Key Points
UBR5 is recurrently mutated in mantle cell lymphoma with a C-terminal cluster of deleterious mutations.
Abstract
We have recently reported the application of RNAseq to mantle cell lymphoma (MCL) transcriptomes revealing recurrent mutations in NOTCH1. Here we describe the targeted resequencing of 18 genes mutated in this discovery cohort using a larger cohort of MCL tumors. In addition to frequent mutations in ATM, CCND1, TP53, and NOTCH1, mutations were also observed recurrently in MEF2B, TRAF2, and TET2. Interestingly, the third most frequently mutated gene was UBR5, a gene encoding a 2799aa protein, with multiple functions, including E3 ligase activity based on a conserved cysteine residue at the C-terminus. Nonsynonymous mutations were detected in 18% (18/102) of tumors, with 61% of the mutations resulting in frameshifts in, or around, exon 58, predicted to result in the loss of this conserved cysteine residue. The recurrence and clustering of deleterious mutations implicate UBR5 mutations as a critical pathogenic event in a subgroup of MCL.
Introduction
Mantle cell lymphoma (MCL) accounts for 7% of non-Hodgkin lymphomas and represents a particularly challenging disease with patient outcomes inferior to most other lymphoma subtypes.1,2 MCL is characterized on the molecular level by the hallmark t(11;14)(q13;q32) translocation that results in overexpression of cyclin D1, and by frequent additional cytogenetic alterations.3 The complete mutational landscape has not been described to date. Using RNAseq as a discovery tool, we recently described recurrent NOTCH1 and CCND1 mutations.4 Herein we describe the targeted resequencing, in a larger cohort of patients, of mutated genes identified by RNAseq. We report frequent mutations in UBR5, a gene encoding an E3 ubiquitin-protein ligase that has not been implicated, thus far, in lymphomagenesis.5
Materials and methods
Sample selection and targeted capture sequencing
RNAseq was performed on the RNA from the tumors of 18 patients and 2 MCL cell lines (Mino and Jeko-1), as reported by Kridel et al.4 We selected 92 diagnostic MCL samples (supplemental Table 1), including 8 samples from patients that were part of the RNAseq discovery set, for targeted resequencing. A capture protocol was used to select specific target regions from a constructed multiplexed library of genomic DNA utilizing complementary DNA (cDNA) clones and PCR amplicons as baits (supplemental Table 2).
For the description of mutational recurrence and patterns, all 102 clinical cases were included. The study was approved by the University of British Columbia–British Columbia Cancer Agency Research Ethics Board and was conducted in accordance with the Declaration of Helsinki.
Variant calling and validation
Short read sequences were aligned using Burrows-Wheeler Aligner.6 To identify mutations, the Genome Analysis Toolkit v3 was used in adherence to the recommended best practices.7 Mutation calls have been validated using Sanger sequencing of genomic and/or cDNA, including constitutional DNA for 37 cases. cDNA sequence flanking exon 58 of UBR5 was analyzed in all 102 cases (supplemental Figure 1).
Results and discussion
As previously reported,4 we performed RNAseq on the MCL tumors from 18 patients and 2 MCL cell lines. In addition to the novel mutations reported for NOTCH1 and CCND1 and known somatic variants in ATM and TP53, mutations were also discovered in other genes involved in cell cycle, proliferation, and DNA damage response (eg, TRAF2, CDKN2A, CHEK2). To determine the incidence and spectrum of mutations in these genes, we performed targeted resequencing of 92 MCL tumors focusing on 18 genes (supplemental Table 2), based on preliminary results from the discovery cohort and proposed gene function. The total capture space was 335 512 bp of genomic DNA including exons, adjacent intronic sequence and untranslated regions (supplemental Table 2). Massively parallel genomic sequencing was performed achieving a mean coverage depth of 50× per sample, with 80% of targets covered at a depth of >16× (supplemental Figure 2).
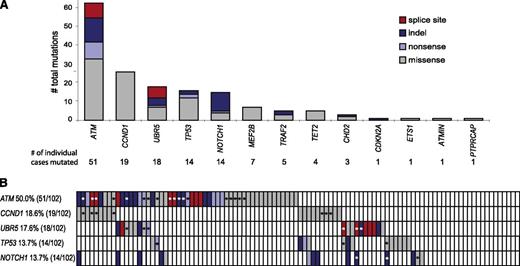
Combining the results from the 92 cases that were part of the targeted capture experiment and 10 additional cases from our RNAseq discovery set, we analyzed sequence alterations in 102 clinical MCL tumors. In 13 genes, we identified a total of 162 nonsilent variants including 32 indels, 102 missense, 13 nonsense, and 15 splice mutations (Figure 1A and supplemental Table 3). Constitutional DNA was available for 37 of the 102 cases and we were able to confirm that all 51 mutations present in these cases were somatic. The mutational frequencies of ATM, TP53, CCND1, and NOTCH1 were consistent with previously published reports.4,8-12 ATM was mutated in 50% (51/102) of MCL cases with a total of 63 nonsilent mutations observed: 9 nonsense mutations, 13 indels, 8 frameshift-inducing splice-site mutations, and 33 missense mutations. In CCND1, 26 nonsynonymous mutations were identified in 19 of 102 cases (19%), all clustering in exon 1 (supplemental Table 3). In addition to those missense mutations, we found 60 synonymous mutations spanning a region of less than 600 bp including the 5′ UTR, exon 1, and an adjacent 150 bp sequence of intron 1 (supplemental Table 4). The proximity of CCND1 to the IGH enhancer locus as a result of the translocation t(11;14), the clustering of variants within 2 kb of the transcriptional start site, and a high ratio of transitions in ratio to transversions (52:34) all suggest that these mutations arose through somatic hypermutation.13 In addition to confirming recurrent somatic mutations in known targets, mutations were observed in genes not previously reported to be mutated in MCL. MEF2B, TRAF2, and TET2 were found to be mutated in 7%, 5%, and 4% of cases, respectively (supplemental Table 3). No mutations were seen in CHEK1, CHEK2, CD14, CUL1, and RB1 in clinical specimens.
Mutations discovered by massive parallel sequencing of 102 MCL cases. (A) Total numbers of nonsynonymous mutations confirmed in 13 target genes analyzed. Next to the well-known and previously described mutations in ATM, TP53, and NOTCH1, UBR5 has the highest percentage of deleterious mutations including 7 splice-site affecting mutations. (B) The schematic represents the status of the 5 most recurrently mutated genes (rows) in MCL in 102 patients (columns) analyzed (red: splice site mutation; dark blue: indel; light blue: nonsense mutation; gray: missense mutation; and white: wild type. Asterisks highlight mutations validated as being somatic by Sanger sequencing.
Mutations discovered by massive parallel sequencing of 102 MCL cases. (A) Total numbers of nonsynonymous mutations confirmed in 13 target genes analyzed. Next to the well-known and previously described mutations in ATM, TP53, and NOTCH1, UBR5 has the highest percentage of deleterious mutations including 7 splice-site affecting mutations. (B) The schematic represents the status of the 5 most recurrently mutated genes (rows) in MCL in 102 patients (columns) analyzed (red: splice site mutation; dark blue: indel; light blue: nonsense mutation; gray: missense mutation; and white: wild type. Asterisks highlight mutations validated as being somatic by Sanger sequencing.
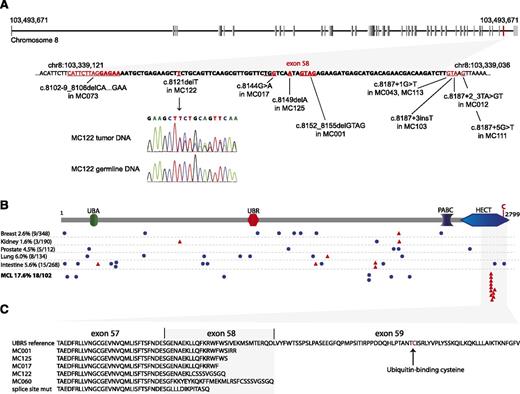
Interestingly, 18% (18/102) of the tumors had mutations in UBR5, a gene not previously described as being mutated in lymphoma, making it the third most frequently mutated gene in MCL. We initially found 1 UBR5 nonsense mutation (W2725Stop) and 1 UBR5 missense mutation (H1932P) in 18 clinical by RNAseq, both validated as being somatic. The capture sequencing approach further identified 2 1-base pair deletions, 5 splice-site, and 6 missense mutations in UBR5. The effect of splice-site mutations was investigated by sequencing corresponding cDNA samples, and all mutations were shown to cause defective splicing leading to frameshift and premature stop codons in the coding sequence. All deleterious (those predicted to produce a truncated protein) mutations in UBR5 gene were clustered within less than 100 bp of genomic sequence including exon 58 (Figure 2A). To identify more complex mutations within this region potentially missed by large scale sequencing approaches, we sequenced the cDNA from all cases. This resulted in the discovery of 3 additional mutations: 1 splice-site mutation, 1 4-bp deletion (missed by previous transcriptome sequencing analysis), and 1 38-bp intron retention causing a frameshift within exon 58. In summary, we found 11 deleterious and 7 missense mutations in UBR5 in 18 of the 102 cases analyzed. Six of 18 mutations (33%) involve splice sites inducing frameshifts, an event almost 3 times higher than comparable mutations in ATM (8/63). The distribution of frequently mutated genes in MCL across all clinical cases is displayed in Figure 1B, highlighting the mutual exclusivity of CCND1 and UBR5 in our cohort. We did not observe any statistically significant differences of patient and disease characteristics between patients harboring wild-type and mutated UBR5 (supplemental Table 1).
Details of UBR5 mutations observed in MCL and mutations reported in other cancers. (A) Top panel: intron-exon structure of UBR5 with Exon 58 highlighted in red. Bottom panel: details of deleterious mutations in the UBR5 gene are shown. All mutations are clustering within the genomic region of exon 58, the second last exon of the gene. The DNA sequence of exon 58 is indicated in bold. The chromatogram is showing a heterozygote 1-base pair deletion in MC122, the corresponding germline DNA sequence confirms this mutation as somatic alteration. (B) UBR5 protein structure with its ubiquitin-associated domain (UBA), UBR box (a zinc finger-like domain), polyadenylate-binding protein C terminus domain (PABC), and a C-terminal HECT domain. Mutations reported in other cancers23 are present in lower frequency and scattered throughout the protein. Blue circles: missense mutations; red triangles: deleterious mutations. All deleterious mutations found in MCL cluster within the C-terminal part of the HECT domain, upstream of the conserved cysteine residue at amino acid position 2768. (C) Protein alignment of truncated UBR5 mutant versions predicting a loss of ubiquitin-binding and transfer function.
Details of UBR5 mutations observed in MCL and mutations reported in other cancers. (A) Top panel: intron-exon structure of UBR5 with Exon 58 highlighted in red. Bottom panel: details of deleterious mutations in the UBR5 gene are shown. All mutations are clustering within the genomic region of exon 58, the second last exon of the gene. The DNA sequence of exon 58 is indicated in bold. The chromatogram is showing a heterozygote 1-base pair deletion in MC122, the corresponding germline DNA sequence confirms this mutation as somatic alteration. (B) UBR5 protein structure with its ubiquitin-associated domain (UBA), UBR box (a zinc finger-like domain), polyadenylate-binding protein C terminus domain (PABC), and a C-terminal HECT domain. Mutations reported in other cancers23 are present in lower frequency and scattered throughout the protein. Blue circles: missense mutations; red triangles: deleterious mutations. All deleterious mutations found in MCL cluster within the C-terminal part of the HECT domain, upstream of the conserved cysteine residue at amino acid position 2768. (C) Protein alignment of truncated UBR5 mutant versions predicting a loss of ubiquitin-binding and transfer function.
The significant recurrence and clustering of deleterious mutations implicate UBR5 mutations in the pathogenesis of MCL. The UBR5 gene is located on chromosome 8q22.3 encoding a 309 kDa homologous to E6-AP carboxyl terminus (HECT)-domain E3 ubiquitin ligase.14 HECT ligases reversibly bind ubiquitin via a conserved cysteine residue at amino acid position 2768 in the UBR5 protein, and directly transfer ubiquitin to the substrate. Several substrates including KATNA1 (katanin p60), TOPBP1, and PAIP2 have been shown to be ubiquitinated specifically by UBR5.15-17 Accumulation of katanin p60 due to knockdown of UBR5 leads to accumulation of tetraploid populations and polyploidy in affected cells.16 In addition to E3 ligase function, multiple protein-protein interactions with UBR5 have been described, suggesting roles in DNA damage response,18 cell-cycle control,19,20 miRNA-mediated gene silencing,21 and chromatin ubiquitination.22 In contrast to UBR5 mutations found in other cancers,23 all deleterious mutations discovered in MCL cluster within the HECT domain, predicted to result in the loss of the conserved cysteine residue (Figure 2B-C). Functional studies are in progress to determine the specific role of these heterozygous truncating mutations in the pathogenesis of MCL.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by a New Frontiers in Cancer Terry Fox program project grant (19001) (M.A.M., J.M.C., and R.D.G.). C.S. is supported by a Career Investigator award by the Michael Smith Foundation for Health Research.
Authorship
Contribution: B.M. and R.K. designed and performed the research, analyzed and interpreted data, and wrote the paper; R.S.L. and S.R. analyzed sequencing data; K.T. designed research; D.W.S. designed experiments and wrote the paper; R.M., A.J.M., and M.A.M. oversaw data collection and analysis; J.M.C. curated the lymphoma database, reviewed the manuscript, and provided editorial input; and C.S. and R.D.G. participated in the design of the original project, reviewed the manuscript, and provided editorial input.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Randy D. Gascoyne, Department of Pathology and Experimental Therapeutics, BC Cancer Agency & BC Cancer Research Centre, 675 W 10th Ave, Vancouver, BC, Canada V5Z 1L3; e-mail: rgascoyn@bccancer.bc.ca.
References
Author notes
C.S. and R.D.G. contributed equally to this study.