression of migration-related genes in CLL cells, thereby reducing their motility and homing within the bone marrow or lymph nodes.
The mode of action of lenalidomide in CLL patients is most likely complex, involving effects on various nonmalignant cells, like T cells, NK cells, or myeloid cells. An alteration of the cytokine milieu induced by this drug seems to be critical. Further, several groups have now independently shown that lenalidomide impacts on the cytoskeleton and on the migratory potential of cells, which might influence migration and homing of CLL cells in vivo and therefore be directly involved in the antitumor activity of the drug.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Daniel Mertens for helpful discussions and Sibylle Ohl for excellent technical assistance.
This work was supported by a grant from the German José Carreras Leukemia-foundation (DJCLS R 10/04) and by the Helmholtz Virtual Institute: Understanding, overcoming resistance to apoptosis, and therapy in leukemia; and by the research project of the German Federal Ministry of Education and Research CancerEpiSys. S.S. was supported by research grants from DFG (SFB1074 project B2) and Celgene (center of excellence). Lenalidomide was generously provided by Celgene (San Diego, CA).
Authorship
Contribution: A.S. designed and performed experiments, analyzed data, and wrote the manuscript; C.D. designed and performed experiments and analyzed data; T.Z., H.D., and S.S. provided patient samples and clinical data and contributed to the writing of the manuscript; P.L. provided logistic and budget support, supervised research, and contributed to the writing of the manuscript; M.S. designed experiments, supervised research, analyzed and approved data, provided budget support, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Martina Seiffert, Division of Molecular Genetics, German Cancer Research Center, Im Neuenheimer Feld 280, 69120 Heidelberg, Germany; e-mail: m.seiffert@dkfz.de.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal