Key Points
The NPM1 mutant affects megakaryocytic development in mice.
NPMc+ mutant mice mimic some features of human NPM1-mutated AML.
Abstract
The NPM1 mutation is the most frequent genetic alteration thus far identified in acute myeloid leukemia (AML). Despite progress in the clinical and biological characterization of NPM1-mutated AML, the role of NPM1 mutation in leukemogenesis in vivo has not been fully elucidated. We report a novel mouse model that conditionally expresses the most common human NPM1 mutation (type A) in the hematopoietic compartment. In Npm1-TCTG/WT;Cre+ mice, the NPM1 mutant localized in the cytoplasm (NPMc+) of bone marrow (BM) cells. The mutant mice developed no AML after 1.5-year follow-up. However, NPMc+ expression determined a significant platelet count reduction and an expansion of the megakaryocytic compartment in the BM and spleen. Serum thrombopoietin levels overlapped in mutant vs control mice, and BM cells from Npm1-TCTG/WT;Cre+ mice formed more megakaryocytic colonies in vitro. Moreover, we demonstrated the up-regulation of microRNAs (miRNAs; miR-10a, miR-10b, and miR-20a) inhibiting megakaryocytic differentiation along with increased expression of HOXB genes. Notably, these findings mimic those of human NPM1-mutated AML, which also exhibits a similar miRNA profile and expansion of the megakaryocytic compartment. Our mouse model provides evidence that the NPM1 mutant affects megakaryocytic development, further expanding our knowledge of the role of NPM1 mutant in leukemogenesis.
Introduction
The NPM1 mutation is the most frequent genetic alteration thus far identified in acute myeloid leukemia (AML), accounting for ∼30% of all cases.1 This mutation is highly stable over time and defines a subgroup of AML with distinctive clinico-pathological features and immunophenotype (including negativity for CD34)2 and a relatively good prognosis in the absence of FMS-like tyrosine kinase 3 internal tandem duplication (FLT3-ITD) mutations.3,4 Moreover, NPM1-mutated AML associates with a unique gene expression profile (including up-regulation of HOX genes)5,6 and microRNA (miRNA) signature (including overexpression of miR10a, miR10b, and miR20a).7-10 These features and the recent demonstration that leukemic stem cells isolated from NPM1-mutated AML patients carry the NPM1 mutation11 point to the latter as a founder genetic event.12 Thus, NPM1-mutated AML is now listed as a provisional entity in the 2008 WHO classification of the lympho-hematopoietic neoplasms.13
The wild-type NPM1 protein shuttles between the nucleus and cytoplasm,14 but, at the immunohistochemistry level, it mainly concentrates in the nucleolus.15 Notably, all molecular variants of the NPM1 mutation12 result in critical changes at the C terminus of NPM116,17 that interfere with its nucleo-cytoplasmic traffic.18 This leads to the aberrant accumulation of nucleophosmin in the cytoplasm of the leukemic cells (thus the term NPM cytoplasmic positive NPMc+ AML).19 The cytoplasmic dislocation of nucleophosmin appears to be critical to its leukemogenic activity.18,20 The increased NPM1 export into the cytoplasm can affect multiple cellular pathways and may drive leukemia by either loss-of-function or gain-of-function mechanisms.18 Indeed, many NPM1 interactors such as p19Arf21 and Fbw7γ22 can be delocalized into the cytoplasm by NPM1c+, and their activity can be significantly impaired. Additionally, the NPM1 mutant has been described to interact and inhibit the cell death activity of caspase 6 and 8 in the cytoplasm.23
Despite the progresses in the biological and clinical characterization of NPM1-mutated AML,2 the role of NPM1 mutations in AML development in vivo is currently under investigation. A transgenic mouse model showed that the NPM1 mutant was implicated in abnormal myelopoiesis but not in progression toward leukemia.24 In zebrafish, ubiquitous NPM1 mutant expression caused primitive myeloid cells expansion and increased the number of definitive erythro-myeloid progenitors cells.25 Recently, Vassiliou et al26 reported a knock-in mouse of human NPM1 mutation A that, combined with insertional mutagenesis in vivo, identified genetic pathways collaborating with the NPM1 mutant in leukemogenesis. Nevertheless, the direct mechanism of action of the NPM1 mutant remains to be fully elucidated, and alternative gene-targeting approaches are still needed to study in vivo novel molecular mechanisms for NPM1-mutated AML.
To this end, we generated and characterized a mouse model that expresses the most frequent human NPM1 mutation (type A) in hematopoietic stem cells. Our results show that the NPM1 mutant affects megakaryocytic development in mice and mimics some features of human NPM1-mutated AML.
Methods
This study was approved by the Institutional Review Board and conducted in accordance with the Declaration of Helsinki.
Generation and genotyping of mice with human NPM1 mutation A
The human NPM1 mutation A mouse was generated in collaboration with genOway (Lyon, France). The targeting vector was obtained by inserting the transgenic cassette expressing the human NPM1 complementary DNA (cDNA) with mutation A into genOway's Rosa26 “quick Knock-in” targeting vector (G220 validation plasmid). This mutation consists of TCTG duplication at positions 956 to 959 of exon 12 of the reference sequence (GenBank accession number NM_002520) and represents the most frequent type of mutations of the NPM1 gene in AML (∼80% of cases).1 The NPM1 mutation cDNA fragment was subcloned into ApaI/HincII sites of a Tg-MSC plasmid containing the human growth hormone polyA. Then a MluI/SmaI fragment containing the NPM1 mutation was cloned into the G220 plasmid. The G220 vector already contained the CAG promoter (a fusion of the cytomegalovirus immediate early enhancer and the chicken β-actin promoter)27 with the downstream loxP flanked combined STOP-neomycin selection cassette.28 Southern blotting and polymerase chain reaction (PCR) analyses were performed to genotype mice. For a record of primers used, see supplemental Materials on the Blood website.
Induction of Mx1-Cre expression in Npm1-TCTG/WT
Mice with the NPM1 mutation A were bred with Mx1-Cre transgenic mice (Jackson Laboratory, Bar Harbor, ME) to generate Npm1-TCTG/WT;Cre+ mice. Expression of Mx1-Cre and excision of PGK-Neo cassette were induced by polyinosinic-polycytidylic acid (pIpC) treatment in vivo. In brief, 8- to 12-week-old Npm1-TCTG/WT;Cre+ mice were injected intraperitoneally with 250 μg/dose pIpC (Sigma-Aldrich, St. Louis, MO) every other day for 3 injections. Mice were analyzed starting 2 months after pIpC induction.
CD41 magnetic cell separation of megakaryocytes
Cells were stained with a fluorescein isothiocyanate (FITC) anti-mouse CD41 and then magnetically labeled with anti-FITC MicroBeads (Miltenyi Biotec, Auburn, CA). The cell suspension was loaded on a magnetic-activated cell sorting column placed in the magnetic field of a magnetic-activated cell sorting separator. The magnetically retained cells were eluted as the positively selected cell fraction. Samples were analyzed on a BD FACSCalibur flow cytometer (Becton Dickinson). Acquisition and data analysis were performed using CellQuest Pro software (Becton Dickinson). CD41+ cells purity was >90%.
qRT-PCR analysis
RNA was extracted with Trizol reagent and reverse-transcribed with RT-kit plus (Nanogene). Quantitative reverse transcriptase-PCR (qRT-PCR) was performed using SYBR Green PCR MasterMix and a 7900 Fast Real-Time PCR System (Applied Biosystems, Foster City, CA). PCR amplifications were performed by incubation at 95°C for 10 minutes, followed by 40 cycles of incubations at 95°C for 15 seconds and 60°C for 1 minute. Data were analyzed according to the comparative Ct method and normalized to glyceraldehyde-3-phosphate dehydrogenase expression levels within each sample. The 2-ΔΔCt method was used to calculate relative expression levels of target genes. Data analysis was performed using the SDS Enterprise Database (Applied Biosystems).
miRNA RT-PCR
Real-time PCR for mature miR-10a (Assay ID 000387), miR-10b (Assay ID 000388), and miR-20a (Assay ID 000580), and immature miR-10a (Assay ID Mm03307058_pri), miR-10b (Assay ID Mm03306401_pri), and miR-20a (Assay ID Mm03307063_pri) was performed using a standard TaqMan PCR kit protocol (Applied Biosystems). The reactions were incubated at 95°C for 10 minutes, followed by 40 cycles of 95°C for 15 seconds and 60°C for 1 minute. miRNA expression data were normalized to snoRNA202 (Assay ID 001232).
Immunoblotting analysis
NPM1 mutant protein was detected on lysates from 1 to 2 × 106 cells by western blot analysis with a rabbit polyclonal antibody against the mutated NPM1 protein.29 Lysates from the OCI/AML3 cell line, expressing the NPM1 mutant A protein,30 were used as control. A monoclonal anti-NPM1 antibody (Zymed) was used to recognize the endogenous mouse Npm1. Nuclear and cytoplasmic extracts were made using the Nuclear Extract Popper kit (Pierce).
Murine peripheral blood counts
Mice were anesthetized with isoflurane followed by retro-orbital bleeding. Blood was taken into glass capillary tubes. Complete blood count was performed using an XE-2100 hematology automated analyzer (Dasit).
Flow cytometry
Flow cytometric analysis was performed on bone marrow (BM) cells and splenocytes using the following monoclonal antibodies: biotin-conjugated antibodies against CD3, CD4, CD8, B220, Ter119, Mac-1, Gr-1, Sca-1, IL7R, CD5, phycoerythrin (PE)-Cy5–conjugated streptavidin, allophycocyanin-eFluor-780–conjugated anti–c-Kit, FITC-conjugated anti-CD41, FITC-conjugated anti–Ly-6A/E(Sca-1), anti-CD135 (eBioscience), allophycocyanin-conjugated anti-CD150, PE/CY7-conjugated anti-CD105 (BioLegend), and PE-conjugated anti-CD16/32 (BD Biosciences). Megakaryocytic progenitors (MKPs) were gated as Lin–c-Kit+Sca-1–CD150+CD41+ cells, and erythromegakaryocytic progenitor cells (Pre-MegE) were gated as Lin–c-Kit+Sca-1–CD41−CD105−CD150+ cells.31 Cell acquisition and analysis were performed on either a Cytomics FC500 cytometer equipped with the CXP analysis software 2.0 (Beckman Coulter) or a FACSCanto flow cytometer using the FACSDiva software (BD Biosciences). Gates were drawn to exclude nonviable cells and debris. Part of the flow cytometry data was analyzed with FlowJo software (Tree Star, Ashland, OR).
In vitro hematopoietic colony-forming assay
For MegaCult-C assays, a total of 105 BM mononuclear cells were used according to the manufacturer’s protocols (Stem-Cell Technologies, Vancouver, Canada); 50 ng/mL human thrombopoietin (TPO), 50 ng/mL human interleukin (IL)-11, 10 ng/mL murine IL-3, and 20 ng/mL human IL-6 (PeproTech, Rocky Hill, NJ) were used in these assays. The cultures were incubated at 37°C with 5% CO2 for 6 to 8 days. Colonies containing >3 megakaryocytes (AchE+ cells) were considered colony forming unit-megakaryocytes (CFU-MKs). Duplicate assays were performed for each mouse.
Serum TPO concentration measurement
The Quantikine murine TPO immunoassay kit (R&D Systems, Minneapolis, MN) was used to quantify TPO concentration in harvested sera.
Megakaryocytes in BM biopsies from NPM1-mutated AML patients
The number of megakaryocytes was assessed by immunohistochemistry in BM biopsies from 97 patients with NPM1-mutated AML. BM biopsies from 10 lymphoma patients that turned out to be normal at histologic examination were used as controls. Megakaryocytes labeling was performed in paraffin sections using a mouse monoclonal antibody against the linker for activation of T cells (LAT) protein (Dako) that is an excellent marker for megakaryocytes in BM biopsies.32 Care was taken to exclude from the count activated T cells that were distinguishable from megakaryocytes because of their weaker LAT positivity and different morphology. The antibody-antigen reaction was detected by an immuno-alkaline phosphatase procedure.19 The adjusted megakaryocytes concentration was determined by 2 independent investigators (PS and BF) with the following formula: (measured megakaryocytes concentration × 100)/percent cellularity.33 Histopathology images were acquired with the ×40/0.85 and ×4/0.16 objectives (Olympus U Plan Apo) of an Olympus BX61 microscope equipped with an Olympus DP71 digital camera, using the Olympus cell^B acquisition software.
Statistical analysis
The statistical significance of differences between different conditions was determined using Prism (GraphPad Software, La Jolla, CA) with a 2-tailed unpaired t test.
Results
Generation of conditional mice for the human NPM1 mutation A
We generated a novel NPM1 mutation mouse model by targeted insertion of the human NPM1 mutation A cDNA into the Rosa26 locus via homologous recombination in embryonic stem (ES) cells (Figure 1A). The targeting vector includes a combined STOP-neomycin selection cassette flanked by two loxP sites that can be removed on Cre induction, allowing for the expression of the NPM1 mutant under the control of a CAG promoter.
Generation of a conditional mouse model with the human NPM1 mutation A. (A) Strategy for targeted insertion of a human NPM1 mutation A cDNA into the murine ROSA26 genomic DNA locus. The figure describes the structure of the mouse ROSA26 locus (top), the targeting vector (upper middle), the targeted allele (lower middle), and Cre-mediated excised allele (bottom). The targeting vector was a 16-kb plasmid containing a transgenic cassette expressing the NPM1 mutant cDNA (NPM1; *position of the TCTG duplication), a combined loxP flanked STOP-Neomycin selection cassette, an inducible CAG promoter, and a negative DTA selection marker. NEO is excised through Cre-mediated recombination between the 2 loxP sites (triangles). Positions of the 5′ and 3′ probes used to confirm the homologous recombination and relevant restriction sites for Southern blot analysis are indicated. (B) Southern blotting analysis confirmed the presence of homologous recombination in Npm1-TCTG/WT mice (top) and ES cells (bottom). (C) PCR genotyping strategy for Npm-TCTG/WT mice to distinguish the wild-type allele (304 bp) from the knocked-in recombined allele (472 bp).
Generation of a conditional mouse model with the human NPM1 mutation A. (A) Strategy for targeted insertion of a human NPM1 mutation A cDNA into the murine ROSA26 genomic DNA locus. The figure describes the structure of the mouse ROSA26 locus (top), the targeting vector (upper middle), the targeted allele (lower middle), and Cre-mediated excised allele (bottom). The targeting vector was a 16-kb plasmid containing a transgenic cassette expressing the NPM1 mutant cDNA (NPM1; *position of the TCTG duplication), a combined loxP flanked STOP-Neomycin selection cassette, an inducible CAG promoter, and a negative DTA selection marker. NEO is excised through Cre-mediated recombination between the 2 loxP sites (triangles). Positions of the 5′ and 3′ probes used to confirm the homologous recombination and relevant restriction sites for Southern blot analysis are indicated. (B) Southern blotting analysis confirmed the presence of homologous recombination in Npm1-TCTG/WT mice (top) and ES cells (bottom). (C) PCR genotyping strategy for Npm-TCTG/WT mice to distinguish the wild-type allele (304 bp) from the knocked-in recombined allele (472 bp).
ES cells that had undergone homologous recombination, as determined by Southern blot analysis (Figure 1B), were microinjected into C57BL/6J blastocysts to generate chimeric mice. Following germ-line transmission from chimeric mice, the resulting heterozygous progeny (Npm1-TCTG/WT) was intercrossed and analyzed by Southern blot to confirm correct targeting of the transgene at the ROSA26 locus (Figure 1B). Npm1-TCTG/WT mice were viable and fertile, exhibited normal survival, and manifested no gross phenotypic abnormalities.
To study the effects of the NPM1 mutation in the hematopoietic compartment, the Npm1-TCTG/WT mice were crossed with transgenic Mx1-Cre mice. The Mx1 promoter was activated by pIpC treatment,34 leading to Cre expression in hematopoietic cells. Cre-mediated excision of the STOP-neomycin selection cassette was detectable in BM cells starting 1 month after pIpC induction (supplemental Figure 1). The offspring of Npm1-TCTG/WT;Cre+ crosses were born at Mendelian ratios.
Cre-dependent expression of NPM1 mutation in the hematopoietic compartment
We next sought to verify that NPM1 mutation A transgene expression could be induced in a Cre-dependent manner. For this purpose, we treated 6- to 12-week-old Npm1-TCTG/WT;Cre+ and Npm1-TCTG/WT;Cre− with pIpC and analyzed the mice 8 weeks later. The expression of the NPM1 mutation A mRNA was detected in BM cells (Figure 2A), spleen, and peripheral blood by quantitative real-time PCR using primers specific for the human NPM1 mutant.35 Moreover, we monitored transgene expression in peripheral blood samples, demonstrating that the NPM1 mutant mRNA levels were stable up to 24-month follow-up. Using an antibody specific for the NPM1 mutant,29 we confirmed that the mutant protein was present in Npm1-TCTG/WT;Cre+ mice in different hematopoietic tissues (Figure 2B). Npm1-TCTG/TCTG;Cre+ mice expressed approximately twofold higher mutant protein levels in BM cells compared with heterozygous Cre+ mice (Figure 2C) and a band intensity similar to the endogenous Npm1. Additional western blot analysis demonstrated the expression of the mutant protein in CD41+ megakaryocytic, Gr-1+ myeloid, and CD3/B220+ T- and B-lymphoid cells in the BM of Npm1-TCTG/WT;Cre+ mice (Figure 2D). Myeloid cells showed higher levels of the NPM1 mutant compared with the endogenous Npm1 protein, whereas CD41+ cells displayed an equal mutant/wild-type ratio. Nevertheless, no conclusions can be made about the absolute ratio of endogenous vs mutant NPM1 proteins, because different antibodies were used in western blotting experiments.
Expression levels of the NPM1 mutation A in mouse hematopoietic tissues. (A) RT-PCR detected transcription of the NPM1 mutant (NPMc+) mRNA in the BM of mutant mice 2 months after pIpC induction of the Cre. (B) Expression of the NPM1 mutant protein in peripheral blood lysates (left) and in hematopoietic tissues (right) using a specific antibody against the mutant. (C) Different expression levels of the NPM1 mutant in BM cells from heterozygous (Npm1-TCTG/WT;Cre+) vs homozygous (Npm1-TCTG/TCTG;Cre+) mice normalized to endogenous Npm1. (D) Western blot of magnetic-activated cell sorting–sorted CD41+ megakaryocytic (Mk), Gr-1+ myeloid (My), and CD3/B220+ T- and B-lymphoid (Ly) cells compared with whole BM of Npm1-TCTG/WT;Cre+ pIpC-induced mice. (E) NPM1 mutant protein expression in 3 different NPM1-mutated AML patients (P1, P2, and P3) compared with heterozygous (HET) and homozygous (HOMO) mutant mice BM. (F) Western blot analysis of NPMc+ expression on nuclear and cytoplasmic fractions of BM cells from heterozygous and homozygous mutant mice. C, cytoplasmic fraction; N, nuclear fraction; TL, total lysate.
Expression levels of the NPM1 mutation A in mouse hematopoietic tissues. (A) RT-PCR detected transcription of the NPM1 mutant (NPMc+) mRNA in the BM of mutant mice 2 months after pIpC induction of the Cre. (B) Expression of the NPM1 mutant protein in peripheral blood lysates (left) and in hematopoietic tissues (right) using a specific antibody against the mutant. (C) Different expression levels of the NPM1 mutant in BM cells from heterozygous (Npm1-TCTG/WT;Cre+) vs homozygous (Npm1-TCTG/TCTG;Cre+) mice normalized to endogenous Npm1. (D) Western blot of magnetic-activated cell sorting–sorted CD41+ megakaryocytic (Mk), Gr-1+ myeloid (My), and CD3/B220+ T- and B-lymphoid (Ly) cells compared with whole BM of Npm1-TCTG/WT;Cre+ pIpC-induced mice. (E) NPM1 mutant protein expression in 3 different NPM1-mutated AML patients (P1, P2, and P3) compared with heterozygous (HET) and homozygous (HOMO) mutant mice BM. (F) Western blot analysis of NPMc+ expression on nuclear and cytoplasmic fractions of BM cells from heterozygous and homozygous mutant mice. C, cytoplasmic fraction; N, nuclear fraction; TL, total lysate.
Finally, we looked at NPMc+ expression in mutant mice compared with blasts from 3 NPMc+ AML patients, and we found that NPMc+ protein levels in the homozygous mice paralleled those of NPMc+ in human AML (Figure 2E). Therefore, our data show that expression of NPM1 mutant protein in BM cells and isolated hematopoietic cells is comparable to that of WT protein encoded by the endogenous locus and increases in a dose-dependent fashion in Npm1-TCTG/TCTG;Cre+.
Nucleophosmin dislocation into cytoplasm is a functional consequence of NPM1 mutation in human AML and appears to be a critical leukemogenic event.18 Evidence of cytoplasmic nucleophosmin in BM cells including megakaryocytes could not be obtained by immunohistochemistry because of the cross-reactivity of the anti-human NPM antibodies with mice tissues. In particular, nonspecific cytoplasmic positivity of megakaryocytes was observed with these antibodies both in control and mutant mice. Thus, we performed western blot analysis on nuclear and cytoplasmic fractions lysates that clearly showed the accumulation of the NPM1 mutant in the cytoplasm of BM cells from Npm1-TCTG/WT;Cre+ mice (Figure 2F).
Platelet numbers are markedly reduced in Npm1-TCTG/WT;Cre+ mice
After pIpC treatment, complete peripheral blood counts were monitored in Npm1-TCTG/WT;Cre+ and age-matched Npm1-TCTG/WT;Cre− littermate controls for 18 months. Strikingly, mice expressing the NPM1 mutation A showed a fully penetrant phenotype in the peripheral blood, characterized by significantly lower platelet counts compared with Npm1-TCTG/WT;Cre− pIpC-treated controls (Figure 3A). Platelets were one-half of the control count in mutants harboring 1 conditional allele and one-quarter in homozygous Npm1-TCTG/TCTG;Cre+ mice (P < .001; Figure 3B). Moreover, the few circulating platelets were considerably larger than normal in mutant mice as shown by the increased mean platelet volume (8.07 ± 0.39 vs 6.250 ± 0.13 femtoliter; n = 4, P < .01; Figure 3Ci). Npm1-TCTG/WT;Cre+ also displayed increased platelet distribution width (8.55 ± 0.98 vs 5.25 ± 0.11 femtoliter; n = 4, P < .05) and platelet-large cell ratio (15.18 ± 2.88% vs 2.42 ± 1.06%; n = 4, P < .01) compared with Npm1-TCTG/WT;Cre− mice (Figure 3Cii-iii). Peripheral blood smears confirmed both the paucity of platelets in Npm1-TCTG/WT;Cre+ relative to littermate Cre− controls and their substantially larger size (Figure 3Civ). Mutant mice showed a tendency to prolonged bleeding times (supplemental Figure 2). Complete blood count analysis revealed no significant changes in white blood cell counts and hemoglobin levels (Figure 3A; supplemental Table 1). Almost all the animals survived to 1.5 years of age without gross evidence of disease.
Thrombocytopenia of Cre-induced mice harboring the mutated NPM1 cDNA transgenic cassette. (A) Peripheral blood cell counts of Npm1-TCTG/WT;Cre+ (n = 9) and Npm1-TCTG/TCTG;Cre+ (n = 8) vs control mice (n = 9; including 3 Npm1-WT/WT;Cre+, 3 Npm1-TCTG/WT;Cre−, and 3 Npm1-TCTG/TCTG;Cre− pIpC-treated mice). (B) Significantly decreased platelets number in peripheral blood of both heterozygous and homozygous conditional mice 1 month after pIpC injection. Results are shown as mean ± standard deviation (error bars) from 13 Npm1-TCTG/WT;Cre+ and 10 Npm1-TCTG/TCTG;Cre+ vs 25 control mice (including Npm1-WT/WT;Cre+ [n = 9], Npm1-TCTG/WT;Cre− [n = 9], and Npm1-TCTG/TCTG;Cre− [n = 7] pIpC-treated mice). (C) Enlarged platelet size in Npm1-TCTG/WT;Cre+ mice (N = 4) is shown as increased (i) mean platelet volume, (ii) platelet distribution width, and (iii) platelet-large cell ratio. (iv) Peripheral blood smears showing both the paucity of platelets and their substantially larger size (arrows) in heterozygous mutant mice vs littermate Cre− pIpC-treated controls. *P < .05, **P < .01, ***P < .001 (unequal-variance t test).
Thrombocytopenia of Cre-induced mice harboring the mutated NPM1 cDNA transgenic cassette. (A) Peripheral blood cell counts of Npm1-TCTG/WT;Cre+ (n = 9) and Npm1-TCTG/TCTG;Cre+ (n = 8) vs control mice (n = 9; including 3 Npm1-WT/WT;Cre+, 3 Npm1-TCTG/WT;Cre−, and 3 Npm1-TCTG/TCTG;Cre− pIpC-treated mice). (B) Significantly decreased platelets number in peripheral blood of both heterozygous and homozygous conditional mice 1 month after pIpC injection. Results are shown as mean ± standard deviation (error bars) from 13 Npm1-TCTG/WT;Cre+ and 10 Npm1-TCTG/TCTG;Cre+ vs 25 control mice (including Npm1-WT/WT;Cre+ [n = 9], Npm1-TCTG/WT;Cre− [n = 9], and Npm1-TCTG/TCTG;Cre− [n = 7] pIpC-treated mice). (C) Enlarged platelet size in Npm1-TCTG/WT;Cre+ mice (N = 4) is shown as increased (i) mean platelet volume, (ii) platelet distribution width, and (iii) platelet-large cell ratio. (iv) Peripheral blood smears showing both the paucity of platelets and their substantially larger size (arrows) in heterozygous mutant mice vs littermate Cre− pIpC-treated controls. *P < .05, **P < .01, ***P < .001 (unequal-variance t test).
Megakaryocytes expansion and block of differentiation in Npm1-TCTG/WT;Cre+ mice
To investigate whether the reduced platelet number in the Npm1-TCTG/WT;Cre+ mice might result from abnormal hematopoiesis, we performed flow cytometric analysis of BM cells. For this purpose, we used CD41 as a marker that is expressed throughout megakaryocyte differentiation from early progenitors to mature megakaryocytes and platelets.36 Interestingly, there was a significant increase of more than twofold in the percentage of CD41+ cells in Npm1-TCTG/WT;Cre+ BM and close to fourfold in Npm1-TCTG/TCTG;Cre+ samples compared with control mice (5.28 ± 3.57% vs 7.94 ± 2.4% vs 2.52 ± 1.4%, respectively; Figure 4A-B). This difference remained significant after adjusting for BM cellularity, which was similar along the 3 groups (Figure 4C). CD41+ cells also accumulated in the spleen of Npm1-TCTG/WT;Cre+ and Npm1-TCTG/TCTG;Cre+ mice, most likely as a result of extramedullary hematopoiesis contributing to a mild splenomegaly (Figure 4D-E). Interestingly, serum TPO levels did not differ between heterozygous, homozygous mutants, and control mice (Figure 4F). Altogether, these findings suggest that the expression of the NPM1 mutant determines a block of megakaryocytes differentiation rather than an increased number of megakaryocytes secondary to platelets destruction.
Expansion of the CD41+ megakaryocytes in Npm1-TCTG/WT;Cre+ and Npm1-TCTG/TCTG;Cre+ mice. (A) Flow cytometric analysis of single-cell suspensions of BM from representative Npm1-TCTG/WT;Cre+, Npm1-TCTG/TCTG;Cre+, and Npm1-TCTG/WT;Cre− pIpC-treated control mice demonstrates an increase in the percentage of CD41+ megakaryocytic cells. (B) Quantification of CD41+ megakaryocytic cells in the BM of age-matched mutant mice analyzed as in panel A: Npm1-TCTG/WT;Cre+ (n = 11) and Npm1-TCTG/TCTG;Cre+ (n = 6); CTRL indicates control mice including Npm1-WT/WT;Cre+ (n = 3), Npm1-TCTG/WT;Cre− (n = 7), and Npm1-TCTG/TCTG;Cre− (n = 2) pIpC-treated mice. (C) BM cellularity in Npm1-TCTG/WT;Cre+ (n = 11), Npm1-TCTG/TCTG;Cre+ (n = 6), and control mice (genotypes and numbers are the same as in panel B). (D) Percentage of CD41+ megakaryocytic cells in the spleen of heterozygous (n = 10) and homozygous (n = 6) mutant mice compared with pIpc-treated Cre− controls (n = 6). (E) Composite data from age-matched littermates of indicated genotypes demonstrating mild hypersplenism in Npm1-TCTG/WT;Cre+ and Npm1-TCTG/TCTG;Cre+ mice. (F) Comparison of serum TPO concentration between Cre− pIpC control mice, Npm1-TCTG/WT;Cre+, and Npm1-TCTG/TCTG;Cre+ (n = 12 per genotype). *P < .05, **P < .01, ***P < .001 (unequal-variance t test). n.s. = not statistically significant.
Expansion of the CD41+ megakaryocytes in Npm1-TCTG/WT;Cre+ and Npm1-TCTG/TCTG;Cre+ mice. (A) Flow cytometric analysis of single-cell suspensions of BM from representative Npm1-TCTG/WT;Cre+, Npm1-TCTG/TCTG;Cre+, and Npm1-TCTG/WT;Cre− pIpC-treated control mice demonstrates an increase in the percentage of CD41+ megakaryocytic cells. (B) Quantification of CD41+ megakaryocytic cells in the BM of age-matched mutant mice analyzed as in panel A: Npm1-TCTG/WT;Cre+ (n = 11) and Npm1-TCTG/TCTG;Cre+ (n = 6); CTRL indicates control mice including Npm1-WT/WT;Cre+ (n = 3), Npm1-TCTG/WT;Cre− (n = 7), and Npm1-TCTG/TCTG;Cre− (n = 2) pIpC-treated mice. (C) BM cellularity in Npm1-TCTG/WT;Cre+ (n = 11), Npm1-TCTG/TCTG;Cre+ (n = 6), and control mice (genotypes and numbers are the same as in panel B). (D) Percentage of CD41+ megakaryocytic cells in the spleen of heterozygous (n = 10) and homozygous (n = 6) mutant mice compared with pIpc-treated Cre− controls (n = 6). (E) Composite data from age-matched littermates of indicated genotypes demonstrating mild hypersplenism in Npm1-TCTG/WT;Cre+ and Npm1-TCTG/TCTG;Cre+ mice. (F) Comparison of serum TPO concentration between Cre− pIpC control mice, Npm1-TCTG/WT;Cre+, and Npm1-TCTG/TCTG;Cre+ (n = 12 per genotype). *P < .05, **P < .01, ***P < .001 (unequal-variance t test). n.s. = not statistically significant.
To further characterize the megakaryocytic compartment in mutant mice, we assessed the presence of early MKPs cells in the BM. Lin–Kit+Sca-1–CD150+CD41+ MKP cells were increased twofold in Npm1-TCTG/WT;Cre+ mice and up to fourfold in Npm1-TCTG/TCTG;Cre+ compared with Cre− controls (0.15 ± 0.07% vs 0.24 ± 0.15% vs 0.05 ± 0.02%, respectively; Figure 5A-B), suggesting that NPM1 mutant A expression drives the expansion of immature megakaryocytes. There was no significant difference in the percentage of Lin–Kit+Sca-1+ (LSK) cells and pre-MegE cells in the BM of the 3 animal groups (Figure 5C). To better characterize the perturbation in megakaryocyte growth, we subsequently conducted in vitro colony-forming assays. Interestingly, Npm1-TCTG/WT;Cre+ BM cells formed significantly more megakaryocytic colonies (CFU-MK) than Cre− control in semisolid media (34.00 ± 5.367 vs 9.0 ± 1.265 CFU-MK/105 BM cells; n = 6, P < .01; Figure 5E), whereas CFU-MK sizes were similar (Figure 5F).
Expansion of the immature megakariocytes in Npm1-TCTG/WT;Cre+ and Npm1-TCTG/TCTG;Cre+ mice. (A) Flow cytometric analysis of single-cell suspensions of BM from representative Npm1-TCTG/WT;Cre+, Npm1-TCTG/TCTG;Cre+, and Npm1-TCTG/WT;Cre− pIpc-treated control mice demonstrates an increase in the percentage of Lin–Kit+Sca-1–CD150+CD41+ megakaryocytic progenitor populations. (B) Quantification of Lin–Kit+Sca-1–CD150+CD41+ MKPs in the BM of age-matched mutant mice analyzed as in panel A (n = 8 per genotype). (C) No differences in the percentage of Lin–Kit+Sca-1+ cells and Lin–Kit+Sca-1–CD41–CD150+FcgR–CD105lo erythromegakaryocytic progenitor cells in mutant Cre+ vs Cre− pIpC-treated control mice (n = 6 per genotype). (D) BM cells from Npm1-TCTG/WT;Cre+, Npm1-TCTG/TCTG;Cre+, and Cre− pIpC-treated mice were plated on M3434 methylcellulose medium (containing stem cell factor, IL-3, IL-6, erythropoietin) and scored for colony formation 7 to 10 days later (BFU-E, burst-forming unit-erythroid; GEMM, granulocyte, erythroid, monocyte, megakaryocyte; GM, granulocyte monocyte; M, monocyte). Results are the average of 3 independent experiments performed in duplicate (mean ± standard deviation are shown). (E) CFU-MK potential from total BM (n = 3 per genotype). (F) Photo of representative colonies (×10 magnification) from (ii) Npm1-TCTG/WT;Cre+ compared with (i) Npm1-TCTG/WT;Cre−. (G) Summary of the alterations in the development of the megakaryocytes after the expression of the NPM mutation A in the hematopoietic compartment. **P < .01; n.s. = not statistically significant.
Expansion of the immature megakariocytes in Npm1-TCTG/WT;Cre+ and Npm1-TCTG/TCTG;Cre+ mice. (A) Flow cytometric analysis of single-cell suspensions of BM from representative Npm1-TCTG/WT;Cre+, Npm1-TCTG/TCTG;Cre+, and Npm1-TCTG/WT;Cre− pIpc-treated control mice demonstrates an increase in the percentage of Lin–Kit+Sca-1–CD150+CD41+ megakaryocytic progenitor populations. (B) Quantification of Lin–Kit+Sca-1–CD150+CD41+ MKPs in the BM of age-matched mutant mice analyzed as in panel A (n = 8 per genotype). (C) No differences in the percentage of Lin–Kit+Sca-1+ cells and Lin–Kit+Sca-1–CD41–CD150+FcgR–CD105lo erythromegakaryocytic progenitor cells in mutant Cre+ vs Cre− pIpC-treated control mice (n = 6 per genotype). (D) BM cells from Npm1-TCTG/WT;Cre+, Npm1-TCTG/TCTG;Cre+, and Cre− pIpC-treated mice were plated on M3434 methylcellulose medium (containing stem cell factor, IL-3, IL-6, erythropoietin) and scored for colony formation 7 to 10 days later (BFU-E, burst-forming unit-erythroid; GEMM, granulocyte, erythroid, monocyte, megakaryocyte; GM, granulocyte monocyte; M, monocyte). Results are the average of 3 independent experiments performed in duplicate (mean ± standard deviation are shown). (E) CFU-MK potential from total BM (n = 3 per genotype). (F) Photo of representative colonies (×10 magnification) from (ii) Npm1-TCTG/WT;Cre+ compared with (i) Npm1-TCTG/WT;Cre−. (G) Summary of the alterations in the development of the megakaryocytes after the expression of the NPM mutation A in the hematopoietic compartment. **P < .01; n.s. = not statistically significant.
Overall, our findings strongly suggest the NPM1 mutant blocks the differentiation of megakaryocytes with a consequent decrease in production of mature platelets (Figure 5G).
Features of myeloproliferation in Npm1-TCTG/WT;Cre+ mice
Flow cytometric and additional pathological analysis revealed that ∼15% of Npm1-TCTG/WT;Cre+ mice displayed features of myeloproliferation in the BM and spleen. In particular, BM samples from Npm1-TCTG/WT;Cre+ mice showed an increased number of mature Gr1+Mac1+ myeloid cells and decreased number of B220 B cells compared with Cre− control mice (84.31 ± 7.55% vs 55.24 ± 14.75% and 4.35 ± 3.62% vs 19.86 ± 6.18%, respectively; supplemental Figure 3A-B). This was further confirmed by histologic analysis of spleen sections, where expansion of myeloid cells was observed in the red pulp of Npm1-TCTG/WT;Cre+ mice (supplemental Figure 4), along with an increased megakaryocyte number. In addition, the number of CFU-GM colonies derived from BM cells of Npm1-TCTG/WT;Cre+ mice showed a trend toward increase compared with Cre− mice that was significant for colonies in Npm1-TCTG/TCTG;Cre+ mice (Figure 5D). However, histologic analysis of BM sections failed to reveal significant morphologic differences between the mutant and control mice. No age-matched Npm1-TCTG/WT;Cre− controls showed signs of disease development. Altogether, these data support evidence of the presence of a myeloid defect in a fraction of NPMc+ mutant mice and are consistent with previous reports.24,26
Overexpression of NPM1 mutant induces miR-10a, miR-10b, and miR-20a in vivo
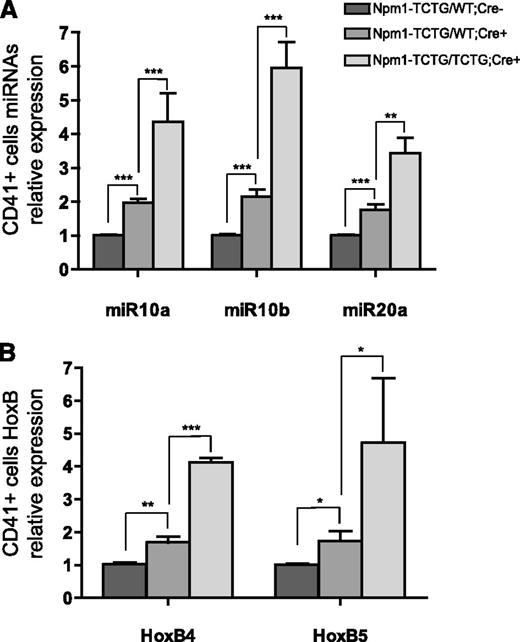
Recent findings suggest a role for miRNAs during megakaryopoiesis where miR-10a, miR-10b, and miR-20a appear to be down-regulated as the cell commits to mature megakaryocytes.37,38 Hypothetically, the down-regulation of miRNAs unblocks the expression of target genes involved in megakaryocyte differentiation. Thus, we analyzed whether the expression of miR-10a, miR-10b, and miR-20a was altered in our mutant mice. Strikingly, both heterozygous and homozygous Cre+ mutant mice displayed significant miRNAs overexpression compared with Cre− controls. The expression of miR-10a, miR-10b and miR-20a in CD41+ cells from Npm1-TCTG/WT;Cre+ vs Npm1-TCTG/TCTG;Cre+ mice increased 1.96-, 2.15-, and 1.69-fold vs 4.36-, 5.96-, and 2.88-fold respectively, compared to Npm1-TCTG/WT;Cre− mice (Figure 6A).
miR-10a, miR10b, and miR20a overexpression correlates with HOXB gene expression. (A) miRNA qRT-PCR for miR-10a, miR-10b, and miR-20a in CD41+ cells from Npm1-TCTG/WT;Cre+ and Npm1-TCTG/TCTG;Cre+ mice vs Cre− controls. (B) Relative gene expression of HoxB4 and HoxB5 in the BM of Npm1-TCTG/WT;Cre+ and Npm1-TCTG/TCTG;Cre+ mice vs Cre− controls. qRT-PCR was performed on pooled CD41+ magnetic-activated cell sorting–sorted cells from ≥3 mice per genotype. Histograms represent mean miRNA expression from triplicate samples obtained from 3 independent experiments. *P < .05, **P < .01, ***P < .001 (unequal-variance t test).
miR-10a, miR10b, and miR20a overexpression correlates with HOXB gene expression. (A) miRNA qRT-PCR for miR-10a, miR-10b, and miR-20a in CD41+ cells from Npm1-TCTG/WT;Cre+ and Npm1-TCTG/TCTG;Cre+ mice vs Cre− controls. (B) Relative gene expression of HoxB4 and HoxB5 in the BM of Npm1-TCTG/WT;Cre+ and Npm1-TCTG/TCTG;Cre+ mice vs Cre− controls. qRT-PCR was performed on pooled CD41+ magnetic-activated cell sorting–sorted cells from ≥3 mice per genotype. Histograms represent mean miRNA expression from triplicate samples obtained from 3 independent experiments. *P < .05, **P < .01, ***P < .001 (unequal-variance t test).
To demonstrate that altered miRNAs expression was directly related to NPM1 shuttling dysfunction, we measured immature to mature miRNAs ratios in Npm1-TCTG/WT;Cre+ mice compared with Cre− controls. The NPM1 mutant is associated with a significant reduction of the immature to mature miR-10a and miR10b ratio because of a decrease in miRNA precursors coupled with a concomitant increase in mature miRNA level (supplemental Figure 5). Moreover, miR10a, 10b, and 20a levels significantly increased 1.55-, 3.07-, and 1.27-fold, respectively, in NIH-3T3 cells overexpressing the NPM1 cDNA mutant (supplemental Figure 6), suggesting that mutant NPM1 expression itself causes an increased miRNA level, which is not simply a consequence of immature megakaryocytes expansion.
It has been reported that in mouse embryonic development, miR-10a and miR-10b expression closely follows the expression of Hox genes in whose cluster they are embedded (miR-10a is located between the HOXB4 and HOXB5 gene in chromosome 17q21 and miR-10b between the HOXD3 and HOXD4 gene in chromosome 2q31).39 To investigate whether miR-10a levels correlated with the expression of its flanking HOXB4 and HOXB5 genes, we measured their expression in CD41+ cells by qRT-PCR. Notably, CD41+ cells displayed a significant up-regulation of both HOXB4 and HOXB5 genes in Npm1-TCTG/WT;Cre+ and Npm1-TCTG/TCTG;Cre+ mice compared with Cre− controls (1.68 ± 0.5 vs 4.12 ± 0.23 vs 1.02 ± 0.15 and 1.73 ± 1.16 vs 4.71 ± 4.79 vs 1.00 ± 0.12, respectively; Figure 6B). No correlation was found between miR-10b and its flanking HOXD3 gene.
BM megakaryocytic expansion in NPM1-mutated AML
To assess whether alterations of the megakaryocytic compartment similar to those observed in our mouse model were detected also in human AML, we reviewed the characteristics of megakaryocytes in BM biopsies from 97 NPM1-mutated AML patients compared with 25 NPM1-unmutated AML and 10 nonleukemic BM samples used as controls. The average megakaryocytic concentration in nonleukemic samples was 8.58 ± 1.86 per ×40 field.
BM samples from NPM1-mutated AML showed an increased number of megakaryocytes (average megakaryocytic concentration per ×40 field: 13.7 ± 13.2). In particular, the LAT-positive megakaryocyte concentration per ×40 field in the 97 NPM1-mutated samples was as follows: <10 in 52/97 (53.6%) cases, >10 in 15/97 (15.4%) cases, >20 in 24/97 (24.7%) cases, and >40 in 6/97 (6.2%) cases (supplemental Table 2).
In contrast, the average megakaryocytic concentration in NPM1-unmutated AML samples was 7.6 ± 13.6 per ×40 field. In particular, only 16% (4/25) of NPM1-unmutated AML patients showed an average megakaryocyte concentration >10. Overall, our data indicated that the percentage of BM samples with megakaryocytosis is increased to 46.4% in NPM1-mutated AML compared with 16% of NPM1-unmutated AML samples. BM samples with a megakaryocytic concentration <10 were more in the NPM1-unmutated than in the NPM1-mutated AML group (84% vs 53.6%, respectively).
Immunohistochemistry usually resulted in a higher scoring of the number of megakaryocytes than histologic examination, because it allowed better detection of dysplastic megakaryocytes and micromegakaryocytes (Figure 7). At immunostaining, most of the megakaryocytes were characterized by cytoplasmic positivity for NPM and nuclear-restricted expression for nucleolin/C23, clearly indicating that they carried the NPM1 mutation. Among the 45 cases with an increased megakaryocyte concentration, 38 presented with thrombocytopenia, 6 cases with a normal number of platelets, and only 1 case with thrombocytosis. Thus, the expansion of megakaryocytic compartment in human NPM1-mutated AML patients mimics that observed in our NPMc+ mice model.
Megakaryocytes expansion in BM biopsy of NPM1-mutated AML patients. (A-B) Representative BM sections from a nonleukemic control were stained with (A) hematoxylin and eosin and (B) an anti-LAT antibody. (C-D) BM trephine sections from an NPM1-mutated AML patient with an increased number of megakaryocytes (∼20 megakaryocytes ×40 field) as assessed by (C) hematoxylin and eosin and (D) immunostaining for human LAT. (E) Nucleus-restricted NPM1 expression in a representative NPM1-unmutated AML patient. (F) Aberrant cytoplasmic expression of NPM1 in dysplastic megakaryocytes (original magnification, ×40) of an NPM1-mutated AML patient. (G) LAT positive megakaryocytes in a representative NPM1-mutated AML patient with a megakaryocytes concentration >40 per ×40 field (original magnification, ×40).
Megakaryocytes expansion in BM biopsy of NPM1-mutated AML patients. (A-B) Representative BM sections from a nonleukemic control were stained with (A) hematoxylin and eosin and (B) an anti-LAT antibody. (C-D) BM trephine sections from an NPM1-mutated AML patient with an increased number of megakaryocytes (∼20 megakaryocytes ×40 field) as assessed by (C) hematoxylin and eosin and (D) immunostaining for human LAT. (E) Nucleus-restricted NPM1 expression in a representative NPM1-unmutated AML patient. (F) Aberrant cytoplasmic expression of NPM1 in dysplastic megakaryocytes (original magnification, ×40) of an NPM1-mutated AML patient. (G) LAT positive megakaryocytes in a representative NPM1-mutated AML patient with a megakaryocytes concentration >40 per ×40 field (original magnification, ×40).
Discussion
In this study, we generated and characterized a new mouse model expressing the most frequent human NPM1 mutation (type A) in the hematopoietic stem cell compartment to investigate the role of the NPM1 mutation in leukemogenesis. Similarly to other animal models for NPM1 mutation,24-26 we found an expansion of the myeloid compartment in a fraction of mice. In addition, we demonstrated for the first time that the NPM1 mutant leads to perturbation of the megakaryocytic compartment in mice. This phenotype mimics some of the features observed in patients with NPM1-mutated AML.
Our observation that megakaryocytes from Npm1-TCTG/WT;Cre+ mice exhibit deregulated growth in vitro and in vivo points to a role of the NPM1 mutant in megakaryocytic differentiation. Mice expressing the NPM1 mutant have decreased platelet counts with a markedly increased mean platelet volume. There is also a remarkable megakaryocytosis in these animals, with a significant increase in megakaryocyte numbers both in BM and spleen. Megakaryocytosis is driven by an increased number of CFU-Meg and a pronounced expansion of early MKP cells that is not accompanied by altered TPO levels. CD41+ megakaryocytes display altered expression of miRNAs directly involved in human and mouse megakaryopoiesis. These findings suggest that the NPM1 mutant is able to induce a block of megakaryocytic differentiation, although we cannot exclude the possibility of a partial contribution on regulating platelet budding.
Notably, the effects of the NPM1 mutant on megakaryopoiesis that we observed in our model was not previously reported by other investigators.26,40 Because cytoplasmic localization of the NPM1 mutant appears to be critical for the development of NPM1-mutated AML,18 it is likely that these conflicting results may be caused by differences in the degree of the relative dosage of the mutated vs normal NPM1 and consequent alterations in the distribution of these proteins across subcellular compartments among different models. In our mice, the transgenic cassette that was used to express the NPM1 mutant cDNA under the control of a CAG promoter may have driven the expression of the NPM1 mutant to an extent to influence the mutated vs normal NPM1 ratio, thus leading to conditions pathophysiologically mimicking more closely those of human NPM1-mutated AML. In contrast, in the first transgenic NPMc+ model,24 the degree of the expression of NPM1 mutant was not as high as that of wild-type NPM1, determining a mild cytoplasmic expression and a nuclear relocalization of the mutant, as it can be simulated in vitro by cotransfection experiments.20 Moreover, in the conditional knock-in mouse model reported by Vassiliou et al,26 the cytoplasmic accumulation of the mutant NPM1 was not clearly shown in BM cells from mutant animals because only total cell lysates were analyzed for the mutant expression. Nevertheless, the same study also described an increase of the mean platelet volume similar to our model, even though blood counts and platelet numbers were not altered in mutant mice.26 Finally, the observation that in our NPM1 homozygous mutant mice platelet counts were further decreased to a half of the heterozygous mice further reinforces the concept that the NPM1 mutant is likely to exert its function and alter platelets formation depending of the mutant to wild-type NPM1 ratio.
The expression of NPM1 mutation in the hematopoietic compartment promoted by the inducible Mx1Cre caused the overexpression of miR10a, miR10b, and miR-20a in CD41+ cells. The relevance of this finding is related to the fact that miRNAs are expressed in megakaryocytes and platelets, where they are likely to regulate lineage development and function. Garzon et al37 showed that differentiation of human CD34+ cells into megakaryocytes is accompanied by down-regulation of numerous miRNAs including miR10a and miR10b. Other investigators found the expression of miR-20a to be reduced more than twofold in mature megakaryocytes compared with hematopoietic progenitors.38 Moreover, miR10a, miR10b, and miR-20a appear to be directly involved in megakaryocytic development by posttranscriptional inhibition of targeted genes expression that allow megakaryocytic precursors to produce platelets.37 Thus, the NPM1 mutant–related increased expression in miR10a, miR10b, and miR-20a may be responsible for the block of differentiation of megakaryocytes in our mouse model. This view is further supported by previous reports showing that the 3 up-regulated miRNAs in our mouse model also contribute to define the distinct microRNA profile of NPM1-mutated AML patients.7,10 The interference of the NPM1 mutant with the megakaryocytic compartment is also consistent with clinico-pathological observations that BM biopsies from a significant number of patients with NPM1-mutated AML show myelodysplastic features affecting ≥2 hemopoietic progenitors, especially megakaryocytes.41,42 In this paper, we further expanded these findings with a more accurate quantitative evaluation of megakaryocytes by immunohistochemistry with the anti-LAT antibody. Notably, we found an increased number of megakaryocytes in NPM1-mutated AML compared with NPM1-unmutated AML and normal controls. These findings are in keeping with the expansion of the megakaryocyte compartment observed in our NPM1c+ mouse model.
Although the mechanism underlying the up-regulation of miR10a, miR10b, and miR-20a in our mice model remains to be elucidated, we demonstrated that NPMc+ specifically promotes this phenomenon, because we were able to reproduce it in a nonhematopoietic cell line using overexpression experiments.
miR10a is located within the HOXB cluster and is coexpressed with HOXB4 and HOXB5 through a common transcriptional mechanism in developing mouse embryos.39 Similarly to patients with NPM1-mutated AML,7 we observed a strong association between up-regulation of miR10a and increased HOXB4 and HOXB5 expression in BM cells from mice harboring the NPM1 mutation A. This finding suggests that expression of this miRNA may be modulated by cis elements that also regulate HOX genes.43 Another possible explanation is that in our setting, the cytoplasmic NPM1 mutant and/or delocalized wild-type NPM1 may increase miR10a, miR10b, and miR20a stabilization, boosting their expression levels. This is not surprising because NPM1 is involved in shuttling RNAs and ribosomal proteins to the cytosol,44 and this shuttling mechanism may be relevant to a possible role in miRNA packaging, export, and stabilization. Interestingly, the NPM1 protein can bind miRNAs and protect them from degradation.45 Indeed, mature cytoplasmic miR10a and miR10b up-regulation was accompanied by reduced miRNA precursor expression in mutant mice. This finding further supports the hypothesis that altered miRNA expression could be directly related to miRNA stabilization promoted by NPMc+ expression rather than a consequence of immature CD41+ megakaryocytes accumulation.
In our mouse model, we did not observe the onset of leukemia after a 1.5-year follow-up, corroborating the idea that the cytoplasmic NPM1 mutant is necessary but not a sufficient genetic lesion to cause AML. These results confirm previous findings in a transgenic NPMc+ mice model24 and are consistent with the concept that, similarly to other human cancers,46 the development of AML results from >1 oncogenic hit.47 For example, in the NPM1-mutated knock-in mouse model reported by Vassiliou et al,26 one-third of mice developed delayed-onset AML requiring cooperating mutations. In leukemic patients, the NPM1 mutant may act synergistically with FLT3-ITD, IDH1, and/or DNMT3A mutations, which frequently associate with NPM1-mutated AML.48-50 In this model, the NPM1 mutation is likely to represent the initiating genetic event, and the other mutations are secondary alterations related to tumor progression.
In summary, we demonstrated for the first time that the expression of the NPM1 mutant results in the direct generation of signals determining a block of megakaryocytic development. Importantly, the phenotype of these mice recalls some features of human NPM1-mutated AML. Thus, our mouse model further expands the knowledge on the role of NPM1 mutant in leukemogenesis and may serve as an additional tool for investigating the cooperation between mutations affecting NPM1 and other genes.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Roberta Pacini and Alessia Tabarrini for performing immunohistochemical staining and Dr Luigi De Angelis for laboratory assistance. The authors also thank Claudia Tibidò for excellent secretarial assistance.
This work was supported by the Associazione Italiana Ricerca Cancro, the Associazione Umbra contro le Leucemie e i Linfomi, Fondazione Cassa di Risparmio di Perugia (grant 2010.011.0391), and Italian Minister of Health Project “Ricerca Finalizzata 2008” (grant RF-UMB-2008-1142331).
Authorship
Contribution: P.S. designed and performed experiments, analyzed and interpreted the data, and wrote the manuscript; E.V. performed experiments, analyzed and interpreted the data, and contributed to the writing of the paper; R.R. performed experiments on protein expression of the NPM1 mutant; O.B. and D.C. performed flow cytometry; I.G. performed experiments on protein expression; N.B. contributed by generating the mouse model and writing the manuscript; T.I., P.Z., and A.M. provided clinical data on AML patients; E.T., M.P.M., F.F., and M.F.M. contributed to the writing of the manuscript; and B.F. led the project, provided expert hematopathologic analysis, supervised the study, and wrote the manuscript.
Conflict-of-interest disclosure: B.F. applied for a patent on the clinical use of NPM1 mutants. The remaining authors declare no competing financial interests.
Correspondence: Brunangelo Falini or Paolo Sportoletti, Institute of Hematology, University of Perugia, Ospedale S. Maria della Misericordia, S. Andrea delle Fratte, 06132 Perugia, Italy; e-mail: faliniem@unipg.it or sportolp@gmail.com.



![Figure 3. Thrombocytopenia of Cre-induced mice harboring the mutated NPM1 cDNA transgenic cassette. (A) Peripheral blood cell counts of Npm1-TCTG/WT;Cre+ (n = 9) and Npm1-TCTG/TCTG;Cre+ (n = 8) vs control mice (n = 9; including 3 Npm1-WT/WT;Cre+, 3 Npm1-TCTG/WT;Cre−, and 3 Npm1-TCTG/TCTG;Cre− pIpC-treated mice). (B) Significantly decreased platelets number in peripheral blood of both heterozygous and homozygous conditional mice 1 month after pIpC injection. Results are shown as mean ± standard deviation (error bars) from 13 Npm1-TCTG/WT;Cre+ and 10 Npm1-TCTG/TCTG;Cre+ vs 25 control mice (including Npm1-WT/WT;Cre+ [n = 9], Npm1-TCTG/WT;Cre− [n = 9], and Npm1-TCTG/TCTG;Cre− [n = 7] pIpC-treated mice). (C) Enlarged platelet size in Npm1-TCTG/WT;Cre+ mice (N = 4) is shown as increased (i) mean platelet volume, (ii) platelet distribution width, and (iii) platelet-large cell ratio. (iv) Peripheral blood smears showing both the paucity of platelets and their substantially larger size (arrows) in heterozygous mutant mice vs littermate Cre− pIpC-treated controls. *P < .05, **P < .01, ***P < .001 (unequal-variance t test).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/121/17/10.1182_blood-2012-08-449553/4/m_3447f3.jpeg?Expires=1763570664&Signature=J3tqGOQtebXtWvqrFRhdy4ZvgqjlAX~XfTmSMJSJaTuPAZde7PGr90L8o6zMvb1XXLW6uUtxP10gtQlH0F~Izsst4mO2mgXrQzIpdOJBsppHjZukwVfQT3WaoXxTIT84v2W8zSRAYXzZqK3NcVZE~dMrugvUf9~mEEViybnrgnUj~MrMXEymQ8d~vgTBHqsmQ-DdWPYzt9PubU6h8zC7yB1wN1YQFJMdB~yxqvwozSwuyQFDlnUf2v34k29qkLQ2QipO2~JwtQ03u2CvV7yKh5-CjpMRDjJdD7Qh292bxvLk5ZzZ1GBgP6polSHPcETLpcXk0G-tUEZzfuottNc6wA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal