Key Points
Serum 2HG analysis by LC-MS can accurately identify patients with AML with and without IDH mutations.
Oncometabolite testing of serum 2HG is indicated as a diagnostic, prognostic, and therapeutic monitoring tool in AML.
Abstract
Cancer-associated isocitrate dehydrogenase (IDH) mutations produce the metabolite 2-hydroxyglutarate (2HG), but the clinical utility of 2HG has not been established. We studied whether 2HG measurements in acute myeloid leukemia (AML) patients correlate with IDH mutations, and whether diagnostic or remission 2HG measurements predict survival. Sera from 223 de novo AML patients were analyzed for 2HG concentration by reverse-phase liquid chromatography–mass spectrometry. Pretreatment 2HG levels ranged from 10 to 30 000 ng/mL and were elevated in IDH-mutants (median, 3004 ng/mL), compared to wild-type IDH (median, 61 ng/mL) (P < .0005). 2HG levels did not differ among IDH1 or IDH2 allelic variants. In receiver operating characteristic analysis, a discriminatory level of 700 ng/mL optimally segregated patients with and without IDH mutations, and on subsequent mutational analysis of the 13 IDH wild-type samples with 2HG levels >700 ng/mL, 9 were identified to have IDH mutations. IDH-mutant patients with 2HG levels >200 at complete remission had shorter overall survival compared to 2HG ≤200 ng/mL (hazard ratio, 3.9; P = .02). We establish a firm association between IDH mutations and serum 2HG concentration in AML, and confirm that serum oncometabolite measurements provide useful diagnostic and prognostic information that can improve patient selection for IDH-targeted therapies.
Introduction
Recurrent somatic mutations in isocitrate dehydrogenase (IDH) enzymes IDH1 and IDH2 have been identified in patients with acute myeloid leukemia (AML) and other myeloid malignancies, with an estimated prevalence of 5% to 30%.1-9 IDH1 and IDH2 mutations occur more frequently in patients with normal karyotype and older age3,6,8,10 and are associated with co-occurring nucleophosmin (NPM1) mutations.2-4,11,12
IDH enzymes function within the citric acid cycle, catalyzing the oxidative decarboxylation of isocitrate to α-ketoglutarate (αKG) while converting nicotinamide adenine dinucleotide phosphate (NADP)+ to reduced NADP (NADPH). Somatic mutations within the IDH1 and IDH2 enzyme–active sites, specifically at codon R132 of IDH1 and codons R140 or R172 of IDH2, lead to a loss of function for the above-described reaction and yield a reverse reaction that reduces αKG to 2-hydroxyglutarate (2HG) via the conversion of NAPDH to NADP+.13-17 2HG can be measured in patient serum, and 2HG levels correlate with the presence of IDH mutations in small cohorts of patients with AML.13,18,19
Despite recent insights into the distinctive pathophysiologic characteristics of IDH mutations, the role of 2HG elevations within neoplastic cells has not been firmly established. Recent studies suggest that increased 2HG levels function through competitive inhibition of αKG-dependent enzymatic activities, including ten-eleven translocation 2 (TET2)–dependent DNA hydroxymethylation as well as histone demethylation, leading to dysregulated epigenetic programming.20,21 Activation of the HIF-1α pathway via inhibition of prolyl hydroxylases has also been implicated.15,22,23
The prognostic significance of IDH mutations remains controversial, perhaps confounded by a differing prognosis conferred by the different allelic mutations and by different therapeutic regimens in different patient cohorts.24-27 IDH2 R140 mutations appear to confer a more favorable outcome, and all IDH mutations were associated with a favorable outcome when present in conjunction with NPM1 mutations.11,27 However, definitive information on prognostic importance is lacking. Improved scientific discernment of this pathway is important, especially given the ongoing development of targeted IDH modulators for clinical use.28,29 Moreover, a specific role for epigenetically targeted therapies, including DNA methyltransferase inhibitors and histone deacetylase inhibitors, may be particularly efficacious in AML subsets characterized by IDH mutations and by mutations in other genes implicated in epigenetic regulation.30,31
Preliminary research has suggested a screening and/or diagnostic role of serum 2HG analysis in myeloid neoplasms.14,32 However, a systematic analysis of 2HG as a predictor of IDH mutation status or to assess clinical response has not been performed. Therefore, we measured 2HG levels in serum from patients with de novo AML, treated on the Eastern Cooperative Oncology Group E1900 trial.
Methods
Patients and treatment
Pretreatment peripheral blood (PB) sera from 223 E1900 patients were analyzed for 2HG levels, in addition to 14 PB serum samples from healthy adult volunteers (>18 years old). All participants provided written informed consent for research. Detailed inclusion and exclusion criteria for E1900 have been published previously.33 Median follow-up of participants in our cohort was 21 months (range, 0.4-81 months).
Serum was obtained from 62 E1900 patients with IDH mutations,11 identified by mutational analysis of coding exons with known somatic mutations using polymerase chain reaction amplification and bidirectional Sanger sequencing as described previously.13 The majority (87%) of patients had intermediate or indeterminate cytogenetics as determined by the Eastern Cooperative Oncology Group/Southwest Oncology Group classification system.34 In 29 of those patients, serum collected within 2 weeks of morphologic complete remission (CR) at postinduction or postconsolidation (including autologous transplantation) time points were also available. Additionally, 161 control samples, selected based on intermediate-risk cytogenetics and IDH wild-type status from the primary mutational studies,11 were analyzed. Analysts were blinded to the IDH mutational status. More than 90% of the patients with intermediate-risk cytogenetics had a normal diploid karyotype.
Additional covariates analyzed within the E1900 cohort included the patient-related variables age and sex; disease-related variables such as white blood cell (WBC) count, platelet count, bone marrow (BM) blast percentage (%), and circulating blast percentage; and treatment covariates such as the treating institution and randomized E1900 treatment allocation. Less than 5% of patients received an allogeneic stem cell transplant during first CR in our E1900 study cohort. The study was approved by the University of Pennsylvania Institutional Review Board. All samples were obtained after informed consent in accordance with the Declaration of Helsinki.
2HG analysis
Serum 2HG levels were measured by reverse-phase liquid chromatography coupled to mass spectrometry (LC-MS) by Agios Pharmaceuticals (Cambridge, MA). Integrated elution peaks were compared with metabolite standard curves for absolute quantification, using established methods.14 A 2HG level <10 ng/mL was defined as below the level of detection using the LC-MS assay (ie, below the quantitation limit) and was analyzed as 10 ng/mL. Similarly, 2HG levels >30 000 ng/mL were considered above the quantitation limit and were analyzed as 30 000 ng/mL, as values above this level were extrapolated.
2HG analysis was repeated for 21 selected samples and demonstrated reproducible 2HG levels with a mean intrasample coefficient of variation of 24.8%. The intrasample variation was most pronounced in samples with markedly elevated 2HG levels and did not affect the interpretation of the IDH mutational status.
Statistical analyses
Nonparametric analyses were performed because of the non-Gaussian distribution of 2HG levels. Categorical variables were compared using χ2 or the Fisher exact test, and continuous variables were compared using the Wilcoxon rank-sum test. 2HG level was evaluated as both a continuous and a categorical variable. Discriminatory cutoff values for 2HG were evaluated to maximize sensitivity, specificity, and create an optimal cutoff point for clinical purposes through receiver operating characteristic curve (ROC) analysis.
Overall survival (OS) was measured as the time from AML diagnosis to death or date of last follow-up (censored). For the remission cohort analysis (n = 29), OS was measured from the time of CR. Leukemia-free survival (LFS) was defined as the time from CR to treatment failure including relapse, death, or date at last follow-up (censored). Kaplan-Meier analysis was used to construct OS and LFS curves, and curves were compared by the log-rank test.
Because of the presence of 2HG values above or below the limits of accurate LC-MS detection, a censored regression model (Tobit) was used, which estimates linear relationships between variables when there is either left- or right-censoring of the dependent variable.35 Adjusted Cox proportional hazards models were constructed to examine the effect of potential confounders. All analyses were 2-sided, with statistical significance established as P < .05 using STATA software, version 12 (College Station, TX).
Results
Patient characteristics
Clinicopathologic characteristics of our E1900 cohort are displayed in Table 1 and in supplemental Table 1, including patient age, sex, treatment group, diagnostic laboratory values, cytogenetic risk group, and the presence of additional somatic mutations. Given the favorable prognosis of IDH2 R140 mutations, characteristics for this subgroup are additionally detailed. No statistically significant differences in age, sex, or treatment group were observed between patients with and without IDH mutations, or by treating institution. Patients with IDH mutations had a higher platelet count (P = .002) and a higher BM blast percentage at diagnosis (P = .009), compared with patients with IDH wild-type mutations. The presence of an IDH mutation correlated with DNMT3A mutations (P = .022) and correlated inversely with FMS-like tyrosine kinase-3 (FLT3) mutations (P < .0005) and CCAAT/enhancer binding protein-α (CEPBA) mutations (P = .023). Although the incidence of TET2 mutations was low overall, no patient with an identified IDH mutation had a co-occurring TET2 mutation, as has been reported previously.11,20
Clinical characteristics and patient outcome of study cohort
| . | IDH wild-type (n = 161) . | IDH mutant (n = 62) . | P value . | IDH2 R140Q mutant (n = 26) . |
|---|---|---|---|---|
| Median age, y (range) | 46 (18-60) | 46 (18-60) | .89 | 47 (27-59) |
| Male/female ratio | 80/81 (1:1) | 26/36 (1:1.4) | .37 | 13/13 (1:1) |
| Treatment group, n (%) | .23 | |||
| DNR 45 mg/m2 | 83 (52) | 26 (42) | 8 (31) | |
| DNR 90 mg/m2 | 78 (48) | 36 (58) | 18 (69) | |
| Median WBC count at dx (×109/L) | 31.8 (0.6- 212.8) | 34.7 (1.0-191.8) | .62 | 44.0 (1.7-191.8) |
| Median PLT count at dx (×109/L) | 67.2 (3.9- 452) | 101.7 (6.2- 650) | .002 | 94.3 (17-304) |
| Median BM blast % at dx (range) | 61.2 (8- 99) | 71.8 (22-100) | .009 | 72.2 (25-100) |
| Median PB blast % at dx (range) | 44.9 (0- 98) | 48.4 (0-97) | .49 | 55.8 (0-97) |
| Cytogenetic risk group, n (%) | <.0005 | |||
| Favorable | 0 (0) | 1 (2) | 0 (0) | |
| Intermediate | 146 (91) | 40 (65) | 16 (62) | |
| Unfavorable | 1 (1) | 7 (11) | 4 (15) | |
| Indeterminate | 14 (9) | 14 (23) | 6 (23) | |
| Mutations present, n (%) | ||||
| FLT3-ITD | 76 (48) | 12 (20) | <.0005 | 6 (23) |
| FLT3-TKD | 12 (8) | 2 (3) | 2 (8) | |
| NPM1 | 74 (47) | 32 (53) | .45 | 16 (62) |
| TET2 | 11 (7) | 0 (0) | .087 | 0 (0) |
| DNMT3A | 44 (28) | 20 (33) | .022 | 4 (15) |
| CEBPA | 20 (13) | 1 (2) | .023 | 0 (0) |
| Achieved CR, n (%) | 103 (64) | 40 (65) | .824 | 19 (73) |
| Median survival duration (mo) | 18.2 | 45.3 | .022 | Not reached |
| . | IDH wild-type (n = 161) . | IDH mutant (n = 62) . | P value . | IDH2 R140Q mutant (n = 26) . |
|---|---|---|---|---|
| Median age, y (range) | 46 (18-60) | 46 (18-60) | .89 | 47 (27-59) |
| Male/female ratio | 80/81 (1:1) | 26/36 (1:1.4) | .37 | 13/13 (1:1) |
| Treatment group, n (%) | .23 | |||
| DNR 45 mg/m2 | 83 (52) | 26 (42) | 8 (31) | |
| DNR 90 mg/m2 | 78 (48) | 36 (58) | 18 (69) | |
| Median WBC count at dx (×109/L) | 31.8 (0.6- 212.8) | 34.7 (1.0-191.8) | .62 | 44.0 (1.7-191.8) |
| Median PLT count at dx (×109/L) | 67.2 (3.9- 452) | 101.7 (6.2- 650) | .002 | 94.3 (17-304) |
| Median BM blast % at dx (range) | 61.2 (8- 99) | 71.8 (22-100) | .009 | 72.2 (25-100) |
| Median PB blast % at dx (range) | 44.9 (0- 98) | 48.4 (0-97) | .49 | 55.8 (0-97) |
| Cytogenetic risk group, n (%) | <.0005 | |||
| Favorable | 0 (0) | 1 (2) | 0 (0) | |
| Intermediate | 146 (91) | 40 (65) | 16 (62) | |
| Unfavorable | 1 (1) | 7 (11) | 4 (15) | |
| Indeterminate | 14 (9) | 14 (23) | 6 (23) | |
| Mutations present, n (%) | ||||
| FLT3-ITD | 76 (48) | 12 (20) | <.0005 | 6 (23) |
| FLT3-TKD | 12 (8) | 2 (3) | 2 (8) | |
| NPM1 | 74 (47) | 32 (53) | .45 | 16 (62) |
| TET2 | 11 (7) | 0 (0) | .087 | 0 (0) |
| DNMT3A | 44 (28) | 20 (33) | .022 | 4 (15) |
| CEBPA | 20 (13) | 1 (2) | .023 | 0 (0) |
| Achieved CR, n (%) | 103 (64) | 40 (65) | .824 | 19 (73) |
| Median survival duration (mo) | 18.2 | 45.3 | .022 | Not reached |
DNR, daunorubicin; dx, diagnosis; FLT3-ITD, FLT3 internal tandem duplication; FLT3-TKD, FLT3 tyrosine kinase domain; PLT, platelet
Elevated 2HG levels in AML are associated with IDH mutations
Pretreatment serum 2HG levels ranged from 10 ng/mL to 30 000 ng/mL, with a median value of 101 ng/mL for all patients tested. In patients with IDH wild-type AML, levels of 2HG ranged from 10 ng/mL to 14 181 ng/mL (median, 61 ng/mL). Serum 2HG levels in patients with IDH mutations ranged from 10 ng/mL to 30 000 ng/mL (median, 3004 ng/mL). 2HG could be detected in 159 (71%) of all samples, including 60 (96.8%) of 62 samples from patients with IDH mutations, and 99 (61.5%) of 161 samples from patients with wild-type IDH. 2HG levels were significantly higher in patients with IDH mutations, compared with patients with IDH wild-type AML (P < .0005) (Figure 1). Serum 2HG levels in volunteers were also obtained (see “Serum 2HG levels in healthy volunteers”) and ranged from 33 ng/mL to 176 ng/mL (median, 48 ng/mL).
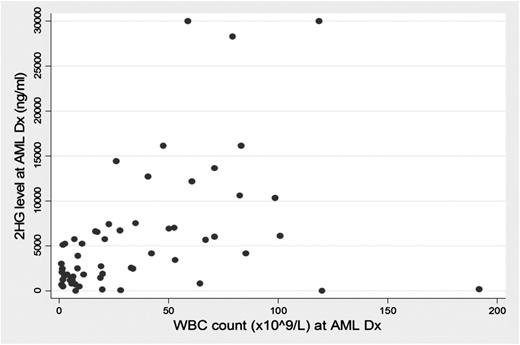
On univariate Tobit regression analysis, an elevated serum 2HG level at diagnosis was associated with an elevated platelet count (P < .0005) and an elevated BM blast percentage at diagnosis (P = .027). We observed an association between an elevated WBC count and serum 2HG level of borderline significance (P = .057); when analyzing only IDH mutations, we identified a strong association between WBC count and elevated level of serum 2HG (P = .007) (Figure 2). No statistically significant association was observed between 2HG level and patient age and sex, treating institution, or study group treatment.
In patients with IDH mutations, serum 2HG level relates to WBC count at AML diagnosis.
In patients with IDH mutations, serum 2HG level relates to WBC count at AML diagnosis.
Next, we evaluated serum 2HG levels in patients with IDH mutations, based on the specific IDH1 or IDH2 mutation present (Figure 3). No significant difference in 2HG levels among patients with IDH mutations was identified based on the specific allelic mutation. Of note, 2HG levels in IDH2 R140 mutations did not differ significantly from the other IDH mutations at baseline or after adjustment for WBC count, BM, or circulating blast percentage.
Serum 2HG levels can identify patients with AML with IDH mutations
Next, we asked whether a “threshold” level of serum 2HG level could serve as a useful screening test to segregate patients with AML with or without IDH mutations. Using ROC analysis, we identified a strong correlation between serum 2HG level and IDH mutations, with an area under the ROC curve between wild-type IDH and IDH mutations of 0.918 (Figure 4), and an optimal cutoff point of 700 ng/mL. When applying this discriminatory 700-ng/mL value, we found that the diagnostic sensitivity and specificity of serum 2HG level for predicting IDH mutations was 86.9% and 90.7%, respectively, with 89.6% of patients correctly classified.
There were 15 IDH wild-type E1900 samples with pretreatment 2HG levels measuring >700 ng/mL. Of those, 13 samples were reanalyzed for IDH1 and IDH2 mutations using identical mutation detection methodology, at the same institution as the initial mutation analysis, with manual inspection of all traces.11 On subsequent testing, 9 of the 13 samples were discovered to have an IDH mutation (2 samples with an IDH1 R132 mutation, 6 with an IDH2 R140Q mutation, and 1 with an IDH2 R172K mutation). In most cases, the mutant allele burden was less than 20%, such that it fell below the level of detection of the analysis pipeline, which used the Mutation Surveyor as a primary screen. A total of 4 samples were again found to be without identifiable IDH1 or IDH2 mutations detectable by Sanger sequencing. Interestingly, however, 2 of these 4 samples had sequencing traces with a potential low-abundance mutant IDH allele. For all further statistical analyses, the 9 IDH wild-type samples with identified IDH mutations on subsequent sequencing were analyzed primarily within the IDH wild-type cohort (akin to an intention-to-treat analysis). Selected exploratory analyses in which these 9 samples were dropped from the analysis, or were analyzed within the IDH-mutant cohort, are detailed in the supplemental Text.
Serum 2HG levels in healthy volunteers
To further investigate the potential of serum 2HG measurements as a diagnostic test, we studied 2HG levels in the serum of 14 healthy volunteers. In these volunteers, the median 2HG level was 48 ng/mL (range, 33-176 ng/mL). No association was observed between 2HG level with patient age or sex. Of note, no significant difference was found between median 2HG level in volunteers (48 ng/mL) with the median value of IDH wild-type AML samples (61 ng/mL; or 50 ng/mL if the 9 samples with identified IDH mutations were excluded). All 14 healthy volunteers had detectable serum 2HG levels by LC-MS.
2HG levels at remission in patients with IDH mutations
Sera from 29 of the E1900 patients with IDH mutations at the time of CR (see supplemental Text for additional clinicopathologic characteristics of the remission cohort) revealed a median 2HG level of 95 ng/mL (range, 28-3149 ng/mL), compared with a median 2HG of 3234 ng/mL (range, 10-13 638 ng/mL) in these 29 patients at AML diagnosis. The median change in 2HG level from diagnosis to remission was 3088 ng/mL (range, +43 to −13 363 ng/mL). 2HG levels decreased in all but 1 patient (this patient had a diagnosis 2HG level below the quantitation limit and 53 ng/mL at CR). Neither the absolute difference in 2HG level nor the percent change in 2HG levels, from diagnosis to CR, was associated with survival duration.
Next, we assessed whether achieving a “normal level” of serum 2HG at CR provided prognostic value. Given that 2HG levels of healthy volunteers ranged from 33 ng/mL to 176 ng/mL, we defined a normal 2HG level as <200 ng/mL. Of the 29 IDH-mutant remission samples, 20 (69%) achieved a remission 2HG level <200 ng/mL (Figure 5A). Patients who did not achieve a 2HG level <200 ng/mL at CR experienced inferior OS (hazard ratio [HR], 3.9; 95% confidence interval [CI], 1.2-12.7; P = .02) and LFS (HR, 3.6; 95% CI, 1.1-11.9; P = .046), which retained significance on multivariable analysis (supplemental Table 2). Median OS was 23.5 months in the group with 2HG levels >200 ng/mL and was not reached in the group with 2HG levels <200 ng/mL (Figure 5B). Because mutational analysis for IDH at the time of CR was not performed, we cannot compare the sensitivity of serum 2HG with molecular tests for minimal residual disease (MRD).
(A) 2HG levels at diagnosis and at CR in 29 IDH-mutant samples (dotted line at 200 ng/mL); (B) Kaplan-Meier survival curve of OS based on remission 2HG.
(A) 2HG levels at diagnosis and at CR in 29 IDH-mutant samples (dotted line at 200 ng/mL); (B) Kaplan-Meier survival curve of OS based on remission 2HG.
Effect of 2HG levels on outcome
There was no impact of elevated serum 2HG level on attainment of CR (P = .24). There was a suggestion of improved survival outcome in patients with an elevated pretreatment 2HG level, although this finding was not statistically significant for OS (HR, 0.72; 95% CI, 0.49-1.06; P = .09) or LFS (HR, 0.66; 95% CI, 0.41-1.06; P = .08).
Interactions were identified between an elevated level of serum 2HG (>700 ng/mL) and mutational status of co-occurring mutations including NPM1 and CEPBA. This trend implies that other genetic abnormalities may alter the magnitude of the relationship between 2HG level and clinical outcome, and is consistent with recent data suggesting that NPM1/IDH double mutations have a favorable outcome. When evaluating pretreatment 2HG levels using our threshold of 700 ng/mL on multivariable analysis (including allocated treatment group), we observed that an elevated 2HG level was associated with improved OS in patients with NPM1 mutations (n = 106) (HR, 0.42; 95% CI, 0.22-0.77; P = .004), and in those without DNMT3A mutations (n = 144) (HR, 0.48; 95% CI, 0.26-0.84; P = .01) or CEPBA mutations (n = 198) (HR, 0.63; 95% CI, 0.43-0.94; P = .02) (supplemental Tables 3-5 and supplemental Figures 2-6).
Finally, the level of serum 2HG was analyzed separately within the 3 IDH allelic mutant cohorts and was split by the median 2HG level observed in IDH mutations (3004 ng/mL). Despite the small cohort size, there was a suggestion of decreased OS in patients with a more elevated 2HG level and IDH2 R172 mutations (HR, 7.29; 95% CI, 0.79-67.1; P = .08). The OS in patients with IDH1 R132 or IDH2 R140 mutations did not appear to be affected by the level of serum 2HG elevation at diagnosis (HR, 1.01; 95% CI, 0.34-2.96; P = .98 and HR, 1.8; 95% CI, 0.59-5.57; P = .28, respectively).
Discussion
Discovery of neomorphic 2HG production as a consequence of recurrent IDH mutations in AML and in other malignancies has strengthened the proposed link between abnormal cellular metabolism, epigenetic reprogramming, and oncogenic transformation. Elevated serum 2HG levels in patients with IDH1 and IDH2 mutations have been described previously in several small AML cohorts13,14,18,36,37 ; however, the relationship between serum 2HG levels, IDH mutations, and patient outcome has not been defined previously. Our study is the first to fully assess pretreatment serum 2HG levels in a cohort of patients with AML treated on a uniform protocol, and to investigate the usefulness of 2HG as a prognostic biomarker.
Our analysis confirms a clear relationship between the presence of an IDH mutation (IDH1 R132, IDH2 R172, or IDH2 R140) and an elevated serum 2HG level, with 97% of patients with IDH mutations having a detectable 2HG level at the time of AML diagnosis, and a median 2HG concentration nearly 50-fold higher than that of patients with IDH wild-type mutations. An association between diagnostic WBC count and serum 2HG levels was identified in patients with IDH mutations, suggesting that circulating WBC count as a measure of “tumor burden” may relate to the level of serum 2HG in these patients. We also found low but detectable levels of serum 2HG in the majority of patients with wild-type IDH AML, as well as in normal volunteers. These data suggest that the LC-MS technique can detect minimal and nonpathologic levels of this metabolite in patients without IDH mutations.
We found that pretreatment serum 2HG levels >700 ng/mL best predict IDH mutational status. Notably, subsequent mutation testing in 13 patients initially characterized as having IDH wild-type mutations and with diagnostic 2HG levels >700 ng/mL reclassified 9 patients as having IDH mutations, albeit at low allelic burden. The predictive usefulness of this value is notable but will require validation in an independent data set. Given that AML is a heterogeneous disease involving distinctive subclones even within the same patient, the ability to detect small IDH-mutant clones undetectable by current molecular techniques, yet leading to 2HG accumulation, must be considered. Whether such clones are of biologic or clinical relevance or represent tractable therapeutic targets is unknown. Additional sequencing and analysis of the 4 samples with 2HG levels >700 ng/mL and without identified IDH mutations are ongoing. It is possible that mutations in the noncoding regions of IDH1 or IDH2, or alterations in other enzymes involved in αKG metabolism, occur in patients without canonic IDH1 or IDH2 mutations.
Our data suggest that diagnostic serum 2HG measurements may allow for rapid, accurate identification of patients with AML with IDH mutations. 2HG testing in patients with glioma, cholangiocarcinoma, and chondrosarcoma may provide similarly useful diagnostic and/or prognostic information. Importantly, our data demonstrate the technical feasibility of this approach. As improved sequencing techniques increase the sensitivity of genetic tests, future studies will be required to compare the sensitivity of 2HG analysis to next-generation testing methodologies.
Drugs targeting mutant IDH are currently being tested in the preclinical setting with promising results,28,29 thus increasing the significance of serum 2HG levels in the selection of patients for such treatment. Serial 2HG measurements may emerge as an important pharmacodynamics marker of IDH-modulating agents and other antileukemic therapies. Nevertheless, analytical validation of the LC-MS technique is warranted before this test can be used for clinical decision making.
Our analysis suggests that the level of serum 2HG alone does not explain the improved survival duration seen in the IDH2 R140 cohort compared with patients with other IDH1 or IDH2 mutations. Recent data demonstrate that IDH+/NPM1+ mutations confer an especially favorable prognosis,11 although the frequency of co-occurring NPM1 mutations within our IDH2 R140 subgroup was not significantly different from the group with IDH mutations overall. It is important to consider that serum 2HG level is only a surrogate measurement for intracellular levels of 2HG in malignant cells, and the true biologic interpretation of serum 2HG levels is not clear at this time. The relationship between intracellular and extracellular 2HG levels, the half-life of serum 2HG, and mechanisms of normal 2HG breakdown and elimination will require further clarification.
In patients with IDH mutations, a decrease of serum 2HG levels to <200 ng/mL at CR was predictive of improved OS (P = .02). This finding suggests that 2HG testing may represent a sensitive means to assess residual leukemic cells after induction chemotherapy, and serve as a marker of MRD with which to guide subsequent therapeutic decisions. Prospective studies are needed to confirm this observation and to inform the use of 2HG measurement as a quantitative tool, compared with other markers of MRD such as flow cytometry and genetic analysis.
Taken together, our data provide a rationale for serum 2HG measurement as a diagnostic, prognostic, and monitoring tool in AML and may establish a role for oncometabolite testing in the clinical setting to improve on therapeutic decisions.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the National Institutes of Health Cancer Clinical Epidemiology Training Grant (T32 CA 009679) (C.D.D.) and by the National Institutes of Health National Cancer Institute Grant (RO1 CA149566) and VA Merit Award 1I01BX000918 (M.C.).
Authorship
Contribution: C.D.D. designed and performed the research, analyzed the data, performed statistical analysis, and wrote the paper; K.J.P. analyzed the data, performed statistical analysis, and wrote the paper; A.W.L. designed the research, analyzed the data, and wrote the paper; E.P. designed the research, contributed vital analytical tools, interpreted the data, and wrote the paper; Z.S. analyzed the data, performed statistical analysis, and wrote the paper; R.L.L. designed the research, contributed vital analytical tools, and wrote the paper; K.S.S. and K.Y. performed the research and contributed vital analytical tools; J.P.P. performed the research, contributed analytical tools, and wrote the paper; S.A. performed the research and contributed vital analytical tools; O.A.-W. performed the research, contributed vital analytical tools, and wrote the paper; A.E.P., M.R.L., J.M.R., H.M.L., and H.F.F. analyzed the data and wrote the paper; D.J.M., M.S.T., and S.M.L. designed the research, analyzed the data, and wrote the paper; and M.C. designed and performed the research, analyzed the data, and wrote the paper.
Conflict-of-interest disclosure: K.S.S., K.Y., and S.A. are employed by Agios Pharmaceuticals. R.L.L. has consulted for Agios Pharmaceuticals and received honoraria. The remaining authors declare no competing financial interests.
Correspondence: Martin Carroll, Division of Hematology/Oncology, Hospital of the University of Pennsylvania, Rm 708 BRB II/III, Philadelphia, PA 19146; email: carroll2@mail.med.upenn.edu.