Key Points
Patients with myocardial infarction have altered platelet miRNA profiles.
Activated platelets release miRNAs that can be taken up by endothelial cells and regulate ICAM-1 gene expression.
Platelets play a crucial role in the pathogenesis of myocardial infarction (MI) by adhering to the site of a ruptured atherosclerotic plaque. The aim of this study was to screen for differences in the micro RNA (miRNA) content of platelets from patients with myocardial infarction and control patients, to investigate a possible release of miRNAs from activated platelets and to elucidate whether platelet-derived miRNAs could act as paracrine regulators of endothelial cell gene expression. Using RNA-seq, we found 9 differentially expressed miRNAs in patients compared with healthy controls, of which 8 were decreased in patients. Of these, miR-22, -185, -320b, and -423-5p increased in the supernatant of platelets after aggregation and were depleted in thrombi aspirated from MI patients, indicating the release of certain miRNAs from activated platelets. To confirm that endothelial cells could take up the released platelet miRNAs, transfer of both fluorescently labeled miRNA and exogenous cel-miR-39 from activated platelets to endothelial cells was shown. Finally, a possible paracrine role of released platelet miR-320b on endothelial cell intercellular adhesion molecule-1 expression was shown. Thus, platelets from patients with MI exhibit loss of specific miRNAs, and activated platelets shed miRNAs that can regulate endothelial cell gene expression.
Introduction
Acute coronary syndrome, including acute myocardial infarction and unstable angina, is the result of destabilization of an atherosclerotic plaque in a coronary artery.1 The subsequent exposure and release of subendothelial factors (eg, collagen and von Willebrand Factor) trigger activation of platelets in the vicinity of the lesion.2 Activated platelets in turn release a multitude of substances affecting the chemotactic, adhesive, and proteolytic properties of endothelial cells.3,4
Although platelets lack nuclei, they do retain a specific set of megakaryocyte messenger RNAs (mRNAs)5,6 and are capable of de novo protein synthesis.7,-9 Protein translation in platelets has been shown to be under posttranscriptional control.9,10 In addition to having protein coding transcripts the authors of a recent report revealed that platelets contain more than 170 different micro RNA (miRNA) species as well as the enzymes required to convert precursor miRNA into mature miRNA.11 miRNAs are short (19-25 nucleotides), noncoding RNA species that play a crucial role in regulating posttranscriptional gene expression through binding to the 3′ untranslated region (3′-UTR) of target mRNAs.12 miR-96 has been shown to regulate the expression of VAMP8, a critical v-SNARE (ie, soluble NSF attachment protein REceptor) involved in platelet granule secretion,13 indicating a functional role of miRNA in regulating platelet mRNA translation.
The concept of exovesicle-mediated transfer of miRNA to the endothelium has been explored in recent studies. For example, the transfer of miR-126 between endothelial cells via apoptotic bodies was shown to limit atherosclerosis, promote incorporation of progenitor cells, and increase plaque stability in a mouse model through the targeting of RGS16 and increasing production of CXCL12.14 Moreover, in a recent study15 authors demonstrated that monocyte-derived exosomes carrying miR-150 were taken up by endothelial cells enhancing cell migration.
The aim of this work was (1) to screen for differences in miRNA content in the platelets of patients diagnosed with ST-elevation myocardial infarction (STEMI) and healthy individuals, (2) to investigate the possible release and transfer of miRNAs from activated platelets to endothelial cells, and (3) to assess the role of platelet-derived miRNAs in the endothelium.
Methods
Patients and samples
Patients diagnosed with STEMI were recruited during 2010 at the coronary care unit at Skane University Hospital. Diagnoses were based on electrocardiographic criteria.
Isolation and preparation of platelets, plasma, and thrombi
The procedure for platelet preparation has been described in detail elsewhere.6 In summary, 100 mL of blood was drawn by self-propagated flow and platelet-poor plasma was passed through a Pall Autostop Leukocyte removal filter (Pall Incorporated, NY) and depleted of additional leukocytes and erythrocytes with the use of dynabeads (Dynal, Oslo, Norway) conjugated with anti-CD235a and anti-CD45 (Becton Dickinson, Franklin Lakes, NJ). Plasma was prepared by centrifugation of whole blood for 15 minutes at 1600g. For a detailed protocol of platelet and thrombi preparation, supplemental Methods at the Blood Web site.
RNA preparation
Cells and plasma/cell supernatants were mixed with Qiazol and TRIzol LS (Life Technologies, Carlsbad, CA), respectively, and RNA was prepared by use of the miRNeasy mini kit (QIAGEN, Hilden, Germany). For miRNA and mRNA analysis, cDNA was synthesized with the use of the miRCURY LNA Universal RT microRNA cDNA kit (Exiqon, Vedbaek, Denmark) and the RevertAid H Minus First Strand cDNA Synthesis Kit (Thermo Scientific, Waltham, MA), respectively.
RNA-seq
Platelet RNA samples from two male patients with STEMI and 2 age- and sex-matched healthy control patients were prepared using the Small RNA Sample Preparation Kit (Illumina, Inc, San Diego, CA). Sequencing was performed on the Illumina GAIIx with a read length of 36 nt. miRNA sequences were identified using miRanalyzer16 based on the bowtie architecture. The number of unique reads were counted, normalized to transcript size, and expressed relative to the total number of reads.
qRT-PCR
miRNA levels were assessed by quantitative reverse-transcription (qRT)-polymerase chain reaction (PCR) with the use of microRNA LNA Primer sets (Exiqon) specific for hsa-miR-16, -22, -126, -185, -320b, 423-5p, U6 small nuclear RNA, and cel-miR-39-5p. mRNA levels for intercellular adhesion molecule (ICAM-1) and cyclophilin were analyzed with TaqMan Assays (Life Technologies). See the supplemental Methods for details.
Confirmation of deep sequencing results by qRT-PCR
Platelet RNA was prepared as described previously from patients with STEMI (n = 10) and healthy control patients (n = 15). The level of U6 RNA was unaltered in patients with STEMI compared with control patients (coefficient of variation 8.02%; supplemental Figure 1) and was therefore considered reliable for the normalization of platelet miRNA data.
Assessment of miRNA release from aggregated platelets
Platelets were preincubated 5 minutes with 0.25 mg/mL fibrinogen before the addition of 1 U/mL of thrombin. Platelets were allowed to aggregate for 5 minutes, then centrifuged, and an aliquot of the supernatant was taken. Before cDNA synthesis an exogenous miRNA spike-in (Exiqon) was added to the RNA preparations. The levels of the different miRNAs in the supernatant were normalized to the exogenous RNA spike-in.
Immunofluorescence staining of coronary thrombi
See supplemental Methods.
Cell culture
See supplemental Methods.
Transfer of miRNA between platelets and endothelial cells
The procedure for transfecting platelets were based on the protocol by Hong et al17 (See supplemental Methods for a detailed description). Platelets were transfected with 40 pM synthetic Caenorhabditis elegans miRNA miR-39 (syn-cel-miR-39) or 400 nM fluorescently labeled scrambled miRNA (miR-Scr-fluorescein isothiocyanate [FITC]) with the use of 6 μL of lipofectamine LTX (Life Technologies) per transfection. Transfection efficiency was determined after 24 hours by flow cytometry as the proportion of FITC-positive events in the platelet gate (mean transfection efficiency 17.4%; SEM 3.3). The effects of transfection on platelet morphology, activation, and apoptosis were deemed minor (supplemental Figure 2). Untreated platelets were completely negative for cel-miR-39 expression (data not shown). To confirm that miRNA was taken up in the platelet cytoplasm, transfected platelets were treated with 20 μg/mL RNase A (Thermo Scientific) and 2 U/μL RNase T1 (Thermo Scientific) for 30 minutes at 37°C (supplemental Figure 3). As a negative control, platelets were transfected with cel-miR-39 pretreated with RNase. The level of cel-miR-39 in transfected platelets was unaffected by RNase-treatment, whereas platelets transfected with degraded cel-miR-39 were completely devoid of cel-miR-39. Moreover, the fluorescence signal from platelets transfected with miR-FITC was unaltered by RNase-treatment. Taken together, these results provide evidence that the transfected miRNA is taken up into the cytoplasm of the platelets.
For assessment of miRNA transfer, we seeded human microvascular endothelial cell-1 (HMEC-1) cells 24 hours before coculture. A total of 0.5-1 × 107 platelets transfected either with syn-cel-miR-39 or Scr-miR-FITC were added per well in the presence or absence of 1 U/mL thrombin or 10 μg/mL brefeldin A. The presence of syn-cel-miR-39 or Scr-miR-FITC in HMEC-1 cells was assessed with qRT-PCR and confocal microscopy, respectively. A detailed protocol can be found in the supplemental Methods.
miRNA levels in HMEC-1 cells cocultured with platelets
HMEC-1 cells were seeded in 96-well plates and cocultured with washed platelets as described previously. HMEC-1 cells were harvested at 0, 1, 3, 8, and 24 hours after the addition of thrombin. RNA was prepared, and the levels of each miRNA relative to miR-16 were determined. miR-16 was unaffected by thrombin during the time course (coefficient of variation 11.09%; supplemental Figure 4).
Assessment of platelet microparticles
Platelets transfected with miR-Scr-FITC were seeded in TAB in a 96-well plates at ∼1 × 105 platelets/well. Then, 10 μg/mL brefeldin A was added, and the platelets were incubated for 30 minutes. 1 U/mL thrombin was added and the platelets were incubated for 20 minutes. Platelets were stained with CD42a-PE, fixed with 0.5% PFA, and run on an Accuri C6 flow cytometer. The microparticle population was defined first on size using 800-nm latex beads (Sigma-Aldrich, St. Louis, MO) and subsequently on the expression of CD42a. A phycoerythrin-conjugated isotype control antibody was used to set the CD42a-gate. Mean fluorescence in FL-1 within the microparticle gate was used to assess FITC+ microparticles.
A screening was performed on 3 compounds reported to inhibit microparticle or exosome release: brefeldin A,18 cyclosporin A,19 and caspase inhibitor.20 A total of 10 μg/mL of brefeldin A (Sigma-Aldrich), 10μM cyclosporin A (Sigma-Aldrich), and 100 μM caspase inhibitor (Ac-DEVD-CHO; Promega) were added to platelet-rich plasma for 30 minutes at 37°C before the addition of 1 U/mL thrombin for 20 minutes. Then, microparticles were detected by flow cytometry.
3′-UTR target plasmid reporter assay
miTarget miRNA Target Sequence 3′-UTR Expression Clones (GeneCopoeia, Rockville, MD) was used for validation of predicted mRNA targets. See the supplemental Methods for a detailed protocol.
Stimulation of HMEC-1 cells with platelet releasate
Platelets were incubated in 25 µM N-terminal thrombin receptor fragment SFLLRN (Bachem, Bubendorf, Switzerland) for 20 minutes at room temperature. Aggregated platelets were pelleted, and the platelet releasate was collected and stored at −80°C.
HMEC-1 cells were seeded in 12-well plates and after 24 hours, fresh medium with or without 20% platelet releasate containing 0 or 100 nM Anti-miR miRNA Inhibitor (Life Technologies) corresponding to miR-22 or -320b was added. After another 24 hours, the cells were harvested by the addition of Qiazol.
Overexpression of miR-320b in HMEC-1 cells
See the supplemental Methods.
Statistical analysis
All statistical analyses were performed in Prism v. 4.0b (GraphPad Software Inc., La Jolla, CA). Data represent the mean ± SEM and were analyzed by Student t test, 1- or 2-way analysis of variance with Tukey post hoc analysis as appropriate. Statistical significance was considered when P < .05.
Ethics
This study was conducted according to the principles of the Declaration of Helsinki and was approved by the local ethics committee of Skåne University Hospital. All patients provided their written approval before participation.
Results
Altered platelet miRNA profile in patients with STEMI
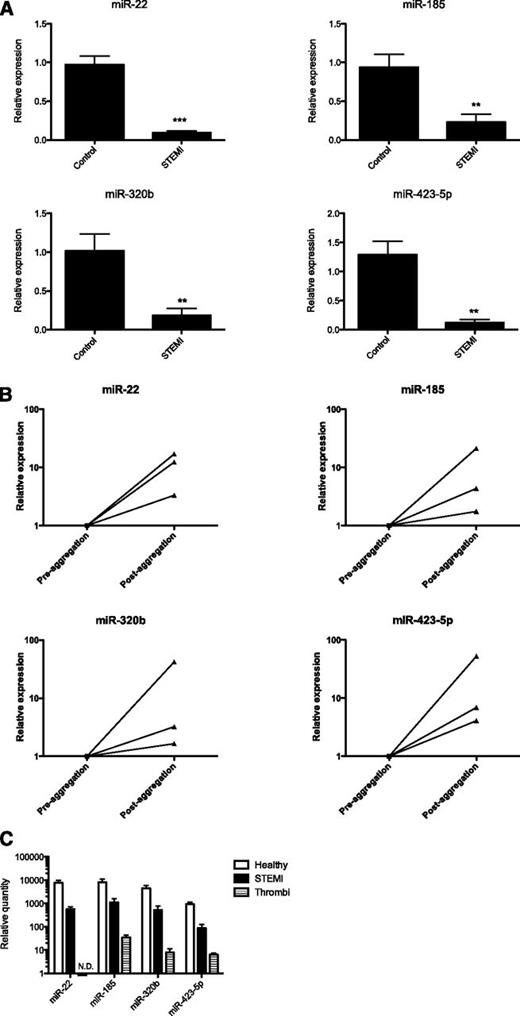
We performed RNA-seq of platelet miRNA in 2 patients with STEMI as well as in 2 age- and sex-matched healthy control patients. The mean number of total transcripts detected was 136 670, and a mean of 109 different miRNA species was detected in the samples. The 20 most highly expressed miRNAs in each individual are presented in Table 1. The complete miRNA profiles can be found in supplemental Table 1. The levels of 9 different miRNA species were significantly altered between patients and controls (supplemental Figure 5). Of these, miR-320a was up-regulated whereas all the other miRNAs were lower in patients with STEMI than in control patients. Possible mRNA targets of these miRNAs were assessed with TargetScan (v. 5.1) and 4 candidate miRNAs (miR-22, -185, -320b, and -423-5p) were selected for further evaluation (Table 2). The criteria for selection were based on: (1) the potential relevance to atherosclerosis or inflammation; (2) the relative abundance in platelets; and (3) the magnitude of the difference between patients and controls. To validate the findings indicated by RNA-seq, in Figure 1A the levels of the 4 selected miRNAs were determined in the platelets from patients with STEMI (n = 10) and from healthy controls (n = 15). The results demonstrated an approximately 70% to 90% decrease of all the 4 miRNAs in STEMI patients (P < .01), thus confirming the results of the RNA-seq.
The 20 most highly expressed miRNAs in the platelets of STEMI and control patients
| miRNA ID . | Transcripts . | Normalized* . | % of total . |
|---|---|---|---|
| Patient 1 | |||
| miR-320a | 44 170 | 42 043.45 | 97.77 |
| miR-423-5p | 180 | 169.43 | 0.40 |
| miR-320b | 139 | 127.00 | 0.31 |
| miR-1274a | 74 | 73.94 | 0.16 |
| miR-1908 | 60 | 55.48 | 0.13 |
| miR-185 | 57 | 54.41 | 0.13 |
| let-7b | 43 | 40.05 | 0.10 |
| miR-320c | 34 | 31.55 | 0.08 |
| let-7c | 33 | 31.23 | 0.07 |
| let-7a | 26 | 24.64 | 0.06 |
| miR-378 | 28 | 23.86 | 0.06 |
| let-7f | 25 | 23.55 | 0.06 |
| miR-103 | 23 | 21.87 | 0.05 |
| miR-221 | 16 | 14.57 | 0.04 |
| miR-1290 | 14 | 13.26 | 0.03 |
| miR-1 | 14 | 13.09 | 0.03 |
| miR-382 | 14 | 13.05 | 0.03 |
| let-7e | 13 | 12.27 | 0.03 |
| miR-483-5p | 13 | 11.91 | 0.03 |
| miR-1224-3p | 11 | 8.95 | 0.02 |
| Patient 2 | |||
| miR-320a | 15 711 | 14 927.68 | 97.97 |
| miR-423-5p | 93 | 87.74 | 0.58 |
| miR-320b | 49 | 44.14 | 0.31 |
| miR-185 | 24 | 22.77 | 0.15 |
| miR-1274a | 19 | 19.00 | 0.12 |
| miR-378 | 13 | 11.29 | 0.08 |
| let-7b | 11 | 10.36 | 0.07 |
| miR-1908 | 11 | 9.95 | 0.07 |
| miR-320c | 11 | 9.85 | 0.07 |
| miR-1274b | 7 | 7.00 | 0.04 |
| miR-1 | 7 | 6.68 | 0.04 |
| let-7a | 7 | 6.64 | 0.04 |
| miR-1307 | 7 | 6.14 | 0.04 |
| miR-422a | 6 | 5.73 | 0.04 |
| miR-33a | 5 | 4.76 | 0.03 |
| miR-375 | 4 | 3.82 | 0.02 |
| miR-765 | 4 | 3.81 | 0.02 |
| miR-107 | 4 | 3.70 | 0.02 |
| miR-576-3p | 4 | 3.68 | 0.02 |
| miR-1308 | 3 | 2.83 | 0.02 |
| Control 1 | |||
| miR-320a | 19 496 | 18 526.55 | 82.81 |
| miR-423-5p | 2020 | 1903.52 | 8.58 |
| miR-185 | 285 | 270.82 | 1.21 |
| let-7a | 191 | 180.86 | 0.81 |
| let-7c | 188 | 177.64 | 0.80 |
| let-7f | 184 | 174.77 | 0.78 |
| let-7b | 112 | 104.32 | 0.48 |
| miR-221 | 102 | 93.61 | 0.43 |
| let-7e | 85 | 80.59 | 0.36 |
| miR-320b | 71 | 64.32 | 0.30 |
| miR-21 | 57 | 54.14 | 0.24 |
| miR-103 | 44 | 41.57 | 0.19 |
| miR-378 | 40 | 36.24 | 0.17 |
| miR-1274a | 32 | 32.00 | 0.14 |
| let-7d | 34 | 32.00 | 0.14 |
| miR-1307 | 32 | 29.77 | 0.14 |
| miR-22 | 30 | 28.41 | 0.13 |
| miR-423-3p | 27 | 25.65 | 0.11 |
| let-7g | 24 | 22.64 | 0.10 |
| Control 2 | |||
| miR-320a | 467 302 | 444 454.86 | 88.44 |
| miR-423-5p | 41 477 | 39 257.65 | 7.85 |
| miR-185 | 6134 | 5839.23 | 1.16 |
| miR-320b | 1591 | 1451.23 | 0.30 |
| miR-221 | 1524 | 1416.91 | 0.29 |
| let-7b | 695 | 653.77 | 0.13 |
| miR-378 | 639 | 570.71 | 0.12 |
| miR-22 | 587 | 558.23 | 0.11 |
| let-7f | 568 | 538.18 | 0.11 |
| miR-1307 | 546 | 505.05 | 0.10 |
| let-7a | 531 | 504.32 | 0.10 |
| miR-1274a | 498 | 497.89 | 0.09 |
| miR-320c | 478 | 441.20 | 0.09 |
| miR-423-3p | 279 | 261.52 | 0.05 |
| miR-103 | 253 | 239.17 | 0.05 |
| miR-744 | 251 | 234.59 | 0.05 |
| miR-139-3p | 229 | 226.64 | 0.04 |
| miR-1908 | 233 | 215.14 | 0.04 |
| miR-584 | 233 | 211.86 | 0.04 |
| miRNA ID . | Transcripts . | Normalized* . | % of total . |
|---|---|---|---|
| Patient 1 | |||
| miR-320a | 44 170 | 42 043.45 | 97.77 |
| miR-423-5p | 180 | 169.43 | 0.40 |
| miR-320b | 139 | 127.00 | 0.31 |
| miR-1274a | 74 | 73.94 | 0.16 |
| miR-1908 | 60 | 55.48 | 0.13 |
| miR-185 | 57 | 54.41 | 0.13 |
| let-7b | 43 | 40.05 | 0.10 |
| miR-320c | 34 | 31.55 | 0.08 |
| let-7c | 33 | 31.23 | 0.07 |
| let-7a | 26 | 24.64 | 0.06 |
| miR-378 | 28 | 23.86 | 0.06 |
| let-7f | 25 | 23.55 | 0.06 |
| miR-103 | 23 | 21.87 | 0.05 |
| miR-221 | 16 | 14.57 | 0.04 |
| miR-1290 | 14 | 13.26 | 0.03 |
| miR-1 | 14 | 13.09 | 0.03 |
| miR-382 | 14 | 13.05 | 0.03 |
| let-7e | 13 | 12.27 | 0.03 |
| miR-483-5p | 13 | 11.91 | 0.03 |
| miR-1224-3p | 11 | 8.95 | 0.02 |
| Patient 2 | |||
| miR-320a | 15 711 | 14 927.68 | 97.97 |
| miR-423-5p | 93 | 87.74 | 0.58 |
| miR-320b | 49 | 44.14 | 0.31 |
| miR-185 | 24 | 22.77 | 0.15 |
| miR-1274a | 19 | 19.00 | 0.12 |
| miR-378 | 13 | 11.29 | 0.08 |
| let-7b | 11 | 10.36 | 0.07 |
| miR-1908 | 11 | 9.95 | 0.07 |
| miR-320c | 11 | 9.85 | 0.07 |
| miR-1274b | 7 | 7.00 | 0.04 |
| miR-1 | 7 | 6.68 | 0.04 |
| let-7a | 7 | 6.64 | 0.04 |
| miR-1307 | 7 | 6.14 | 0.04 |
| miR-422a | 6 | 5.73 | 0.04 |
| miR-33a | 5 | 4.76 | 0.03 |
| miR-375 | 4 | 3.82 | 0.02 |
| miR-765 | 4 | 3.81 | 0.02 |
| miR-107 | 4 | 3.70 | 0.02 |
| miR-576-3p | 4 | 3.68 | 0.02 |
| miR-1308 | 3 | 2.83 | 0.02 |
| Control 1 | |||
| miR-320a | 19 496 | 18 526.55 | 82.81 |
| miR-423-5p | 2020 | 1903.52 | 8.58 |
| miR-185 | 285 | 270.82 | 1.21 |
| let-7a | 191 | 180.86 | 0.81 |
| let-7c | 188 | 177.64 | 0.80 |
| let-7f | 184 | 174.77 | 0.78 |
| let-7b | 112 | 104.32 | 0.48 |
| miR-221 | 102 | 93.61 | 0.43 |
| let-7e | 85 | 80.59 | 0.36 |
| miR-320b | 71 | 64.32 | 0.30 |
| miR-21 | 57 | 54.14 | 0.24 |
| miR-103 | 44 | 41.57 | 0.19 |
| miR-378 | 40 | 36.24 | 0.17 |
| miR-1274a | 32 | 32.00 | 0.14 |
| let-7d | 34 | 32.00 | 0.14 |
| miR-1307 | 32 | 29.77 | 0.14 |
| miR-22 | 30 | 28.41 | 0.13 |
| miR-423-3p | 27 | 25.65 | 0.11 |
| let-7g | 24 | 22.64 | 0.10 |
| Control 2 | |||
| miR-320a | 467 302 | 444 454.86 | 88.44 |
| miR-423-5p | 41 477 | 39 257.65 | 7.85 |
| miR-185 | 6134 | 5839.23 | 1.16 |
| miR-320b | 1591 | 1451.23 | 0.30 |
| miR-221 | 1524 | 1416.91 | 0.29 |
| let-7b | 695 | 653.77 | 0.13 |
| miR-378 | 639 | 570.71 | 0.12 |
| miR-22 | 587 | 558.23 | 0.11 |
| let-7f | 568 | 538.18 | 0.11 |
| miR-1307 | 546 | 505.05 | 0.10 |
| let-7a | 531 | 504.32 | 0.10 |
| miR-1274a | 498 | 497.89 | 0.09 |
| miR-320c | 478 | 441.20 | 0.09 |
| miR-423-3p | 279 | 261.52 | 0.05 |
| miR-103 | 253 | 239.17 | 0.05 |
| miR-744 | 251 | 234.59 | 0.05 |
| miR-139-3p | 229 | 226.64 | 0.04 |
| miR-1908 | 233 | 215.14 | 0.04 |
| miR-584 | 233 | 211.86 | 0.04 |
miRNA, microRNA; STEMI, ST-elevation myocardial infarction.
Normalized to transcript size.
Putative mRNA targets of candidate miRNAs
| miRNA . | Target . |
|---|---|
| miR-22 | ICAM-1 |
| miR-185 | eNOS VEGFA |
| miR-320b | ICAM-1 |
| miR-423-5p | VEGFB eNOS |
| miRNA . | Target . |
|---|---|
| miR-22 | ICAM-1 |
| miR-185 | eNOS VEGFA |
| miR-320b | ICAM-1 |
| miR-423-5p | VEGFB eNOS |
Possible targets were analyzed using TargetScan v. 5.1.
eNOS, endothelial nitric oxide synthase; ICAM-1, intercellular adhesion molecule; mRNA, messenger RNA; miRNA, microRNA; VEGF, vascular endothelial growth factor.
miRNA is released from activated platelets. (A) Platelet miRNA levels in patients with STEMI (n = 10) and control patients (n = 15) are expressed relative to U6 small nuclear RNA and normalized to the mean of the control samples. **P < .01; ***P < .001. (B) miRNA levels in the supernatant of pelleted platelets from 3 individual donors before and after aggregation with 1 U/mL thrombin (n = 3). Data are expressed relative to an exogenous RNA spike-in and normalized against the control samples (before aggregation). (C) Levels of miR-22, -185, -320b, and -423-5p in platelets of healthy individuals (n = 15), patients with STEMI (n = 10), and thrombi aspirated from patients with STEMI (n = 2) relative to U6 RNA. N.D. indicates not detected.
miRNA is released from activated platelets. (A) Platelet miRNA levels in patients with STEMI (n = 10) and control patients (n = 15) are expressed relative to U6 small nuclear RNA and normalized to the mean of the control samples. **P < .01; ***P < .001. (B) miRNA levels in the supernatant of pelleted platelets from 3 individual donors before and after aggregation with 1 U/mL thrombin (n = 3). Data are expressed relative to an exogenous RNA spike-in and normalized against the control samples (before aggregation). (C) Levels of miR-22, -185, -320b, and -423-5p in platelets of healthy individuals (n = 15), patients with STEMI (n = 10), and thrombi aspirated from patients with STEMI (n = 2) relative to U6 RNA. N.D. indicates not detected.
Platelets release miRNAs upon aggregation
The fact that of the 9 differentially expressed miRNAs, 8 showed decreased levels in patients led us to hypothesize that the greater level of platelet activation in patients with STEMI resulted in release of platelet miRNA. To examine this, we used qRT-PCR to measure the levels of the 4 candidate miRNAs in the supernatant of resting platelets and platelets aggregated with 1 U/mL thrombin with qRT-PCR (Figure 1B). After aggregation, the levels of all 4 miRNAs increased in the supernatant. Although the magnitude of the increase varied between donors, from 3.1-fold (SEM 0.49) to 21-fold (SEM 7.9), the increase was significant in all of them.
To further investigate the idea of miRNA release from activated platelets, we compared the miRNA profiles of platelets from healthy control patients, STEMI patients, and from coronary thrombi retrieved from the occlusion causing the STEMI (Figure 1C). The results confirmed the previous findings because the levels of all 4 miRNAs were even lower in thrombi than in STEMI platelets, indicating an activation-generated depletion of certain miRNAs. The presence of platelets in thrombi was confirmed by confocal microscopy (supplemental Figure 6).
The levels of these miRNAs were also analyzed in the circulation of a subset of patients. miR-22 and miR-423-5p were undetectable, miR-185 was unaltered, and miR-320b was slightly elevated (P = .31; supplemental Figure 7).
miRNA is transferred from platelets to endothelial cells
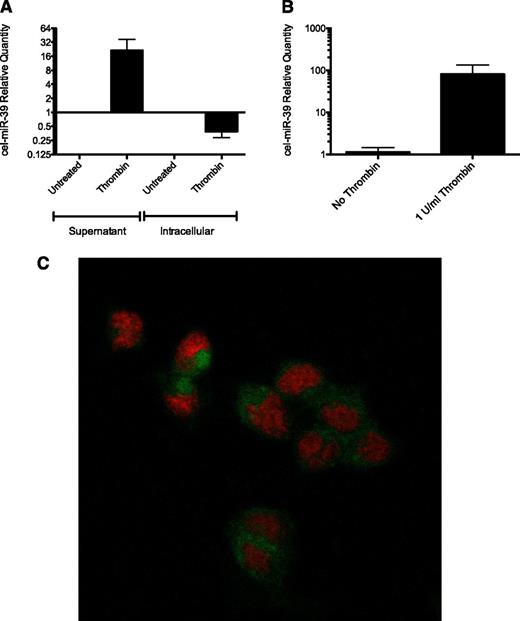
We hypothesized that the miRNAs released from activated platelets could be taken up by endothelial cells. To test this idea, platelets were transfected with a synthetic exogenous miRNA, syn-cel-miR-39. The efficiency of the transfection was determined by absolute qRT-PCR and although untransfected platelets were completely negative for cel-miR-39 expression, transfected platelets contained ∼2000 copies/platelet. Activation of transfected platelets with 1 U/mL Thrombin resulted in a 21-fold increase of cel-miR-39 in the supernatant and a corresponding 60% decrease in intracellular cel-miR-39 (Figure 2A). In the next experiment, transfected platelets were cocultured with HMEC-1 cells for either 1 or 3 hours in the presence or absence of 1 U/mL thrombin. After we removed the platelets and washed the monolayer, we assessed cel-miR-39 levels with qRT-PCR (Figure 2B). We found that although Cel-miR-39 was undetectable in HMEC-1 cells after 1 hour of coculture (data not shown), 3 hours of coculture produced levels that were readily detectable in HMEC-1 cells. Furthermore, the transfer was clearly dependent on the platelet activation status because cells cocultured with activated platelets contained 80-fold more cel-miR-39 than cells cocultured with resting platelets.
Activated platelets transfer miRNA to endothelial cells in vitro. (A) Relative quantity of intracellular and secreted cel-miR-39 from resting and activated platelets is shown. Expression data are normalized to an exogenous miRNA spike in for the supernatant and to U6 RNA for the intracellular levels (n = 3). (B) Relative quantity of cel-miR-39 in HMEC-1 cells cocultured with transfected platelets after 3 hours of coculture in the presence or absence of 1 U/mL thrombin (n = 3). Levels are presented relative to miR-16 and normalized against the mean of the cells without thrombin. (C) Representative confocal image of HMEC-1 cells cocultured with miR-Scr-FITC transfected platelets in the presence of 1 U/mL thrombin. Nuclei are stained red.
Activated platelets transfer miRNA to endothelial cells in vitro. (A) Relative quantity of intracellular and secreted cel-miR-39 from resting and activated platelets is shown. Expression data are normalized to an exogenous miRNA spike in for the supernatant and to U6 RNA for the intracellular levels (n = 3). (B) Relative quantity of cel-miR-39 in HMEC-1 cells cocultured with transfected platelets after 3 hours of coculture in the presence or absence of 1 U/mL thrombin (n = 3). Levels are presented relative to miR-16 and normalized against the mean of the cells without thrombin. (C) Representative confocal image of HMEC-1 cells cocultured with miR-Scr-FITC transfected platelets in the presence of 1 U/mL thrombin. Nuclei are stained red.
To further confirm miRNA transfer from platelets to endothelial cells, platelets were transfected with a scrambled, fluorescently labeled miRNA (miR-Scr-FITC). Transfected platelets were cocultured with HMEC-1 for 3 hours in the presence of 1 U/mL of thrombin. After removal of the platelets, the HMEC-1 cells were analyzed with confocal microscopy. The cytoplasm of HMEC-1 was clearly fluorescent, indicating uptake of miRNA from activated platelets (Figure 2C; supplemental Figure 8).
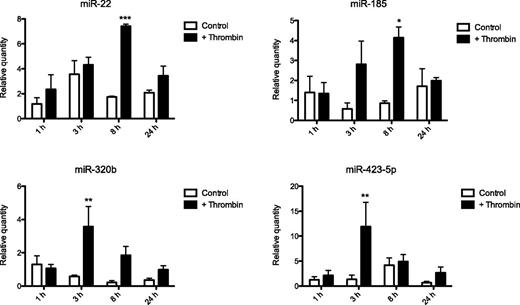
After these proof-of-concept experiments, we wanted to examine whether there was an endothelial uptake of the 4 miRNAs that were shown to be released from platelets (miR-22, -185, -320b, and -423-5). Platelets and endothelial cells were again cocultured in the presence or absence of thrombin and the time course for the uptake process was measured using qRT-PCR (Figure 3). In the presence of thrombin-activated platelets, there was a distinct but transient increase of all 4 miRNAs in the endothelial cells, peaking at either 3 or 8 hours depending on the miRNA species. However, when the experiment was repeated using resting platelets, the miRNA levels were unaffected. Expression of the endothelial cell-enriched miRNA miR-12621 was unaffected by the presence of activated platelets (supplemental Figure 9). Thrombin itself did not have any effect on miRNA levels in the endothelial cells (supplemental Figure 10).
Relative miRNA levels in endothelial cells cocultured with platelets. Levels of miR-22, -185, -320b, and 423-5p in HMEC-1 cells cocultured with platelets in the presence or absence of 1 U/mL thrombin at indicated time points. Levels are expressed relative to miR-16 and normalized to the mean baseline expression in each group (n = 3). Two-way analysis of variance with Bonferroni post hoc test was used to assess the effect of platelet activation on the levels of miRNA in HMEC-1 cells (*P < .05; **P < .01; ***P < .001).
Relative miRNA levels in endothelial cells cocultured with platelets. Levels of miR-22, -185, -320b, and 423-5p in HMEC-1 cells cocultured with platelets in the presence or absence of 1 U/mL thrombin at indicated time points. Levels are expressed relative to miR-16 and normalized to the mean baseline expression in each group (n = 3). Two-way analysis of variance with Bonferroni post hoc test was used to assess the effect of platelet activation on the levels of miRNA in HMEC-1 cells (*P < .05; **P < .01; ***P < .001).
Platelet miRNA is released and transferred by a vesicle-dependent mechanism
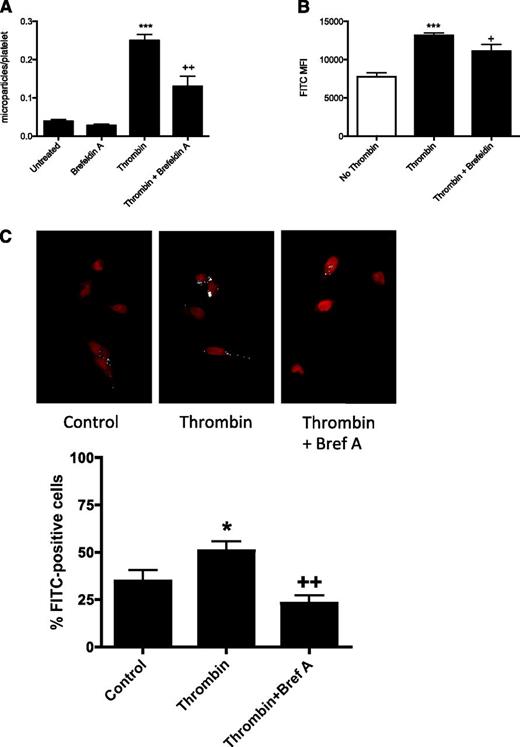
Considering the recent reports of microvesicle-dependent cell-to-cell miRNA transfer and the well-established notion of microparticle release from platelets, we hypothesized that the platelet miRNAs were released in microparticles. After a screening of several compounds reported to inhibit microparticle formation, we found that brefeldin A was the most effective microparticle inhibitor (Figure 4A; supplemental Figure 11). We then transfected platelets with fluorescently labeled miR-Scr-FITC and stimulated with thrombin in the presence or absence of brefeldin A. Fluorescent labeling of the generated microparticle fraction was then assessed with flow cytometry. There was a significant enrichment of miR-Scr-FITC in the CD42a+ microparticle population after stimulation with thrombin. This effect was partly inhibited with the addition of 10 μg/mL brefeldin A (Figure 4B). There was also a small but significant decrease in fluorescence intensity in transfected platelets, indicating release of miR-FITC upon activation (supplemental Figure 12).
miRNA is transferred to endothelial cells by a vesicle-dependent mechanism. (A) The effect of brefeldin A on platelet microparticle release was assessed with flow cytometry. Brefeldin A (10 μg/mL) was added to platelet-rich plasma 30 minutes before the addition of 1 U/mL thrombin. The microparticle population was defined based on size and expression of CD42a. ***P < .001 comparing untreated with thrombin-stimulated, ++P < .01 comparing thrombin vs thrombin + brefeldin A, n = 3. (B) Mean fluorescence in the FL1 channel within the microparticle population was used to assess miR-Scr-FITC content (***P < .001 comparing no thrombin vs thrombin-stimulated; +P < .05 comparing thrombin versus thrombin + brefeldin A, n = 6). (C) Quantification of FITC-labeled miRNA in HMEC-1 cells. Pixels of intensity greater than background staining were pseudocolored white. In each image the percentage of cells positive for white pixels were counted manually (*P < .05 comparing HMEC-1 cells cocultured with platelets in the presence or absence of thrombin; ++P < .01 comparing HMEC-1 cells cocultured with platelets in the presence of thrombin alone or with brefeldin A).
miRNA is transferred to endothelial cells by a vesicle-dependent mechanism. (A) The effect of brefeldin A on platelet microparticle release was assessed with flow cytometry. Brefeldin A (10 μg/mL) was added to platelet-rich plasma 30 minutes before the addition of 1 U/mL thrombin. The microparticle population was defined based on size and expression of CD42a. ***P < .001 comparing untreated with thrombin-stimulated, ++P < .01 comparing thrombin vs thrombin + brefeldin A, n = 3. (B) Mean fluorescence in the FL1 channel within the microparticle population was used to assess miR-Scr-FITC content (***P < .001 comparing no thrombin vs thrombin-stimulated; +P < .05 comparing thrombin versus thrombin + brefeldin A, n = 6). (C) Quantification of FITC-labeled miRNA in HMEC-1 cells. Pixels of intensity greater than background staining were pseudocolored white. In each image the percentage of cells positive for white pixels were counted manually (*P < .05 comparing HMEC-1 cells cocultured with platelets in the presence or absence of thrombin; ++P < .01 comparing HMEC-1 cells cocultured with platelets in the presence of thrombin alone or with brefeldin A).
To further test this, we cocultured miR-Scr-FITC−transfected platelets with HMEC-1 cells and activated the platelets with thrombin in the presence or absence of brefeldin A. After 3 hours of coculture, the proportion of FITC+ HMEC-1 was then assessed with confocal microscopy (Figure 4C). In the presence of activated platelets, ∼50% of the cells were FITC+ (a 1.6-fold increase compared with cells cultured with untreated platelets, P < .05). The addition of brefeldin A significantly decreased the proportion of FITC+ HMEC below the baseline, indicating that vesicle formation is required for an efficient release and transfer of miRNA from platelets to endothelial cells.
Endothelial cell gene expression is altered by activated platelets via a miRNA-dependent mechanism
Finally, our aim was to investigate whether the miRNAs released from activated platelets could confer any effect on target gene expression in the endothelial cells. The first step was to confirm the predicted mRNA targets of miR-22, -185, -320b, and -423-5p (see Table 1). HEK293 cells were cotransfected with luciferase reporter plasmids containing the 3′-UTR of ICAM-1, endothelial nitric oxide synthase, vascular endothelial growth factors A and B, and pre-miRNA corresponding to each of the 4 miRNA candidates. Overexpression of miR-22 and miR-320b caused significant dose-dependent quenching of the ICAM1 reporter signal compared with cells transfected with a scrambled pre-miR, indicating interaction of miRNA and mRNA (Figure 5A). The remaining miRNA-target interactions could not be confirmed (data not shown), so we chose to focus the rest of the study on the interaction of miR-22 and miR-320b with ICAM1.
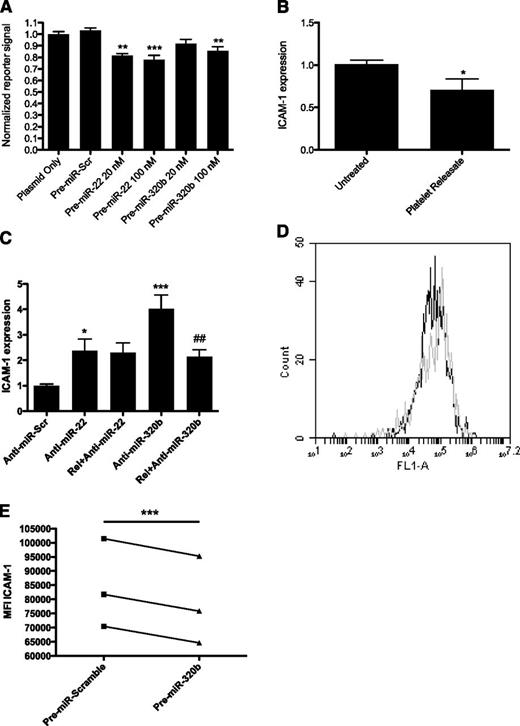
Function of miR-320b in endothelial cells. (A) Reporter gene signal from HEK293 cells transfected with 10 ng of a reporter plasmid containing the 3′-UTR of ICAM-1 and 20 or 100nM precursor miRNA corresponding to miR-22 or miR-320b and 10 ng of a reporter plasmid containing the 3′-UTR of ICAM-1. The reporter signal (Firefly luciferase) was normalized to Renilla luciferase to account for differences in transfection efficiency and expressed relative to the mean of the cells transfected with plasmid alone. Data come from 3 separate experiments with each sample run in triplicates. **P < .01; ***P < .001. (B) The level of ICAM-1 expression in HMEC-1 cells cultured for 24 hours in the presence of 20% platelet releasate. Gene expression was measured by qRT-PCR and is presented relative to the expression of cyclophilin A and normalized against the mean of the untreated control samples. Data from 3 separate experiments using triplicates are presented. *P < .05. (C) The levels of ICAM-1 in HMEC-1 cells upon treatment with 100nM scrambled anti-miRNA (anti-miR-Scr), anti-miR-22, or -320b in the presence or absence of 20% platelet releasate. Expression data are handled as mentioned previously (n = 6). *P < .05; ***P < .001 comparing cells treated with anti-miR-scr and anti-miR in the absence of platelet releasate; ##P < .01, comparing cells treated with anti-miR in the absence of releasate with cells treated with a combination of anti-miR and releasate. (D) Representative FACS plot of HMEC-1 cells transfected with 100nM pre-miR-320b (black line) or scrambled control pre-miRNA (gray line) and stained with a FITC-conjugated monoclonal antibody to ICAM-1. FL-1 fluorescence intensity reflects the surface expression of ICAM-1. (E) Surface expression of ICAM-1 in HMEC-1 (n = 3) transfected with 100nM pre-miR-320b or scrambled control pre-miRNA assessed with flow cytometry. ICAM-1 levels are expressed as the mean fluorescence intensity (MFI).***P < .001.
Function of miR-320b in endothelial cells. (A) Reporter gene signal from HEK293 cells transfected with 10 ng of a reporter plasmid containing the 3′-UTR of ICAM-1 and 20 or 100nM precursor miRNA corresponding to miR-22 or miR-320b and 10 ng of a reporter plasmid containing the 3′-UTR of ICAM-1. The reporter signal (Firefly luciferase) was normalized to Renilla luciferase to account for differences in transfection efficiency and expressed relative to the mean of the cells transfected with plasmid alone. Data come from 3 separate experiments with each sample run in triplicates. **P < .01; ***P < .001. (B) The level of ICAM-1 expression in HMEC-1 cells cultured for 24 hours in the presence of 20% platelet releasate. Gene expression was measured by qRT-PCR and is presented relative to the expression of cyclophilin A and normalized against the mean of the untreated control samples. Data from 3 separate experiments using triplicates are presented. *P < .05. (C) The levels of ICAM-1 in HMEC-1 cells upon treatment with 100nM scrambled anti-miRNA (anti-miR-Scr), anti-miR-22, or -320b in the presence or absence of 20% platelet releasate. Expression data are handled as mentioned previously (n = 6). *P < .05; ***P < .001 comparing cells treated with anti-miR-scr and anti-miR in the absence of platelet releasate; ##P < .01, comparing cells treated with anti-miR in the absence of releasate with cells treated with a combination of anti-miR and releasate. (D) Representative FACS plot of HMEC-1 cells transfected with 100nM pre-miR-320b (black line) or scrambled control pre-miRNA (gray line) and stained with a FITC-conjugated monoclonal antibody to ICAM-1. FL-1 fluorescence intensity reflects the surface expression of ICAM-1. (E) Surface expression of ICAM-1 in HMEC-1 (n = 3) transfected with 100nM pre-miR-320b or scrambled control pre-miRNA assessed with flow cytometry. ICAM-1 levels are expressed as the mean fluorescence intensity (MFI).***P < .001.
To investigate whether activated platelets could affect ICAM1 gene expression, HMEC-1 cells were cultured in the presence of platelet releasate. After 24 hours ICAM1 expression was down-regulated by ∼30% (P < .05; Figure 5B), supporting the hypothesis that the effect is mediated by miRNA.
To further confirm the interaction between miR-22/-320b and ICAM1 mRNA, we knocked down endogenous miRNA by using anti-miRNAs corresponding to miR-22 and miR-320b and measured ICAM1 gene expression using qRT-PCR (Figure 5C). As expected, knockdown of these miRNAs caused a significant increase in target mRNA, by two fold (P < .05) and four fold (P < .001), respectively, compared with cells treated with a scrambled Anti-miRNA. Then, our aim was to examine whether the effect on ICAM-1 expression observed when adding platelet releasate was caused by miRNAs. Platelet releasate was added to HMEC-1 cells, where miR-22 and miR-320b had been suppressed using anti-miRNA. Interestingly, the effect of anti-miR-320b, but not anti-miR-22, on ICAM-1 expression was rescued by the addition of platelet releasate to the endothelial cells (P < .01). This finding indicates an efficient transfer of specific functional miRNA species from the platelet releasate to the endothelial cells.
miR-320b regulates cell-surface ICAM-1 expression on endothelial cells
To confirm that miR-320b also can confer an effect on ICAM1 on the protein level, HMEC-1 cells were transfected with synthetic precursor miRNA or scrambled pre-miRNA and cell surface−bound ICAM-1 protein was quantified with flow cytometry after 48 hours. Overexpression of miR-320b was confirmed by qRT-PCR (supplemental Figure 13). The ICAM-1 mean fluorescence intensity decreased 7% (P < .001) in cells overexpressing miR-320b compared with scrambled control (Figure 5D-E), indicating a functional role of miR-320b on ICAM-1 in endothelial cells.
Discussion
The discovery of miRNA22,23 and the enzymatic infrastructure required to convert precursor-miRNA into mature miRNA11 in platelets was reported recently. A few studies mapping the full platelet miRNA repertoire have been conducted since then,24,25 but to our knowledge, no one has studied the platelet miRNA transcriptome in patients with myocardial infarction. In an effort to address this question we conducted RNA-seq of highly purified platelet preparations from patients with STEMI and from healthy individuals. The dominating miRNA species in both patients and controls were miR-320a, comprising >80% of total miRNA transcripts in healthy individuals. miR-320a has been detected in platelets in previous studies but not in the same proportions as here. Overall, the platelet miRNA profiles reported so far are quite disparate. Apart from differences in method used (microarray, qRT-PCR, RNA-seq), this might relate to differences in platelet preparation, varying degrees of platelet activation or presence of contaminating leukocyte or erythrocyte miRNAs. It is worth mentioning that RNA-seq, unlike for example microarray, is not a hybridization-based technique and the results are therefore considered more unbiased.26
We found 9 differentially expressed miRNAs in patients compared with controls, of which 8 were down-regulated in patients. This prompted us to investigate whether general platelet activation after myocardial infarction leads to a release of miRNA from platelets. The notion of miRNA release from activated or apoptotic cells has been proposed by several groups recently. Exosome-mediated transfer of miRNA have been shown in activated mast cells27 and from T cells to antigen-presenting cells.18 In a recent report, apoptotic bodies carrying miR-126 between endothelial cells caused up-regulation of CXCL12 in recipient cells.14 A very recent report shows that smooth muscle cells provide an atheroprotective signal to endothelial cells through vesicular transfer of miR-143/145.28
We could confirm release of miR-22, -185, 320b, and -423-5p from ex vivo−aggregated platelets and also noticed a depletion of these miRNAs in thrombi aspirated from patients with STEMI. The release of platelet miRNAs was not reflected in increased overall levels in the circulation of patients. It could be argued that the release of miRNAs from activated platelets is likely to be a local event at the site of the ruptured plaque and would therefore not be detectable in the periphery. Regarding the results from the thrombi, it is possible that the relative decrease in platelet miRNAs is partly attributable to the dilution of platelets with other cell types in the thrombus. This may exaggerate the decline in platelet miRNAs somewhat but does not account for the total lack of miR-22, for example.
We were able to demonstrate efficient transfer of both fluorescently labeled miRNA and exogenous C elegans miRNA from activated platelets to an endothelial cell line. Coculture of activated platelets and endothelial cells caused a transient increase of all 4 platelet miRNAs, but not the endothelial cell-enriched miR-126, in HMEC-1 cells. Considering that the miRNA transfer could be completely abolished with the addition of brefeldin A, which we show to be an inhibitor of platelet microparticle release, it is likely that the miRNA release is vesicle-dependent. The effect of brefeldin A is probably best explained by its well documented ability to disrupt vesicular transport processes.29 Brefeldin A has recently been reported to inhibit release of exosome-like vesicles from cells30 and to suppress exosome-dependent transfer of miRNA between immune cells.18
However, we cannot rule out that brefeldin A also affects the endothelial uptake of miRNA. In a recent report, Diehl et al31 showed that platelet microparticles were enriched for distinct miRNA species, which suggests that secretion of miRNAs from platelets is a selective process. Although the exact mechanism of release and uptake requires further research, recent reports on exovesicle/microparticle-mediated cell-to-cell transfer of miRNA and the well-researched mechanism of platelet microparticle release, do strengthen this hypothesis.
We observed a decreased expression of ICAM-1 in the presence of platelet releasate and hypothesized that this effect might be mediated by released miRNA. De-repression of ICAM-1 mediated by anti-miR-320b, but not anti-miR-22, was almost completely abolished by the addition of platelet releasate. This selective effect rules against the possibility that some factor in the platelet releasate causes a general effect on the endothelium, independent of miRNA.
In contrast to the results presented herein, previous authors have reported increased endothelial ICAM-1 expression in the presence of platelet releasate through an interleukin-1−dependent mechanism.32,33 The discrepancy might be attributed to differences in the dynamics of ICAM-1 regulation. Gawaz et al33 reported a maximal ICAM-1 expression after 16 hours. In the present study, ICAM-1 expression was evaluated after 24 hours. It is possible that regulation of ICAM-1 expression by external miRNAs might represent an additional, regulatory mechanism, counteracting that of interleukin-1 in the long-term. One might speculate that the role of platelet-derived miRNA in the endothelium might be to regulate and fine-tune the effects of the proinflammatory mediators in the releasate. Indeed, several recent articles have been published in which authors propose miRNAs as fine-tuners of the inflammatory response.34,35
In conclusion, we report that the platelet miRNA content of patients with STEMI is distinctly different from that of healthy patients, that miRNAs are shed from activated platelets, and that platelet-derived miRNAs can affect endothelial cell gene expression.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Karin Wallmark and Helén Brogren of the Wallenberg Laboratory for Cardiovascular Research, Institute of Medicine, University of Gothenburg for preparation of platelet releasate, and Siv Svensson of the Department of Cardiology, Lund University, for assistance with blood sampling and preparation of platelets.
This work was supported by the Swedish Heart and Lung Foundation, Swedish Scientific Research Council (521-2009-2276), governmental funding of clinical research within the Swedish National Health Service, and Lund University Hospital funds.
Authorship
Contribution: O.G. designed and performed experiments, analyzed data, and wrote the manuscript; M.v.d.B. designed and performed experiments, analyzed data, and edited the manuscript; J.Ö. designed and performed experiments, analyzed data, and edited the manuscript; P.G. provided vital samples and edited the manuscript; B.O. designed experiments and edited the manuscript; and C.W. and D.E. designed the study and edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for M.v.d.B. is Genentech, South San Francisco, CA.
Correspondence: Olof Gidlöf, Department of Cardiology, Lund University Hospital, SE-221 85 Lund, Sweden; e-mail: olof.gidlof@med.lu.se.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal