Key Points
Rituximab causes a polarization of B cells, involving a reorganization of CD20, intercellular adhesion molecule 1, and moesin, and orientation of the microtubule organizing center.
The polarization of B cells induced by rituximab augments its therapeutic role in triggering ADCC by effector NK cells.
Abstract
Rituximab, which binds CD20 on B cells, is one of the best-characterized antibodies used in the treatment of B-cell malignancies and autoimmune diseases. Rituximab triggers natural killer (NK)-cell–mediated antibody-dependent cellular cytotoxicity (ADCC), but little is known about the spatial and temporal dynamics of cell-cell interactions during ADCC or what makes rituximab potent at triggering ADCC. Here, using laser scanning confocal microscopy, we found that rituximab caused CD20 to cap at the B-cell surface independent of antibody crosslinking or intercellular contact. Unexpectedly, other proteins, including intercellular adhesion molecule 1 and moesin, were selectively recruited to the cap of CD20 and the microtubule organizing center became polarized toward the cap. Importantly, the frequency at which NK cells would kill target cells via ADCC increased by 60% when target cells were polarized compared with when they were unpolarized. Polarized B cells were lysed more frequently still when initial contact with NK cells occurred at the place where CD20 was capped. This demonstrates that the site of contact between immune cells and target cells influences immune responses. Together, these data establish that rituximab causes a polarization of B cells and this augments its therapeutic function in triggering NK-cell–mediated ADCC.
Introduction
Depletion of malignant or autoreactive B cells plays an important role in the treatment of B-cell lymphomas and autoimmune diseases.1,2 Rituximab is a chimeric human-mouse antibody that targets CD20, a pan–B-cell surface marker, and mediates depletion of these cells.3 CD20 is highly expressed on the surface of B cells as well as the majority of B-cell lymphomas4 but absent from hematopoietic stem cells, differentiated plasma cells, and other healthy tissues, allowing for a specific targeting of desired cells. Furthermore, it is not shed or internalized from the surface of the cells upon antibody binding,5 making it a good target for efficient induction of effector mechanisms that mediate depletion of B cells.
The exact mechanism of rituximab-mediated B-cell depletion in patients is not fully understood. Rituximab can potentially trigger 3 effector functions: programmed cell death6 ; induction of complement-dependent cytotoxicity; or activation of immune cells, including natural killer (NK) cells, which express Fc γ receptor III (CD16) to mediate antibody-dependent cellular cytotoxicity (ADCC).7,8 The respective importance of these mechanisms may vary in different environments. Evidence that ADCC is important for the activity of rituximab in vivo is that mice deficient in activating Fc receptors responded poorly to antibody treatment.7 Similarly for humans, patients with high-affinity CD16 polymorphism responded better to rituximab treatment than those with low-affinity receptor.8 There is also evidence that macrophages and neutrophils conjugate with antibody-opsonized targets, forming ADCC synapses in vivo in mice.9 In humans, NK cells are considered to be the main mediators of ADCC and, indeed, NK cells efficiently kill B cells opsonized with rituximab in vitro and in vivo.10-13
However, few studies have used microscopy to visualize what happens during ADCC.14 Here, we employed high-resolution microscopy to study the sequence of events when NK cells attach to and then kill target cells opsonized by rituximab. Unexpectedly, we found that rituximab induces polarization of CD20, intercellular adhesion molecule 1 (ICAM-1), myosin, and the microtubule organizing center (MTOC); such polarized cells are preferentially killed by effector NK cells, especially in interactions where NK cells initially contact B cells where CD20 has been capped. These data are important in establishing the properties that a therapeutic antibody should have to be optimal in triggering ADCC.
Methods
Cells and constructs
Daudi, 721.221, Raji, and Jurkat cells were maintained in RPMI-1640 supplemented with 10% fetal calf serum, 50 U/mL penicillin, 50 µg/mL streptomycin, and 2 mM l-glutamine (all from Invitrogen) (referred to later as “complete medium”). The plasmid encoding CD20 and green fluorescent protein (GFP) was a kind gift from J. Deans (Calgary, Canada). CD20-GFP was subcloned as an AgeI/BamHI fragment into the retroviral pIB2 vector, a gift from M. Purbhoo, Imperial College London.
Peripheral blood NK cells and B cells were isolated by negative selection from healthy donor lymphocyte cones purchased from the National Blood Service or fresh blood using magnetic beads (NK cell isolation kit, B cell isolation kit; Miltenyi Biotec). All fresh-blood donors were healthy and gave informed consent for their blood to be used in accordance with the Declaration of Helsinki (with ethics approved by The National Research Ethics Service, Ref 05/Q0401/108). Cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% human serum (type AB; Sigma-Aldrich), 30% nutrient mixture F-12, 2 mM l-glutamine, 1× nonessential amino acids, 1 mM sodium pyruvate, 50 µM 2-ME, 50 U/mL penicillin, and 50 µg/mL streptomycin (all from Invitrogen). Clinical-grade rituximab (Rituxan, Roche) was used at 10 µg/mL for 1 hour unless indicated otherwise. Where indicated, cells were pretreated with 20 µg/mL CD32 blocking monoclonal antibody (mAb) (Clone IV.3, Stem Cell Technologies) or 10 µM nocodazole (Sigma) in complete medium for 30 minutes at 37°C prior to incubation with rituximab. Drugs were then maintained in the medium during incubation with rituximab.
Immunostaining and imaging
For colocalization experiments, Daudi or primary human B cells were incubated with 10 µg/mL 2H7 or rituximab for 1 hour and fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS). For intracellular staining, cells were then permeabilized with 0.05% saponin/PBS (Sigma) and stained with mAb. For live cell imaging, 5 × 104 Daudi cells were preincubated with 0.01 to 10 µg/mL rituximab for 1 hour. A total of 1 × 105 primary NK cells were preincubated with 1 µL/mL LysoTracker red DND-99 (Molecular Probes) for 1 hour. Cells were then mixed together and imaged in 8-well chambers (Chambered Borosilicate Coverglass, Nunc) precoated with 10 μg/mL fibronectin/PBS (Sigma). Cells were imaged in the presence of 0.5 μM Sytox blue (Invitrogen) to visualize cell death. Bright-field and fluorescence images were obtained by confocal microscopy (Leica SP5 RS) with a 63× water immersion lens (NA 1.2) with live cell samples maintained at 37°C with 5% (vol/vol) CO2. Time-lapse imaging was performed over 40 to 60 minutes with confocal stacks being acquired every 30 to 40 seconds. For quantification of target cell killing, no killing was scored if target and effector cells parted without target cell death or if the 2 cells stayed in contact for at least 20 minutes until the end of the acquisition. Images were analyzed (Volocity, Improvision, and ImageJ, National Institutes of Health) and colocalization between 2 fluorescence channels assessed by calculating the Pearson’s correlation coefficient (image correlation analysis plug-in for ImageJ15 ). Brightness and contrast were changed in some images for presentation of the figures shown, but analysis used raw images. For flow-based microscopy, Daudi/CD20-GFP cells were incubated with rituximab for 1 hour and fixed with 4% paraformaldehyde/PBS, imaged using a multispectral imaging flow cytometer (ImageStream100, Amnis), and CD20 capping analyzed (IDEAS software, Amnis).
Statistical analysis
Column statistics were performed (GraphPad software, Prism), and unless specified otherwise, mean values and standard error of the mean (SEM) are shown. Data were analyzed by a 1-way analysis of variance (ANOVA) test with Bonferroni adjustment. To analyze the MTOC polarization, a Kolmogorov-Smirnov test was performed using an application available online (http://www.physics.csbsju.edu/stats/KS-test.html).
Results
Rituximab triggers capping of CD20 at the surface of B cells independent of crosslinking
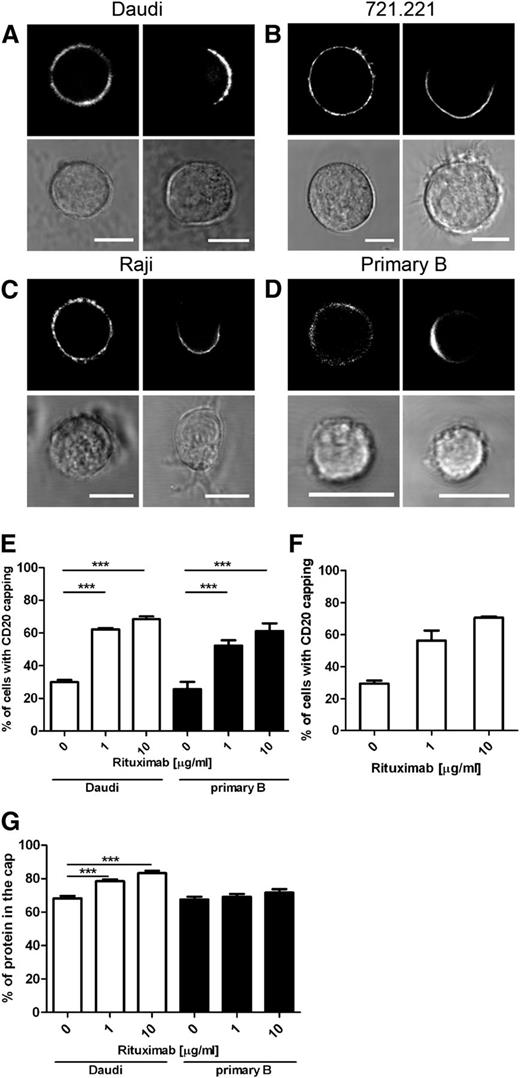
To investigate the effect of opsonization with rituximab on the organization of the B-cell surface, 3 B-cell lines—the Burkitt lymphoma B cell lines, Daudi and Raji, and the EBV-transformed B cell line 721.221—were transfected to express CD20 with an N-terminal GFP tag. Levels of CD20 expression in these transfectants were somewhat higher but comparable to the levels of expression of CD20 in WT cells (supplemental Figure 1). CD20-GFP was frequently distributed evenly (on a micrometer scale) around the cell surface of all 3 cell lines prior to the addition of rituximab (Figure 1A-C left panels). Incubation with 10 µg/mL rituximab induced CD20 to frequently cap on 1 side of all 3 cell lines tested (Figure 1A-C right panels). Importantly, crosslinking of rituximab by a secondary antibody was not required to observe this effect.
Rituximab enhances capping of CD20 on the B-cell surface. Fluorescent and bright-field images of B-cell lines Daudi (A), 721.221 (B), and Raji (C) expressing CD20-GFP or primary B cells (D) untreated or incubated with 10 μg/mL rituximab (left and right panels, respectively). Primary B cells were additionally labeled with anti–CD20-AF647 antibody recognizing the intracellular portion of the protein. Scale bars represent 10 μm. (E) Capping of CD20 was quantified on images of Daudi/CD20-GFP and primary B cells incubated in the absence or presence of 1 μg/mL and 10 μg/mL rituximab. Graph represents mean ± SEM of 3 independent experiments. (F) Capping of CD20 in Daudi/CD20-GFP cells was quantified by flow microscopy. Cells were incubated alone or with 1 μg/mL or 10 μg/mL rituximab and then fixed and analyzed by the ImageStream multispectral imaging flow cytometer. Graph represents mean ± SEM of 2 independent experiments. (G) Quantification of the fraction of CD20 localized in the cap in Daudi/CD20-GFP and primary B cells. A total of 30 to 32 cells were analyzed per condition. Data were analyzed by 1-way ANOVA with Bonferroni post-test. ***P < .001.
Rituximab enhances capping of CD20 on the B-cell surface. Fluorescent and bright-field images of B-cell lines Daudi (A), 721.221 (B), and Raji (C) expressing CD20-GFP or primary B cells (D) untreated or incubated with 10 μg/mL rituximab (left and right panels, respectively). Primary B cells were additionally labeled with anti–CD20-AF647 antibody recognizing the intracellular portion of the protein. Scale bars represent 10 μm. (E) Capping of CD20 was quantified on images of Daudi/CD20-GFP and primary B cells incubated in the absence or presence of 1 μg/mL and 10 μg/mL rituximab. Graph represents mean ± SEM of 3 independent experiments. (F) Capping of CD20 in Daudi/CD20-GFP cells was quantified by flow microscopy. Cells were incubated alone or with 1 μg/mL or 10 μg/mL rituximab and then fixed and analyzed by the ImageStream multispectral imaging flow cytometer. Graph represents mean ± SEM of 2 independent experiments. (G) Quantification of the fraction of CD20 localized in the cap in Daudi/CD20-GFP and primary B cells. A total of 30 to 32 cells were analyzed per condition. Data were analyzed by 1-way ANOVA with Bonferroni post-test. ***P < .001.
To test whether or not CD20 endogenously expressed in primary B cells also caps, peripheral blood B cells were isolated and left untreated or incubated with 10 µg/mL rituximab and then fixed and stained with an antibody targeting the cytoplasmic portion of CD20. Rituximab caused CD20 to commonly cap to 1 side of primary B cells (Figure 1D). Without rituximab, the frequency at which CD20 was capped was 25.7% ± 4.5% and 29.9% ± 1.4% for primary B cells and Daudi/CD20-GFP cells, respectively (Figure 1E). Upon treatment with 10 µg/mL rituximab, 61.2% ± 4.7% of primary B cells and 68.5% ± 1.6% of Daudi transfectants had CD20 capped to 1 side of the cell (Figure 1E). Additionally, quantification of CD20 polarization in Daudi/CD20-GFP cells was assessed by flow microscopy, allowing unbiased automated measurements of CD20 capping in a large number of cells (>350 cells per condition per experiment). Data obtained by flow microscopy were in agreement with the results from single-cell quantification; capping of CD20 was observed in 29.6% ± 1.9% of untreated cells and increased to 70.6% ± 0.8% in cells incubated with 10 µg/mL rituximab (Figure 1F).
Quantification of the confocal microscope images taken throughout the volume of the cell revealed that the majority of the CD20 was accumulated in the cap. A total of 68% ± 2% to 83% ± 1% and 68% ± 2% to 72% ± 2% of total CD20 was accumulated in the cap within Daudi and primary B cells, respectively (Figure 1G). The amount of protein accumulated in the cap slightly (but significantly) increased in Daudi cells treated with rituximab but not significantly in primary B cells. These differences may indicate quantitative differences in the effects of rituximab on primary cells and immortal cell lines.
To investigate whether or not capping of CD20 was specific for rituximab or could be triggered by any mAb against CD20, we compared treatment with 10 µg/mL 2H7, a mAb that targets a similar epitope within CD20 as rituximab.16 In contrast to rituximab, incubation with 2H7 did not increase the number of cells with polarized CD20 (Figure 2A). One possible explanation for antibodies to vary in their ability to cause protein capping would be if they were internalized differentially. However, flow cytometric analysis confirmed that neither rituximab nor 2H7 was internalized to a significant extent over the time frame of these experiments (supplemental Figure 2A). Taken together, these data establish that a large fraction of cell-surface CD20 is capped to 1 side of a B cell upon treatment with rituximab specifically.
Enhancement of CD20 capping is specific for rituximab and B cells. (A) Daudi/CD20-GFP cells were incubated alone; or in the presence of 10 μg/mL rituximab (Rtx) or 10 μg/mL mouse CD20-targeting antibody 2H7; or preincubated with 20 µg/mL CD32-blocking antibody followed by incubation with 10 μg/mL rituximab (Rtx+CD32); or incubated with 20 μg/mL of a monovalent version of rituximab (Rtx mono). Number of cells with polarization of CD20 was scored. Graph represents mean ± SEM of 3 independent experiments. More than 160 cells were analyzed per condition. (B) Fluorescent and bright-field images of Jurkat/CD20-GFP cells untreated (left panels) or incubated with 10 μg/mL rituximab with uniform distribution of CD20 or CD20 capped on 1 side (middle and right panels, respectively). (C) Capping of CD20 was quantified on images of Jurkat/CD20-GFP cells incubated in the absence or presence of 10 μg/mL rituximab. Graph represents mean ± SEM of 3 independent experiments. A total of 112 to 118 cells were analyzed per condition. (D) Amount of CD20 accumulated in the cap in Jurkat/CD20-GFP cells pretreated with 10 μg/mL rituximab was compared with the amount of CD20 accumulated in the cap in primary B cells pretreated with 10 μg/mL rituximab. Data were analyzed by unpaired t test (2-tailed). A total of 21 to 31 cells from 3 or 4 experiments were analyzed per condition. (E) Daudi/CD20-GFP (CD20 WT) or Daudi cells expressing a mutant form of CD20-GFP (CD20 mut) were left untreated or incubated with rituximab and then fixed and analyzed by laser scanning confocal microscopy. The proportion of cells with polarized CD20 is shown. Graph represents mean ± SEM of 3 independent experiments. Data were analyzed by unpaired t test (2-tailed). More than 200 cells were analyzed per condition. (F) The amount of CD20 accumulated in the cap in Daudi cells expressing a mutant form of CD20-GFP (CD20 mut) pretreated with 10 μg/mL rituximab was compared with the amount of CD20 accumulated in the cap in Daudi/CD20-GFP (CD20 WT) cells pretreated with 10 μg/mL rituximab. A total of 31 cells from 2 or 3 experiments were analyzed per condition. Data were analyzed by unpaired t test (2-tailed). **P < .01; ***P < .001. Ab, antibody.
Enhancement of CD20 capping is specific for rituximab and B cells. (A) Daudi/CD20-GFP cells were incubated alone; or in the presence of 10 μg/mL rituximab (Rtx) or 10 μg/mL mouse CD20-targeting antibody 2H7; or preincubated with 20 µg/mL CD32-blocking antibody followed by incubation with 10 μg/mL rituximab (Rtx+CD32); or incubated with 20 μg/mL of a monovalent version of rituximab (Rtx mono). Number of cells with polarization of CD20 was scored. Graph represents mean ± SEM of 3 independent experiments. More than 160 cells were analyzed per condition. (B) Fluorescent and bright-field images of Jurkat/CD20-GFP cells untreated (left panels) or incubated with 10 μg/mL rituximab with uniform distribution of CD20 or CD20 capped on 1 side (middle and right panels, respectively). (C) Capping of CD20 was quantified on images of Jurkat/CD20-GFP cells incubated in the absence or presence of 10 μg/mL rituximab. Graph represents mean ± SEM of 3 independent experiments. A total of 112 to 118 cells were analyzed per condition. (D) Amount of CD20 accumulated in the cap in Jurkat/CD20-GFP cells pretreated with 10 μg/mL rituximab was compared with the amount of CD20 accumulated in the cap in primary B cells pretreated with 10 μg/mL rituximab. Data were analyzed by unpaired t test (2-tailed). A total of 21 to 31 cells from 3 or 4 experiments were analyzed per condition. (E) Daudi/CD20-GFP (CD20 WT) or Daudi cells expressing a mutant form of CD20-GFP (CD20 mut) were left untreated or incubated with rituximab and then fixed and analyzed by laser scanning confocal microscopy. The proportion of cells with polarized CD20 is shown. Graph represents mean ± SEM of 3 independent experiments. Data were analyzed by unpaired t test (2-tailed). More than 200 cells were analyzed per condition. (F) The amount of CD20 accumulated in the cap in Daudi cells expressing a mutant form of CD20-GFP (CD20 mut) pretreated with 10 μg/mL rituximab was compared with the amount of CD20 accumulated in the cap in Daudi/CD20-GFP (CD20 WT) cells pretreated with 10 μg/mL rituximab. A total of 31 cells from 2 or 3 experiments were analyzed per condition. Data were analyzed by unpaired t test (2-tailed). **P < .01; ***P < .001. Ab, antibody.
Fc receptors are not involved in rituximab-mediated capping of CD20
B cells express FcγRIIb (CD32) on their surface, and therefore binding of the Fc portion of rituximab to CD32 could potentially be involved in capping CD20. Indeed, it has been previously suggested that rituximab can crosslink CD20 and CD32.17 To test this possibility, Daudi cells were pretreated with a blocking mAb against CD32 for 1 hour prior to incubation with rituximab. Blocking CD32 did not influence the frequency at which cells were capped by rituximab, indicating that the interaction between the Fc portion of the antibody and CD32 does not play a role in mediating CD20 polarization (Figure 2A). To assess whether or not the bivalency of rituximab was required to induce CD20 capping, cells were also incubated with a monovalent version of rituximab immunoglobulin G (IgG) (described in “supplemental Methods”) that bound efficiently to CD20 (supplemental Figure 2B). Monovalent rituximab IgG was not able to increase the frequency at which CD20 was capped in B cells (Figure 2A). Taken together, these data demonstrate that rituximab triggers the capping of CD20 independent of crosslinking but requiring the bivalency of the mAb.
Rituximab does not efficiently cap CD20 in T-cell transfectants
To investigate whether or not capping of CD20 was specific to B cells or could occur in other cell types, Jurkat T cells were transfected to express CD20-GFP (Jurkat/CD20-GFP). The level of expression of CD20 in Jurkat/CD20-GFP and Daudi/CD20-GFP was comparable (supplemental Figure 3A). Surprisingly, CD20 was never polarized in untreated Jurkat/CD20-GFP and rituximab treatment induced capping of CD20 in only 12.1% ± 1.6% of cells (Figure 2B-C). In the cells where CD20 was capped, the amount of protein in the cap (58.1% ± 2.5%) was significantly less than in primary B cells (Figure 2D). The interaction of rituximab with CD20 was preserved in Jurkat/CD20-GFP cells as they became susceptible to NK-cell–mediated ADCC, albeit to a relatively low extent (supplemental Figure 3B). These data indicate that rituximab-mediated capping of CD20 is especially pronounced for B cells, likely requiring cellular proteins that are absent from T cells. This adds further evidence that rituximab does more than merely bind CD20 passively at the cell surface.
Redistribution to lipid rafts is not essential for rituximab-mediated CD20 capping
Rituximab has previously been shown to redistribute CD20 into lipid rafts.18 To test whether or not recruitment to lipid rafts caused capping of CD20, Daudi B cells were transfected to express GFP attached to a mutant variant of CD20 (CΔ219-225) that lacks a membrane-proximal sequence previously established to be important for lipid-raft redistribution of the protein.19 Translocation of this mutated version of CD20 to the lipid rafts upon antibody binding is reduced by 75% as compared with the wild-type (WT) protein.19 Here, cells expressing the WT or mutated version of CD20-GFP were treated with rituximab and the localization of the fluorescent CD20 was compared (Figure 2E). Both the mutant and WT CD20 were equally frequently capped in Daudi transfectants upon treatment with rituximab (70.4% ± 5.6% vs 66.9% ± 6.4%, respectively), and the amount of mutant CD20 localized in the cap (77.9% ± 1.4%) was significantly lower than the amount observed for WT CD20, though the difference was very small (Figure 2F). Thus, redistribution of CD20 to lipid rafts is not essential for capping of this protein caused by rituximab.
Other proteins selectively colocalize with CD20 in the cap
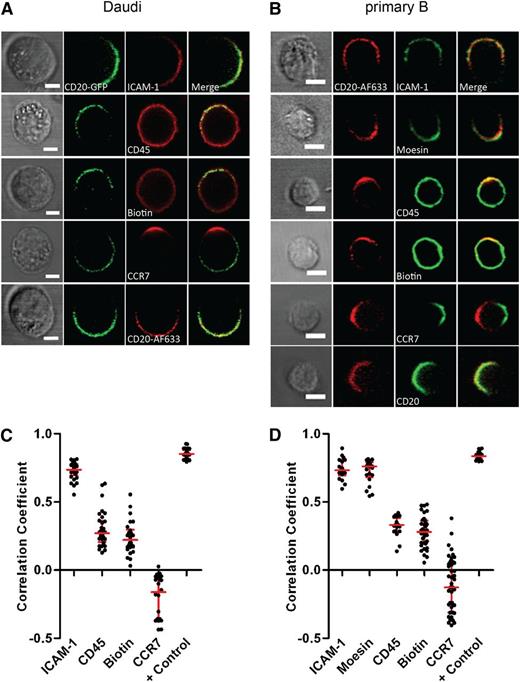
We next set out to test whether or not other proteins colocalized within the cap of CD20 after rituximab treatment. Primary B cells or Daudi were incubated with rituximab and then fixed and stained for the localization of other proteins: (1) the integrin ICAM-1; (2) moesin, a member of the ezrin-radixin-moesin family that is involved in crosslinking plasma membrane proteins such as ICAM-1 with the actin cytoskeleton20 ; (3) CD45, a protein tyrosine phosphatase abundantly expressed on the surface of B cells; or (4) surface proteins in general, marked with biotin (Figure 3A-B). Colocalization of proteins relative to CD20 was analyzed by confocal microscopy and Pearson’s correlation coefficients calculated. Pearson’s correlation coefficients are between 1 and −1, where 1 indicates high colocalization and −1 indicates anticorrelation. The adhesion molecule ICAM-1 strongly colocalized with capped CD20 both in Daudi (Figure 3A,C) and primary B cells (Figure 3B,D), with correlation coefficient values of 0.73 ± 0.01 and 0.74 ± 0.02, respectively. Staining for moesin was very weak in Daudi, but in primary B cells this protein also colocalized with CD20 (correlation coefficient: 0.72 ± 0.02) (Figure 3B,D).
Other proteins colocalize with CD20 in the cap. (A) Daudi/CD20-GFP and (B) primary B cells were incubated with 10 μg/mL rituximab or rituximab-AF633, respectively, for 1 hour and then fixed and stained for ICAM-1, CD45, CCR7, moesin, or biotinylated as indicated followed by incubation with secondary fluorescently labeled antibodies. As a positive control in Daudi/CD20-GFP cells, CD20 was targeted by rituximab AF633 and colocalization between green and red fluorescent channels was calculated (A). As a positive control, primary B cells were additionally stained for CD20 using an antibody recognizing the cytoplasmic portion of the protein followed by incubation with secondary fluorescently labeled antibody (B). Scale bars represent 5 μm. (C-D) Pearson’s correlation coefficients calculated for colocalization of CD20 and other cell components as indicated in individual Daudi (C) or primary B cells (D) are shown. A total of 18 to 46 cells were analyzed per condition.
Other proteins colocalize with CD20 in the cap. (A) Daudi/CD20-GFP and (B) primary B cells were incubated with 10 μg/mL rituximab or rituximab-AF633, respectively, for 1 hour and then fixed and stained for ICAM-1, CD45, CCR7, moesin, or biotinylated as indicated followed by incubation with secondary fluorescently labeled antibodies. As a positive control in Daudi/CD20-GFP cells, CD20 was targeted by rituximab AF633 and colocalization between green and red fluorescent channels was calculated (A). As a positive control, primary B cells were additionally stained for CD20 using an antibody recognizing the cytoplasmic portion of the protein followed by incubation with secondary fluorescently labeled antibody (B). Scale bars represent 5 μm. (C-D) Pearson’s correlation coefficients calculated for colocalization of CD20 and other cell components as indicated in individual Daudi (C) or primary B cells (D) are shown. A total of 18 to 46 cells were analyzed per condition.
However, in contrast, CD45 did not cap with CD20 and remained uniformly distributed throughout the plasma membrane (correlation coefficient: 0.30 ± 0.02 and 0.32 ± 0.02 for Daudi and primary B, respectively). Likewise, surface proteins in general, visualized by biotinylation followed by staining with fluorescently labeled streptavidin, remained homogeneously distributed around the cell surface and the level of colocalization with CD20 was not high (correlation coefficient: 0.24 ± 0.02 and 0.28 ± 0.02 for Daudi and primary B, respectively) (Figure 3A-D). Thus, the enrichment of surface proteins in the cap was selective.
The localization of surface proteins in rituximab-treated cells is reminiscent of polarized lymphocytes characterized by a differential localization of proteins at the leading edge and uropod, rather than cells in which the antibody has merely capped its ligand. ICAM-1 and moesin are known to be strongly enriched in the uropod of lymphocytes.21 Conversely, CD45 has been reported to be uniformly distributed on the cell surface of polarized lymphocytes.22 The chemokine receptor CCR7 is a surface protein known to localize specifically to the leading edge of migrating lymphocytes, and thus we assessed the localization of CCR7 in relation to the cap of CD20 in cells treated with rituximab. CCR7 did not colocalize with CD20 and commonly accumulated at the opposite side of the cell to the cap of CD20, both in primary B cells and in Daudi, evidenced by a negative correlation coefficient (−0.11 ± 0.03 and −0.20 ± 0.03 for primary B and Daudi, respectively) (Figure 3A-D). Taken together, these data suggest that rituximab causes B cells to adopt a polarized phenotype.
The MTOC polarizes toward the CD20 cap
A characteristic consequence of cellular polarization is a specific orientation for the MTOC. Here, to assess the localization of the MTOC relative to the CD20-rich cap, Daudi and primary B cells were incubated for 1 hour with rituximab and then fixed and stained for α-tubulin (Figure 4A-B). The relative distance between the MTOC and the center of CD20-enriched cap was measured and a polarity index calculated as the ratio between distance of the MTOC to the CD20 cap and the cell diameter (Figure 4B). A total of 97% of Daudi and 92% of primary B cells had a polarity index below 0.5, which indicated that the MTOC was almost always polarized toward the cap of CD20 (Figure 4B).
The microtubule network is involved in rituximab-mediated CD20 polarization. (A) Fluorescent and bright-field images of Daudi/CD20-GFP cells incubated with rituximab and labeled for α-tubulin (red). The MTOC is identified as the brightest spot in the red channel. Scale bar represents 10 μm. (B, left) Schematic representation of a cell in which MTOC polarization toward the CD20-enriched region was assessed by calculating the polarity index values corresponding to the ratio between the distance from the MTOC to the CD20 cap (a) and the cell diameter (b). Distribution of polarity indexes in primary B cells (B, top right) and Daudi cells (B, bottom right) incubated with rituximab is shown. A total of 74 Daudi and 84 primary B cells were analyzed. Proportion of Daudi (C) and primary B cells (D) with polarized CD20 in untreated cells or after incubation with rituximab and nocodazole. Graphs represent mean ± SEM of 3 independent experiments. More than 100 cells were analyzed per condition. Data were analyzed by 1-way ANOVA with Bonferroni post-test. *P < .05; **P < .01; ***P < .001.
The microtubule network is involved in rituximab-mediated CD20 polarization. (A) Fluorescent and bright-field images of Daudi/CD20-GFP cells incubated with rituximab and labeled for α-tubulin (red). The MTOC is identified as the brightest spot in the red channel. Scale bar represents 10 μm. (B, left) Schematic representation of a cell in which MTOC polarization toward the CD20-enriched region was assessed by calculating the polarity index values corresponding to the ratio between the distance from the MTOC to the CD20 cap (a) and the cell diameter (b). Distribution of polarity indexes in primary B cells (B, top right) and Daudi cells (B, bottom right) incubated with rituximab is shown. A total of 74 Daudi and 84 primary B cells were analyzed. Proportion of Daudi (C) and primary B cells (D) with polarized CD20 in untreated cells or after incubation with rituximab and nocodazole. Graphs represent mean ± SEM of 3 independent experiments. More than 100 cells were analyzed per condition. Data were analyzed by 1-way ANOVA with Bonferroni post-test. *P < .05; **P < .01; ***P < .001.
An intact microtubule network is required for CD20 capping
To determine whether the microtubule network was involved in facilitating the reorganization of cell-surface proteins, cells were treated with the microtubule-perturbing drug nocodazole and analyzed for the extent to which CD20 was capped after treatment with rituximab. The activity of the drug was confirmed since after nocodazole treatment the MTOC was visually undetected when cells were stained with an anti–α-tubulin mAb (supplemental Figure 4). The frequency at which rituximab caused CD20 to cap in cells was 64.3% ± 3.5% and 66.7% ± 1.8% for Daudi and primary B cells, respectively, but this reduced to 31.3% ± 2.5% and 42.3% ± 2.7% when cells were also treated with nocodazole (Figure 4C-D). Nocodazole did not affect the frequency at which cells not treated with rituximab sometimes exhibited a cap of CD20 (21.9% ± 1.3% and 29.7% ± 4.2% in untreated Daudi and primary B cells, respectively), and for the same cells treated with nocodazole, this was 18.3% ± 3.6% and 22.7% ± 8.1%, respectively. This indicates that an intact microtubule network is important for rituximab-mediated CD20 reorganization.
Polarization of B cells by rituximab influences the efficiency of target cell killing
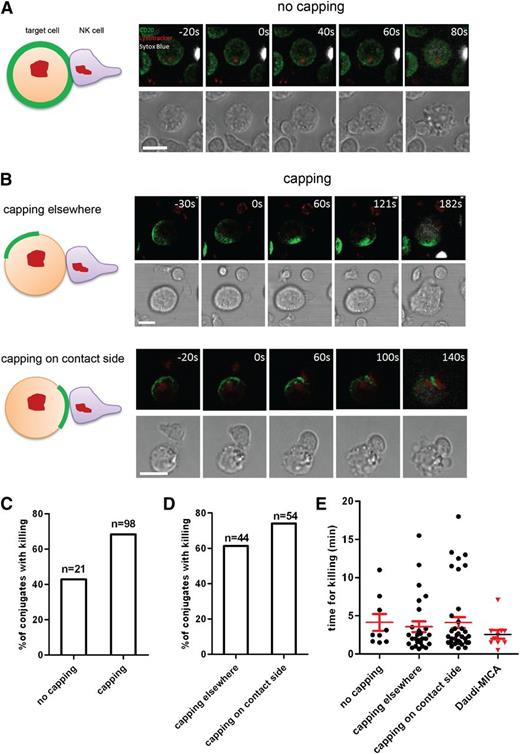
Treatment with rituximab had a profound impact on the organization of proteins at the plasma membrane of B cells, and we hypothesized that these changes could influence the outcome of interactions with NK cells and the efficiency of ADCC. To assess this, Daudi cells were pretreated with rituximab prior to coincubation with primary NK cells for imaging by laser scanning confocal microscopy at 37°C. Target cell death was visualized by positive staining for DNA (Sytox Blue). Killing of target cells by primary unstimulated NK cells in the presence of rituximab was mostly due to ADCC, as in the absence of rituximab almost no killing was observed and blocking the Fc receptor CD16 inhibited killing via rituximab (supplemental Figure 5). To investigate whether or not the organization of CD20 influenced the efficiency of target cell killing, we characterized each cell-cell interaction according to the organization of CD20 on Daudi cells and the site of initial contact with NK cells (Figure 5A-B and supplemental Videos 1-3).
Conjugates could be assessed as to whether or not CD20 was uniformly distributed in the plasma membrane in target cells upon contact with an NK cell. In 82.4% of all conjugates scored (n = 119), CD20 was polarized. We then assessed whether or not target cells were killed by NK cells and found that unpolarized target cells, with a uniform surface distribution of CD20, were killed much less efficiently. Specifically, 42.9% of contacts between NK cells and unpolarized target cells lead to target cell death within 20 minutes, while 68.4% of contacts with polarized cells, in which CD20 was capped, lead to lysis (Figure 5C). Thus, polarized B cells were killed more efficiently by NK cells.
Surface organization of CD20 influences efficiency of target cell killing. Daudi/CD20-GFP cells were preincubated with 10 µg/mL rituximab and then mixed with freshly isolated primary NK cells and imaged for 40 to 60 minutes. (A) A schematic representation (left panel) and 3D reconstructed snapshots from live cell microscopy (right panels) of Daudi cells with unpolarized CD20 interacting with and being killed by an NK cell (CD20 is green, LysoTracker used to visualize lytic granules within NK cells is red, Sytox blue used to visualize dead cells is white). (B) A schematic representation (left) and 3-dimensional reconstructed snapshots from live cell microscopy (right) show examples of target cell killing in the context of CD20 organization when CD20 is capped: cells with CD20 capping away from the site of contact with an NK cell (top panels) and cells with CD20 enriched on the side of the initial contact with an NK cell (bottom panels). Scale bars represent 10 μm. (C) Proportion of conjugates with or without capping of CD20 in which killing of a target cell took place. (D) Proportion of conjugates with capping of CD20 away from the initial contact or at the contact side in which killing of a target cell took place. (E) Time of conjugation between Daudi and NK cell leading to killing of the target was quantified for each category as well as for Daudi-MICA in conjugates with NK cells.
Surface organization of CD20 influences efficiency of target cell killing. Daudi/CD20-GFP cells were preincubated with 10 µg/mL rituximab and then mixed with freshly isolated primary NK cells and imaged for 40 to 60 minutes. (A) A schematic representation (left panel) and 3D reconstructed snapshots from live cell microscopy (right panels) of Daudi cells with unpolarized CD20 interacting with and being killed by an NK cell (CD20 is green, LysoTracker used to visualize lytic granules within NK cells is red, Sytox blue used to visualize dead cells is white). (B) A schematic representation (left) and 3-dimensional reconstructed snapshots from live cell microscopy (right) show examples of target cell killing in the context of CD20 organization when CD20 is capped: cells with CD20 capping away from the site of contact with an NK cell (top panels) and cells with CD20 enriched on the side of the initial contact with an NK cell (bottom panels). Scale bars represent 10 μm. (C) Proportion of conjugates with or without capping of CD20 in which killing of a target cell took place. (D) Proportion of conjugates with capping of CD20 away from the initial contact or at the contact side in which killing of a target cell took place. (E) Time of conjugation between Daudi and NK cell leading to killing of the target was quantified for each category as well as for Daudi-MICA in conjugates with NK cells.
Among cells in which CD20 was polarized, there was the possibility that (1) the cap of CD20 was away from the initial site of the contact with the NK cell (44.9% of contacts; n = 98) or (2) NK cells initially contacted target cells where CD20 was capped (55.1%) (Figure 5B; Table 1). When CD20 was capped away from the site of initial contact with the effector cell, 61.4% of cells were subsequently killed (Figure 5D). However, in conjugates where the NK cell initially contacted the target cells precisely where CD20 was capped, the frequency of target cell lysis increased to 74.1%.
Quantification of target cell killing dependent on CD20 organization
| Profile of capping . | Percent of conjugates . | Conjugates with killing . |
|---|---|---|
| No capping | 17.6% (21/119)* | 42.9% (9/21) |
| Capping | 82.4% (98/119)* | 68.4% (67/98) |
| Localization of cap | ||
| Capping elsewhere | 44.9% (44/98)† | 61.4% (27/44) |
| Capping on contact side | 55.1% (54/98)† | 74.1% (40/54) |
| Profile of capping . | Percent of conjugates . | Conjugates with killing . |
|---|---|---|
| No capping | 17.6% (21/119)* | 42.9% (9/21) |
| Capping | 82.4% (98/119)* | 68.4% (67/98) |
| Localization of cap | ||
| Capping elsewhere | 44.9% (44/98)† | 61.4% (27/44) |
| Capping on contact side | 55.1% (54/98)† | 74.1% (40/54) |
Percent of conjugates.
Percent of conjugates with CD20 capping.
From the movies, we also quantified whether or not the time required by the NK cell to kill the target differed according to the topology of the cell-cell interaction and whether or not target cells were polarized. The average time from initial contact to target cell death (indicated by staining with a DNA dye) was between 3.5 and 4.1 minutes in all circumstances, and no significant differences were observed (Figure 5E). This time scale for NK-cell–mediated cytotoxicity was similar for lysis of MICA-expressing target cells killed through engagement of the NKG2D receptor on NK cells (ie, independent of ADCC, which took 2.6 ± 0.6 minutes) (Figure 5E). Thus, the time for NK-cell–mediated killing is likely relatively fixed by the time needed for cell biological processes, including cytolytic granule release by the NK cell and apoptosis in the target cell. Together, these data establish that polarization of B cells does not alter the time needed for NK cells to kill target cells but, importantly, does increase the probability that the outcome of NK cell surveillance will be target cell lysis.
Discussion
Rituximab was the first therapeutic mAb accepted for therapy of non-Hodgkin lymphomas over 15 years ago,3 and following this success, it has been since introduced to treatments of other diseases where B-cell depletion is desirable.1,2 However, much debate remains as to its mechanism of action—perhaps involving multiple lines of attack—one of which is ADCC. Here, we investigated the sequence of events taking place during the process of rituximab-mediated ADCC from the initial opsonization of B cells to the eventual killing by NK cells.
First, we found that rituximab mediated capping of CD20 such that the protein accumulated at 1 pole of the B cell. This was true across multiple B-cell lines and in primary B cells isolated from healthy donors, which suggests that this phenomenon is common, though the process remains to be tested in cells isolated from primary tumors such as lymphomas. Capping of CD20 was also observed in untreated cells, albeit with a lower frequency, and it remains to be established whether or not rituximab enhanced this process occurring naturally within B cells or triggered CD20 through an alternative route.
Surprisingly, capping of CD20 occurred independent of antibody crosslinking by a secondary antibody, as is usually required for protein capping. It has been previously reported that rituximab induces or strengthens the association of CD20 with lipid rafts,14,18,23,24 and this process has been suggested to be at least partially responsible for rituximab effectiveness in mediating B-cell depletion, especially through complement-dependent cytotoxicity. The capping of CD20 observed here, however, was not dependent on an association with lipid rafts, since a mutant variant of CD20 that does not associate with lipid rafts was still capped by rituximab. In addition, it has been shown that mAb 2H7 as well as its monovalent Fab fragment are both able to mediate the association of CD20 with the lipid rafts.23 Here, 2H7 or a monovalent version of rituximab were not able to cause CD20 to cap. Thus, association of CD20 with lipid rafts is not sufficient to cause its polarization.
Unexpectedly, we found that the action of rituximab was not to merely cluster its CD20 ligand but to more generally rearrange several proteins at the B-cell surface. Specifically, ICAM-1 and moesin colocalized with CD20 in the cap while others such as CCR7 segregated away from CD20. Furthermore, intracellular cellular changes are caused by rituximab, as evidenced by a specific orientation of the MTOC toward the CD20-rich cap. Indeed, an intact microtubule network was required for CD20 capping. Thus, rituximab mediates a polarization of B cells.
The mechanism by which this occurs is an important unknown, but we can speculate a model based on our findings and previous research. CD20 has been previously shown to reside in the plasma membrane of cells as homotetramers.24,25 Thus, binding to CD20 rituximab may interact with neighboring tetramers, crosslink them, and bring them together.24 Multiple tetramers could be then assembled in a form of lattice by rituximab leading to the creation of clusters. An additional process, dependent on the microtubule network, may aid the coalescence of CD20 clusters into a cap and, more broadly, trigger cellular polarization, which is likely responsible for ICAM-1 and moesin being recruited to the cap of CD20 while CCR7 is excluded. It has been previously shown that rituximab is able to induce signals resembling B-cell receptor (BCR) stimulation,26 perhaps due to a functional association of CD20 with the BCR.26,27 Thus, it is possible that rituximab causes B-cell polarization in manner that involves the BCR. Consistent with this hypothesis, CD20 was not readily capped by rituximab when expressed in Jurkat T cells (which obviously lack the BCR).
These data are functionally important because the reorganization of the B-cell surface by rituximab influences the efficiency of target cell killing by ADCC. Polarized cells are killed up to 60% more frequently than those with uniform distribution of CD20; the most efficient killing of target cells takes place when the NK cell contacts the target directly on the side where CD20 is accumulated.
Many factors must be considered in the rational design of antibodies for use in ADCC. These include their affinity to target antigen, little or no internalization into target cells, and efficient engagement of Fc receptors on effector cells. Here, we describe yet another factor that could be taken into account: changes to the cell-surface organization of the target cell. It may be important to consider screening putative therapeutic antibodies for their ability to trigger protein clustering and cellular polarization.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank J. P. Deans (University of Calgary) and M. Purbhoo (Imperial College London) for DNA constructs, M. Mehrabi for isolation of PBMCs, M. Spitaler in the Facility of Imaging by Light Microscopy for help with imaging, and B. Kemp and the IgG Purification Team at MedImmune Ltd for production and QC of reagents.
This work was supported by the Medical Research Council, a studentship from the Manchester Collaborative Centre for Inflammation Research (A.O.), a Wolfson Royal Society research merit award (D.M.D.), and a Marie Curie European reintegration grant (D.R.).
Authorship
Contribution: D.R. and A.O. performed experiments and analyzed data; D.K.F. and M.A.S. helped design the experiments and edited the manuscript; I.S. and D.R. performed experiments on ImageStream and analyzed data; D.J.S. and D.C.L. created and provided a reagent; D.R., M.A.S., and D.M.D. conceived the project and designed experiments; and D.R. and D.M.D. wrote the manuscript.
Conflict-of-interest disclosure: D.K.F., I.S., D.J.S., D.C.L., and M.A.S. are employees of MedImmune Ltd, a wholly owned subsidiary of the Astrazeneca. The remaining authors declare no competing financial interests.
Correspondence: Daniel M. Davis, Manchester Collaborative Center for Inflammation Research, CTF Building, 46 Grafton Street, Manchester M13 9NT, United Kingdom; e-mail: daniel.davis@manchester.ac.uk.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal