Key Points
Proteomic analysis of the leukemia cell surface reveals new leukemia-associated features with a potential to improve diagnostics.
The ALL surfaceome is a resource for systematic functional exploration.
Abstract
A better description of the leukemia cell surface proteome (surfaceome) is a prerequisite for the development of diagnostic and therapeutic tools. Insights into the complexity of the surfaceome have been limited by the lack of suitable methodologies. We combined a leukemia xenograft model with the discovery-driven chemoproteomic Cell Surface Capture technology to explore the B-cell precursor acute lymphoblastic leukemia (BCP-ALL) surfaceome; 713 cell surface proteins, including 181 CD proteins, were detected through combined analysis of 19 BCP-ALL cases. Diagnostic immunophenotypes were recapitulated in each case, and subtype specific markers were detected. To identify new leukemia-associated markers, we filtered the surfaceome data set against gene expression information from sorted, normal hematopoietic cells. Nine candidate markers (CD18, CD63, CD31, CD97, CD102, CD157, CD217, CD305, and CD317) were validated by flow cytometry in patient samples at diagnosis and during chemotherapy. CD97, CD157, CD63, and CD305 accounted for the most informative differences between normal and malignant cells. The ALL surfaceome constitutes a valuable resource to assist the functional exploration of surface markers in normal and malignant lymphopoiesis. This unbiased approach will also contribute to the development of strategies that rely on complex information for multidimensional flow cytometry data analysis to improve its diagnostic applications.
Introduction
The identification of informative cell surface protein markers has markedly improved disease classification and monitoring of a therapeutic response in childhood acute lymphoblastic leukemia (ALL). Over the past 2 decades, flow cytometry (FCM) has been developed based on studies that identified a limited number of leukemia cell surface markers to distinguish them from normal hematopoietic cells.1-4 A significant advantage of FCM is the possibility to detect informative changes in different subpopulations at the single-cell level. Besides its central role for leukemia diagnosis, FCM is now often used to monitor response to treatment, with most accuracy during initial induction therapy.5,6 Yet currently used markers do not always allow us to unequivocally discriminate malignant cells from the background of regenerating marrow, which limits sensitivity and specificity of FCM for the detection of leukemia cells.
So far, FCM-based discrimination of leukemic subpopulations in clinical samples is done based on expression patterns of markers determining lineage and developmental stage of hematopoietic differentiation.1 Some of these markers, including CD19, CD20, and CD33, have also been used to target malignant cells with antibody-derived therapeutics in hematologic malignancies.7-10 However, broader knowledge about possible clinically informative markers present on the surface of leukemia cells is still very limited. The reason for this gap is the lack of technologies for the unbiased and direct measurement of the surfaceome and the limited availability of primary patient material. Gene expression analysis can provide comprehensive information on transcription profiles of a particular cell population but does not allow us to infer about the subcellular location of the correspond proteins. Antibody-based approaches provide only a fragmented view of the leukemia cell surface protein composition, depending on the availability of validated reagents. The recently developed Cell Surface Capture (CSC) technology11-13 enables a discovery-driven exploration of the leukemia surfaceome using chemoproteomic tagging of proteins on the surface of living cells. However, the cell quantities that are required for comprehensive proteomic screening are often not available from diagnostic patient samples. This limitation can be circumvented by expansion of leukemia cells using xenograft mouse models.14-16 The leukemia cells generated in this approach preserve phenotypic and genotypic characteristics of the disease and reflect the original disease in its clonal composition.15,17
Here we analyzed the surfaceome composition of xenografts from 19 B-cell precursor ALL (BCP-ALL) patients using the CSC technology. This discovery-driven approach led to the identification of 713 cell surface–exposed proteins including 181 CD-annotated proteins. We report a strategy to preselect candidate leukemia surface markers for better discrimination of the malignant cell subpopulation from normal hematopoietic cells in patients at different stages of disease treatment.
Methods
Patient samples
Primary human ALL cells used for xenoamplification and subsequent mass spectrometric studies were obtained from cryopreserved bone marrow aspirates from patients enrolled in the ALL-BFM 2000 trial. Patient material for prospective FCM validation was obtained from bone marrow of patients enrolled in ALL-BFM 2000 and 2009 trials (Switzerland), ALL-IC BFM 2009 trial (Poland), and ALL-BFM 2009 and ALL REZ BFM 2002 trials (Czech Republic) or from healthy donors. Informed consent was obtained from patients or legal guardians in accordance with the Declaration of Helsinki, and the local Institutional Review Boards of each participating center approved these studies.
Xenoamplification of primary human material
CSC technology
The Cys-Glyco-CSC, Glyco-CSC, or Lys-CSC technology was applied as described.12 Briefly, glycoproteins on the surface of viable cells were oxidized and biotinylated using biocytin hydrazide (Cys-Glyco-CSC/Glyco-CSC) or directly biotinylated with the sulfo-NHS-SS-biotin (Lys-CSC). Cells were lysed in the presence of iodoacetamide, and the protein mixture from the membrane fraction enriched by differential centrifugation was digested overnight with trypsin. The resulting peptides were incubated with streptavidin-coated beads; unspecific peptides were removed by stringent washing; and biotinylated peptides were eluted either by chemical reduction (cysteine- and lysine-containing peptides) or enzymatic cleavage (glycosylated peptides), desalted, and analyzed individually by liquid chromatography–mass spectrometry (LC-MS).
Mass spectrometry analysis
Each peptide sample was analyzed in duplicates by liquid chromatography–tandem mass spectrometry on an LTQ Orbitrap, LTQ Orbitrap XL, or LTQ FT (Thermo Scientific, Waltham, MA). Data were searched with Sorcerer-SEQUEST against a concatenated human and mouse protein database of the UniProtKB/Swiss-Prot Protein Knowledgebase (version 56.9). Statistical analysis of each search result for each LC-MS analysis was performed using the Trans-Proteomic Pipeline TPP v4.3.19
Multiparametric FCM
Combinations of multiple directly conjugated antibodies against cell surface proteins were used in all cases. For immunophenotyping of xenoamplified human ALL cells, antibodies against 26 proteins presented in Figure 2B were integrated into clinical antibody panels used for leukemia diagnosis. Antibodies against 9 proteins used for primary human material screening were combined with the backbone antibodies against CD10, CD19, CD20, CD34, and CD45. Detailed staining procedures and antibody specification are provided in the supplemental Methods (see the Blood Web site). Data acquisition was performed using FACS Canto II or LSRII (Becton Dickinson, Franklin Lakes, NJ) flow cytometers and analyzed using FACSDiva version 6.1.2 (Becton Dickinson) or FlowJo (TreeStar, Ashland, OR) software.
Statistical analysis
Median fluorescence intensity (MFI) for each cell population of interest was calculated by FACSDiva analytical software. All statistical tests were performed using GraphPad Prism version 4.0 (GraphPad Software, La Jolla, CA).
Results
Chemoproteomic cell surface capturing provides unprecedented information of the ALL surfaceome
For the generation of a comprehensive ALL surfaceome data set, we selected 19 well-characterized samples from BCP leukemia patients (supplemental Table 1). Xenograft ALL cells were harvested from the spleen of euthanized animals, shown to contain 94% to 99% of human ALL cells by FCM, and directly subjected to the CSC workflow (Figure 1). This discovery-driven CSC screen of glycosylated and lysine-containing cell surface proteins resulted in the identification of 713 bona fide surface proteins including general hematopoietic and B-cell–specific markers (eg, CD45, HLA, CD34, CD19, and CD10), as well as proteins that were not yet described to be present on leukemia (supplemental Table 2). This provides an unprecedented view of the leukemia cell surface and a resource for further research.
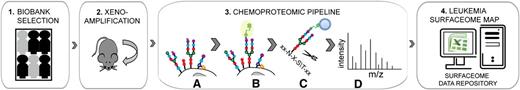
Chemoproteomic workflow for childhood ALL surfaceome mapping. Relevant human samples were selected from the biobank of cryopreserved bone marrow aspirates collected at diagnosis (1) and expanded in immunodeficient mice (2). Subsequently, selective chemical tagging was used to enrich for cell surface–exposed glycoproteins (3). Mild oxidation of glycan residues on the surface of intact cells (A), followed by their biotinylation (B), membrane disruption, and protein digestion, allowed for affinity enrichment of selectively tagged glycopeptides (C). The peptide fraction stripped from glycan residues by enzymatic elution from streptavidin-coated beads was analyzed by mass spectrometry, resulting in protein identification (D). The established leukemia surfaceome data repository includes detailed information on identified cell surface–exposed proteins and respective peptides (4).
Chemoproteomic workflow for childhood ALL surfaceome mapping. Relevant human samples were selected from the biobank of cryopreserved bone marrow aspirates collected at diagnosis (1) and expanded in immunodeficient mice (2). Subsequently, selective chemical tagging was used to enrich for cell surface–exposed glycoproteins (3). Mild oxidation of glycan residues on the surface of intact cells (A), followed by their biotinylation (B), membrane disruption, and protein digestion, allowed for affinity enrichment of selectively tagged glycopeptides (C). The peptide fraction stripped from glycan residues by enzymatic elution from streptavidin-coated beads was analyzed by mass spectrometry, resulting in protein identification (D). The established leukemia surfaceome data repository includes detailed information on identified cell surface–exposed proteins and respective peptides (4).
CSC recapitulates the immunophenotype of ALL samples
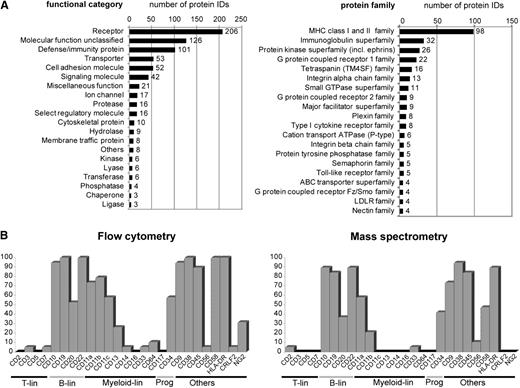
To evaluate the quality of the surfaceome data, we categorized the proteins by their function (Figure 2A and supplemental Table 2). Most proteins could be assigned to protein families that are consistent with a membrane location, with immunoglobulin, integrin, and cytokine receptor families. From the 389 CD molecules listed by the Gene Nomenclature committee, 181 were represented in the ALL surfaceome (supplemental Table 2, highlighted red). These included 23 CD molecules that are commonly used in FCM panels for leukemia diagnostics (Figure 2B). We compared the detection of these surface markers by CSC with flow cytometric data from the corresponding ALL samples (Figure 2B). Detection of the markers was concordant in 84% and discordant in only 16% of all individual comparisons. The majority of the discordant cases (61% of all discordances) corresponded to weak positive FCM signals of markers that could not be detected in CSC data (supplemental Table 3A-B). The markers that constitute the backbone for BCP-ALL detection by FCM (ie, CD10, CD19, CD20, CD45, and CD34) were consistently identified by CSC. The comparison of 4 samples with translocation t(1;19) and 3 samples with high hyperdiploid cytogenetic features retrieved, among other interesting markers, the receptor tyrosine kinase ROR1 to be associated with t(1;19)-positive ALL (supplemental Table 3C). ROR1, which was recently proposed as a therapeutic target for this ALL subgroup,20 was also detected in the t(17;19)-positive ALL case, as suggested by Bicocca et al20 (supplemental Table 2). These observations provide a first validation of the ALL surfaceome data, supporting the notion that the CSC analysis may expand our knowledge about the ALL cell surface composition.
CSC technology captures the surface phenotype of ALL cells. (A) Cell surface proteome map of ALL cells: 713 identified membrane-associated proteins were categorized based on their biological function assigned by the PANTHER algorithm (left); the 20 most abundant protein families and superfamilies are displayed (right; protein family assignment according to UniProt KB annotation). (B) Comparison between immunophenotypes described using 2 different technologies. ALL xenograft samples (n = 19) included in the proteomic pipeline were analyzed by FCM for expression of 26 selected glycosylated cell surface proteins (with an exception of nonglycosylated protein CD20 detected here with the Lys-CSC strategy), which are commonly used in clinical diagnostic panels. Shown is the percentage of cases positive for a given marker by FCM and mass spectrometry identification. Proteins with at least 5 independent spectra acquired for a given sample were taken into account.
CSC technology captures the surface phenotype of ALL cells. (A) Cell surface proteome map of ALL cells: 713 identified membrane-associated proteins were categorized based on their biological function assigned by the PANTHER algorithm (left); the 20 most abundant protein families and superfamilies are displayed (right; protein family assignment according to UniProt KB annotation). (B) Comparison between immunophenotypes described using 2 different technologies. ALL xenograft samples (n = 19) included in the proteomic pipeline were analyzed by FCM for expression of 26 selected glycosylated cell surface proteins (with an exception of nonglycosylated protein CD20 detected here with the Lys-CSC strategy), which are commonly used in clinical diagnostic panels. Shown is the percentage of cases positive for a given marker by FCM and mass spectrometry identification. Proteins with at least 5 independent spectra acquired for a given sample were taken into account.
Integration of ALL surfaceome and transcriptome data from sorted hematopoietic differentiation states (DMAP) identifies candidate leukemia markers
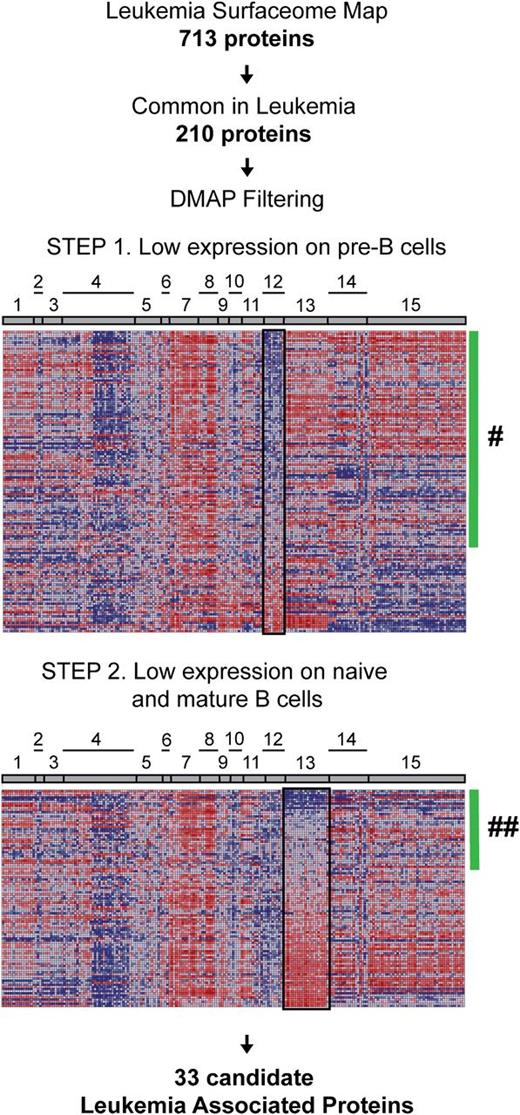
To filter the surfaceome for putative leukemia markers, we sought to detect proteins that are not abundantly expressed on normal human lymphoid progenitors. Given the limitations of small sample size for CSC, we took advantage of a recent transcriptome analysis of sorted human hematopoietic states (differentiation map portal [DMAP]) to interrogate our surfaceome data set21 (Figure 3); 401 surfaceome members were represented in the preprocessed DMAP data set that includes genes differently regulated between 38 sorted hematopoietic states (supplemental Table 4). We ranked the corresponding genes according to their expression in sorted BCP cells. As expected, high messenger RNA expression levels were detected in normal BCPs for typical B-lymphoid markers identified in the surfaceome data set (CD79B, CD9, CD10, CD19, HLA-DR, CXC chemokine receptor 4). Among transcripts underrepresented in this compartment, we detected 13 markers (SELL, CD44, ITGB7, IL6ST, ICAM2, ITGA6, TNFRSF1A, CD86, IL3RA, CD300A, CD99, CD97, and CD69) shown recently by Coustan-Smith et al22 as being differently regulated between leukemia and the CD10+ CD19+ cell subset isolated from bone marrow of healthy donors. Thus, we hypothesized that filtering of surfaceome data based on the transcriptomic information will reveal further markers aberrantly detected on leukemia cells.
Integrated proteomic and transcriptomic investigation for the identification of new leukemia-associated markers. The leukemia surfaceome data set was reduced to 210 proteins identified in more than half of investigated leukemia cases (at least 11 of 19 cases; 58%) and probed against published gene expression signatures of B cells at different maturation stages in normal hematopoiesis (DMAP). One hundred eighteen genes represented in DMAP corresponding to the proteins of interest were ranked based on average expression in precursor B-cell populations (step 1). Eighty-three genes with average expression below 0 (green bar; #) were further ranked based on average gene expression in the cluster of B cells from later ontogeny stages (step 2) resulting in 33 gene IDs with expression below 0 (green bar; ##). Cell populations and lineages: 1, hematopoietic stem cells; 2, common myeloid progenitors; 3, megakaryocyte/erythroid progenitors; 4, erythroid cells; 5, megakaryocytes; 6, granulocyte/monocyte progenitors; 7, granulocytes; 8, monocytes; 9, eosinophils; 10, basophils; 11, dendritic cells; 12, precursor B cells; 13, naive and mature B cells; 14, natural killer cells; 15, T cells.
Integrated proteomic and transcriptomic investigation for the identification of new leukemia-associated markers. The leukemia surfaceome data set was reduced to 210 proteins identified in more than half of investigated leukemia cases (at least 11 of 19 cases; 58%) and probed against published gene expression signatures of B cells at different maturation stages in normal hematopoiesis (DMAP). One hundred eighteen genes represented in DMAP corresponding to the proteins of interest were ranked based on average expression in precursor B-cell populations (step 1). Eighty-three genes with average expression below 0 (green bar; #) were further ranked based on average gene expression in the cluster of B cells from later ontogeny stages (step 2) resulting in 33 gene IDs with expression below 0 (green bar; ##). Cell populations and lineages: 1, hematopoietic stem cells; 2, common myeloid progenitors; 3, megakaryocyte/erythroid progenitors; 4, erythroid cells; 5, megakaryocytes; 6, granulocyte/monocyte progenitors; 7, granulocytes; 8, monocytes; 9, eosinophils; 10, basophils; 11, dendritic cells; 12, precursor B cells; 13, naive and mature B cells; 14, natural killer cells; 15, T cells.
To select candidates for further validation, we restricted the initial data set to the proteins that were consistently identified in more than half of the cases investigated by CSC. This resulted in 118 proteins for which the transcripts were represented on the DMAP platform. In the first step, we filtered the surfaceome protein data set against the DMAP data set for proteins for which respective transcripts had low expression in pre–B cells. This step should reduce the number of proteins that would usually be abundant in the corresponding normal differentiation stage and possibly enrich for features that are aberrantly expressed on leukemic cells. Next, we filtered the resulting set of 86 surface markers for proteins with respective transcripts having low expression levels in naive and mature B cells, to reduce the number of proteins that would be commonly expressed in more mature B-cell differentiation states. This resulted in a list of 33 candidate leukemia-associated proteins (LAPs) for further analysis (Table 1). For prevalidation studies, we selected a subset of the candidates for which FCM-validated reagents were available, including ITB2 (CD18), CD63, ICAM2 (CD102), BST2 (CD317), IL17RA (CD217), LAIR1 (CD305), PECAM1 (CD31), CD97, and BST1 (CD157). We also included as a reference CD58, which was previously established as a useful marker for detection of minimal residual disease (MRD) by FCM.23
List of 33 candidate LAPs
| UniProt ID . | CD protein . | ENTREZ gene ID . | Description . | Avg. expression pre-B* . | Avg. expression B others† . |
|---|---|---|---|---|---|
| Proteins selected for validation | |||||
| ITB2_HUMAN | CD18 | 3689 | Integrin β-2 | −1.428 | −0.416 |
| CD63_HUMAN | CD63 | 967 | CD63 antigen | −1.170 | −0.716 |
| ICAM2_HUMAN | CD102 | 3384 | Intercellular adhesion molecule 2 | −0.525 | −0.084 |
| BST2_HUMAN | CD317 | 684 | Bone marrow stromal antigen 2 | −0.327 | −0.141 |
| I17RA_HUMAN | CD217 | 23 765 | Interleukin-17 receptor A | −0.106 | −0.152 |
| LAIR1_HUMAN | CD305 | 3903 | Leukocyte-associated immunoglobulin–like receptor 1 | −0.072 | −0.204 |
| PECA1_HUMAN | CD31 | 5175 | Platelet endothelial cell adhesion molecule | −0.064 | −0.550 |
| CD97_HUMAN | CD97 | 976 | CD97 antigen | −0.029 | −0.090 |
| BST1_HUMAN | CD157 | 683 | Adenosine 5'-diphosphate-ribosyl cyclase 2 | −0.001 | −1.512 |
| Proteins not selected for initial validation | |||||
| ITAL_HUMAN | CD11a | 3683 | Integrin α-L | −1.695 | −0.266 |
| SEM4D_HUMAN | CD100 | 10 507 | Semaphorin-4D | −1.391 | −0.071 |
| AT2B4_HUMAN | na | 493 | Plasma membrane calcium-transporting adenosine triphosphatase 4 | −1.312 | −1.624 |
| S39AE_HUMAN | na | 23 516 | Zinc transporter ZIP14 | −1.169 | −0.082 |
| ITAV_HUMAN | CD51 | 3685 | Integrin α-V | −1.126 | −0.745 |
| SERC3_HUMAN | na | 10 955 | Serine incorporator 3 | −1.084 | −0.522 |
| S29A1_HUMAN | na | 2030 | Equilibrative nucleoside transporter 1 | −0.976 | −0.462 |
| AT1A1_HUMAN | na | 476 | Sodium/potassium-transporting ATPase subunit α-1 | −0.891 | −0.210 |
| CD59_HUMAN | CD59 | 966 | CD59 glycoprotein | −0.590 | −0.430 |
| IL7RA_HUMAN | CD127 | 3575 | Interleukin-7 receptor subunit α | −0.558 | −1.234 |
| FLT3_HUMAN | CD135 | 2322 | fms-related tyrosine kinase 3 | −0.438 | −0.750 |
| PLXD1_HUMAN | na | 23 129 | Plexin-D1 | −0.434 | −0.460 |
| CTR1_HUMAN | na | 6541 | High-affinity cationic amino acid transporter 1 | −0.399 | −0.015 |
| S43A3_HUMAN | na | 29 015 | Solute carrier family 43 member 3 | −0.361 | −0.736 |
| SIRB1_HUMAN | CD172b | 10 326 | Signal-regulatory protein β-1 | −0.354 | −0.946 |
| GTR1_HUMAN | na | 6513 | Solute carrier family 2, facilitated glucose transporter member 1 | −0.212 | −0.089 |
| SREC_HUMAN | na | 8578 | Endothelial cells scavenger receptor | −0.211 | −1.823 |
| TSN14_HUMAN | na | 81 619 | Tetraspanin-14 | −0.172 | −0.586 |
| IL3RA_HUMAN | CD123 | 3563 | Interleukin-3 receptor subunit α | −0.110 | −0.073 |
| CLM8_HUMAN | CD300a | 11 314 | CMRF35-like molecule 8 | −0.108 | −0.917 |
| CD99_HUMAN | CD99 | 4267 | CD99 antigen | −0.103 | −0.166 |
| JAM1_HUMAN | CD321 | 50 848 | Junctional adhesion molecule A | −0.098 | −0.163 |
| MEGF9_HUMAN | na | 1955 | Multiple epidermal growth factor–like domains 9 | −0.038 | −0.695 |
| LFA3_HUMAN | CD58 | 965 | Lymphocyte function-associated antigen 3 | −0.014 | −0.623 |
| UniProt ID . | CD protein . | ENTREZ gene ID . | Description . | Avg. expression pre-B* . | Avg. expression B others† . |
|---|---|---|---|---|---|
| Proteins selected for validation | |||||
| ITB2_HUMAN | CD18 | 3689 | Integrin β-2 | −1.428 | −0.416 |
| CD63_HUMAN | CD63 | 967 | CD63 antigen | −1.170 | −0.716 |
| ICAM2_HUMAN | CD102 | 3384 | Intercellular adhesion molecule 2 | −0.525 | −0.084 |
| BST2_HUMAN | CD317 | 684 | Bone marrow stromal antigen 2 | −0.327 | −0.141 |
| I17RA_HUMAN | CD217 | 23 765 | Interleukin-17 receptor A | −0.106 | −0.152 |
| LAIR1_HUMAN | CD305 | 3903 | Leukocyte-associated immunoglobulin–like receptor 1 | −0.072 | −0.204 |
| PECA1_HUMAN | CD31 | 5175 | Platelet endothelial cell adhesion molecule | −0.064 | −0.550 |
| CD97_HUMAN | CD97 | 976 | CD97 antigen | −0.029 | −0.090 |
| BST1_HUMAN | CD157 | 683 | Adenosine 5'-diphosphate-ribosyl cyclase 2 | −0.001 | −1.512 |
| Proteins not selected for initial validation | |||||
| ITAL_HUMAN | CD11a | 3683 | Integrin α-L | −1.695 | −0.266 |
| SEM4D_HUMAN | CD100 | 10 507 | Semaphorin-4D | −1.391 | −0.071 |
| AT2B4_HUMAN | na | 493 | Plasma membrane calcium-transporting adenosine triphosphatase 4 | −1.312 | −1.624 |
| S39AE_HUMAN | na | 23 516 | Zinc transporter ZIP14 | −1.169 | −0.082 |
| ITAV_HUMAN | CD51 | 3685 | Integrin α-V | −1.126 | −0.745 |
| SERC3_HUMAN | na | 10 955 | Serine incorporator 3 | −1.084 | −0.522 |
| S29A1_HUMAN | na | 2030 | Equilibrative nucleoside transporter 1 | −0.976 | −0.462 |
| AT1A1_HUMAN | na | 476 | Sodium/potassium-transporting ATPase subunit α-1 | −0.891 | −0.210 |
| CD59_HUMAN | CD59 | 966 | CD59 glycoprotein | −0.590 | −0.430 |
| IL7RA_HUMAN | CD127 | 3575 | Interleukin-7 receptor subunit α | −0.558 | −1.234 |
| FLT3_HUMAN | CD135 | 2322 | fms-related tyrosine kinase 3 | −0.438 | −0.750 |
| PLXD1_HUMAN | na | 23 129 | Plexin-D1 | −0.434 | −0.460 |
| CTR1_HUMAN | na | 6541 | High-affinity cationic amino acid transporter 1 | −0.399 | −0.015 |
| S43A3_HUMAN | na | 29 015 | Solute carrier family 43 member 3 | −0.361 | −0.736 |
| SIRB1_HUMAN | CD172b | 10 326 | Signal-regulatory protein β-1 | −0.354 | −0.946 |
| GTR1_HUMAN | na | 6513 | Solute carrier family 2, facilitated glucose transporter member 1 | −0.212 | −0.089 |
| SREC_HUMAN | na | 8578 | Endothelial cells scavenger receptor | −0.211 | −1.823 |
| TSN14_HUMAN | na | 81 619 | Tetraspanin-14 | −0.172 | −0.586 |
| IL3RA_HUMAN | CD123 | 3563 | Interleukin-3 receptor subunit α | −0.110 | −0.073 |
| CLM8_HUMAN | CD300a | 11 314 | CMRF35-like molecule 8 | −0.108 | −0.917 |
| CD99_HUMAN | CD99 | 4267 | CD99 antigen | −0.103 | −0.166 |
| JAM1_HUMAN | CD321 | 50 848 | Junctional adhesion molecule A | −0.098 | −0.163 |
| MEGF9_HUMAN | na | 1955 | Multiple epidermal growth factor–like domains 9 | −0.038 | −0.695 |
| LFA3_HUMAN | CD58 | 965 | Lymphocyte function-associated antigen 3 | −0.014 | −0.623 |
Validation in an independent patient cohort reveals differential abundance of selected candidates in leukemia and residual normal B cells
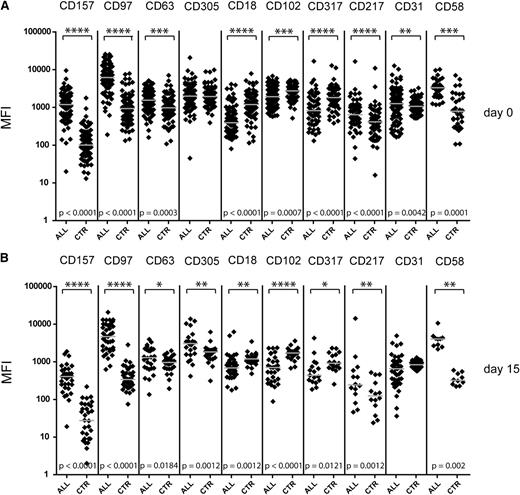
First, we validated the presence of all 9 selected markers at the cell surface of the leukemia xenograft samples that were used for CSC screening (data not shown). Next, we tested the cell surface expression of these 9 candidate proteins on ALL cells from biobanked diagnostic cases (n = 86). The comparison with the remaining nonmalignant CD19+ B-cell populations in the same sample revealed on average higher levels of CD157, CD97, and CD217 in leukemic cells. In contrast, the expression of CD18, CD102, and CD317 was decreased in leukemia cells in comparison with the nonmalignant (mainly mature) B-cell population (Figure 4A). A similar pattern of marker expression was present in samples that were acquired prospectively at day 15 of induction (Figure 4B).
High levels of candidate marker expression help to identify the blast population at diagnosis and early treatment time points. Comparison of the marker expression in leukemic cell populations (ALL) and the residual nonmalignant CD19+ population (in-sample control, CTR). Shown are the MFI measured for indicated markers in newly diagnosed leukemia samples (n = 86) (A) and MRD samples at day 15 (n = 45) of the treatment (B). Represented are data from 3 independent patient cohorts; for simultaneous representation, the median value for each cohort was adjusted. Values for a population size <100 events at diagnosis or <30 events in MRD samples were excluded. Each point represents a single measurement, and gray bars represent the median value for the group. The asterisk indicates a statistically significant difference in marker expression between paired measurements by Wilcoxon matched pairs test. No difference was detected between ALL and control samples for CD31 and CD305.
High levels of candidate marker expression help to identify the blast population at diagnosis and early treatment time points. Comparison of the marker expression in leukemic cell populations (ALL) and the residual nonmalignant CD19+ population (in-sample control, CTR). Shown are the MFI measured for indicated markers in newly diagnosed leukemia samples (n = 86) (A) and MRD samples at day 15 (n = 45) of the treatment (B). Represented are data from 3 independent patient cohorts; for simultaneous representation, the median value for each cohort was adjusted. Values for a population size <100 events at diagnosis or <30 events in MRD samples were excluded. Each point represents a single measurement, and gray bars represent the median value for the group. The asterisk indicates a statistically significant difference in marker expression between paired measurements by Wilcoxon matched pairs test. No difference was detected between ALL and control samples for CD31 and CD305.
Day 15 of induction therapy has been established as an informative time point for risk assessment based on FCM MRD analysis.6 Here we compared the results of FCM MRD assessment at day 15 for 20 available cases with the standard MRD panel used in the clinical setting (combination of backbone antibodies CD10, CD19, CD34, CD20, CD45 with CD58, CD38, CD11a, and/or CD11b) and the combination of the same backbone with our experimental markers incorporated in the bright fluorochrome positions. As shown in Figure 5A, leukemia cell quantification with both antibody panels yielded comparable results. These findings show that in particular CD157, CD97, CD18, and CD102 may have the potential to serve as additional markers to increase the confidence of residual leukemia cell detection and quantification at early treatment time points.
Prospective analysis of LAPs in patient samples. (A) Correlation of leukemia detectability based on gating performed with standard MRD markers and panels including experimental markers. To gate the leukemic population, 9 candidate markers were used in combination with CD19 and CD45 antibodies for a total of 69 tests performed on 20 MRD cases from day15. The percentage of leukemic cells was calculated in respect to all nucleated events as determined by Syto 41 or Syto16 stainings and compared with levels of leukemia determined by standard gating based on CD19, CD45, and CD10 expression. R indicates Pearson correlation score. (B) Differential expression of experimental markers between leukemia cells and residual normal lymphoid cells at diagnosis and in paired MRD cases. The difference in MFI (arbitrary units) between leukemia cell population and nonmalignant CD19+ cells within the same sample measured at diagnosis (Dx) and in MRD cases at day 15 of treatment (M) is represented as a color-coded plot. In total, 187 stainings were performed for experimental markers at both time points in 34 matched ALL cases from 2 cohorts. Depending on material availability, different numbers of markers were assigned to a given sample (number of patients screened for each individual marker at both time points: CD157, 24; CD97, 27; CD63, 19; CD305, 15; CD18, 27; CD102, 22; CD317, 12; CD217, 10; and CD31, 31). Cases in which the experimental marker expression in both leukemic and nonmalignant cell population was below the positivity threshold established for each cohort are not displayed in the graph. Differential expression of CD58 is presented for 9 matched cases as a control.
Prospective analysis of LAPs in patient samples. (A) Correlation of leukemia detectability based on gating performed with standard MRD markers and panels including experimental markers. To gate the leukemic population, 9 candidate markers were used in combination with CD19 and CD45 antibodies for a total of 69 tests performed on 20 MRD cases from day15. The percentage of leukemic cells was calculated in respect to all nucleated events as determined by Syto 41 or Syto16 stainings and compared with levels of leukemia determined by standard gating based on CD19, CD45, and CD10 expression. R indicates Pearson correlation score. (B) Differential expression of experimental markers between leukemia cells and residual normal lymphoid cells at diagnosis and in paired MRD cases. The difference in MFI (arbitrary units) between leukemia cell population and nonmalignant CD19+ cells within the same sample measured at diagnosis (Dx) and in MRD cases at day 15 of treatment (M) is represented as a color-coded plot. In total, 187 stainings were performed for experimental markers at both time points in 34 matched ALL cases from 2 cohorts. Depending on material availability, different numbers of markers were assigned to a given sample (number of patients screened for each individual marker at both time points: CD157, 24; CD97, 27; CD63, 19; CD305, 15; CD18, 27; CD102, 22; CD317, 12; CD217, 10; and CD31, 31). Cases in which the experimental marker expression in both leukemic and nonmalignant cell population was below the positivity threshold established for each cohort are not displayed in the graph. Differential expression of CD58 is presented for 9 matched cases as a control.
Prospective investigation of matched samples reveals treatment-induced changes in LAP expression levels
Antigenic shifts are sometimes observed during the initial treatment phase with glucocorticoids,24,25 which can provide useful information, as illustrated by the occasionally striking increase of CD20 expression.26 To assess the stability of the detected phenotypes over time, we analyzed the differential expression of the 9 candidates between leukemia cells and in-sample lineage controls in 34 matched diagnostic and MRD-positive day 15 samples (Figure 5B). In order to define the degree to which a given marker distinguishes between the leukemic cell population and residual nonmalignant B cells, we calculated the difference of the MFI between the individual populations in the same sample. We defined a cutoff value of 450 (in arbitrary units) for this MFI difference to consider it informative for detection of the leukemic population. This comparison of the MFI changes revealed that although CD97 and CD58 features did not appear to be modulated at all by chemotherapy, others were clearly reduced (CD102 and CD317) or increased (CD305 and CD63) in several cases. Although this modulation resulted in the loss of discriminating power for a given marker in a few cases (indicated by white color), the downmodulation of CD102 and especially a consistent increase in the relative abundance of CD305 and CD63 in a subset of the samples could additionally contribute to differentiating between malignant clones and normal B cells during disease follow-up.
Multiparametric FCM analysis demonstrates different expression levels of candidate LAP in leukemic cells and their normal counterparts
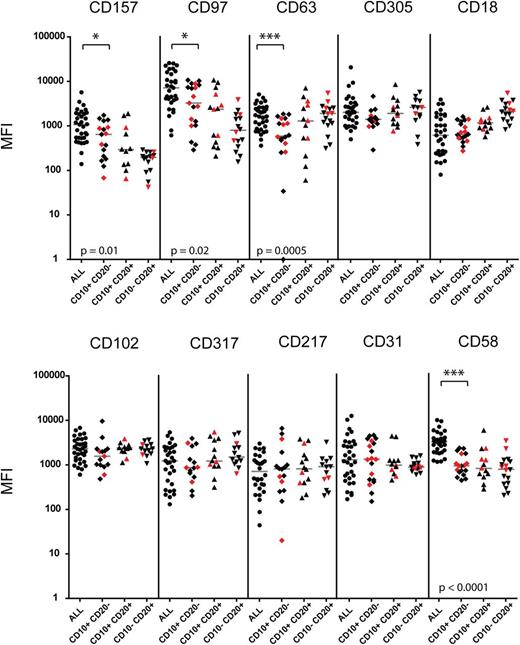
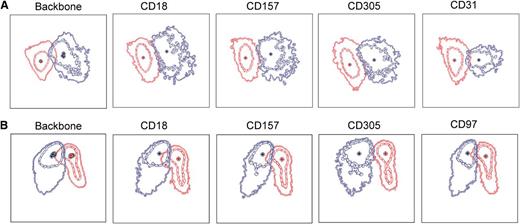
For a potential clinical application, proteins with differential expression between disease and healthy state need to be identified to allow distinction of malignant cell populations on the background of regenerating normal lymphocytes. To elucidate if the 9 investigated markers may contribute to improved distinction between malignant and normal populations, we compared their expression level on ALL cells with the levels detected in normal or MRD-negative remission marrows (n = 20) including postinduction therapy samples with increased numbers of hematogones (Figure 6). The experimental markers were included in bright positions into a backbone antibody panel in order to visualize the major stages in normal B-cell ontogeny in the bone marrow. These included BCP-I CD10+ CD20–, BCP-II/III CD10+ CD20+, and BCP-IV CD10– CD20+ populations. Besides CD58 serving as an internal control, and CD97, which was recently proposed as an MRD marker by Coustan-Smith et al,22 we identified 2 additional markers, CD157 and CD63, with significantly higher expression on ALL cells compared with normal B-cell populations in the bone marrow. Although not statistically significant, we also observed differences in CD305 and CD18 expression levels between ALL cases and their normal counterparts in some samples. A subset of ALL cases had much lower levels of CD18 compared with their normal counterparts, suggesting that this marker could also serve to discriminate leukemia from normal cells when expressed at low levels on leukemia. Importantly, differential expression of CD63, CD157, CD97, CD18, and CD305 on ALL cells compared with normal precursor B cells was confirmed in an independent cohort of patients investigated prospectively in a second study center (supplemental Figure 1). An example for the visualization of these markers in the context of normal bone marrow is provided with the analysis of a patient presenting with an isolated ALL relapse in the central nervous system and a submicroscopic involvement of the bone marrow (supplemental Figure 2). In this case, increased detection of CD63 on the surface of ALL cells was particularly evident compared with nonmalignant early B cells. To visualize the effect of additional surface markers on the separation of malignant from normal cell populations, we used a bidimensional principal component analysis (PCA)27 (Figure 7). We compared the immunophenotype of leukemia cells from presentation with leukemia-free marrows from 2 patients for which bone marrows after 78 days on their treatment plan were available, a time point at which regeneration with normal lymphoid progenitors is usually prominent. This analysis indicates that several markers contributed to a better separation of the normal and malignant cell populations, and that, as expected from our observations in general, the contributions of individual markers were different in these 2 patients.
Differential expression of candidate markers between leukemic blasts and B-cell subsets from nonmalignant controls. Shown are MFI values measured for indicated markers in malignant populations of newly diagnosed leukemia samples (ALL, n = 34) and in subpopulations representing different B-cell maturation stages (CD10+ CD20–; CD10+ CD20+; and CD10– CD20+) from normal bone marrow and remission samples with no evidence of disease (n = 20). Samples from postinduction therapy time point (day 78) are labeled in red. MFI values measured for cell population size <100 events were excluded. Each point represents a single measurement, and gray bars represent the median value for the group. The asterisk indicates a statistically significant difference in marker expression by the Mann-Whitney test with specified P value.
Differential expression of candidate markers between leukemic blasts and B-cell subsets from nonmalignant controls. Shown are MFI values measured for indicated markers in malignant populations of newly diagnosed leukemia samples (ALL, n = 34) and in subpopulations representing different B-cell maturation stages (CD10+ CD20–; CD10+ CD20+; and CD10– CD20+) from normal bone marrow and remission samples with no evidence of disease (n = 20). Samples from postinduction therapy time point (day 78) are labeled in red. MFI values measured for cell population size <100 events were excluded. Each point represents a single measurement, and gray bars represent the median value for the group. The asterisk indicates a statistically significant difference in marker expression by the Mann-Whitney test with specified P value.
Bidimensional PCA of the effect of additional surface markers on the separation of normal and malignant cell populations. Bone marrow samples obtained at diagnosis and after induction therapy (day 78) were obtained from 2 pediatric ALL patients (A-B). Samples were stained with the usual backbone of antibody combinations including CD19, CD34, CD45, CD10, and CD20 in all tubes and supplemented with either CD18 and CD157, CD31 and CD97, CD63, CD84, CD100, CD102, or CD305. After data acquisition, .fcs files were merged using the Infinicyt software.27 ALL cells at diagnosis (red) and normal regenerating BCPs at day 78 in MRD-negative samples (blue) were identified based on the backbone markers and plotted in a bidimensional PCA. The first principal component (PC) is shown on the x-axis, and the second PC on the y-axis, using the Automatic Population Separator (APS) graphical representation of the Infinicyt software.27 Data represent the mean and first and second standard deviation of the 2 populations. Backbone: APS plots using backbone markers only. Both populations show some overlap, which impacts the resolution of the analysis. The effect of the addition of 1 additional marker to this backbone as indicated in the figure legends is visualized by PCA. The separation between the ALL and normal BCPs improves when some of these markers are added. The relative contribution of each marker varies in the 2 samples.
Bidimensional PCA of the effect of additional surface markers on the separation of normal and malignant cell populations. Bone marrow samples obtained at diagnosis and after induction therapy (day 78) were obtained from 2 pediatric ALL patients (A-B). Samples were stained with the usual backbone of antibody combinations including CD19, CD34, CD45, CD10, and CD20 in all tubes and supplemented with either CD18 and CD157, CD31 and CD97, CD63, CD84, CD100, CD102, or CD305. After data acquisition, .fcs files were merged using the Infinicyt software.27 ALL cells at diagnosis (red) and normal regenerating BCPs at day 78 in MRD-negative samples (blue) were identified based on the backbone markers and plotted in a bidimensional PCA. The first principal component (PC) is shown on the x-axis, and the second PC on the y-axis, using the Automatic Population Separator (APS) graphical representation of the Infinicyt software.27 Data represent the mean and first and second standard deviation of the 2 populations. Backbone: APS plots using backbone markers only. Both populations show some overlap, which impacts the resolution of the analysis. The effect of the addition of 1 additional marker to this backbone as indicated in the figure legends is visualized by PCA. The separation between the ALL and normal BCPs improves when some of these markers are added. The relative contribution of each marker varies in the 2 samples.
Collectively, our results demonstrate that LAPs with significant differences in abundance between leukemic cells and nonmalignant precursor B cells can be identified by systematic exploration based on the CSC ALL surfaceome data set. Here we have shown a strategy for selection and prequalification of disease-associated markers for further clinical validation and provided data to support the use of multidimensional bioinformatic approaches to improve FCM-based detection of leukemia. Furthermore, our approach will also be useful for the analysis of specific leukemia subgroups, for which appropriate leukemia-associated markers are lacking, such as early thymic precursor or mature T-cell ALL.
Discussion
We report here for the first time an expanded description of the leukemia surfaceome, based on a chemoproteomic analysis of primary cells derived from xenotransplanted patient samples. The direct combination of the CSC technology and the possibility to expand leukemia cells reliably in immunodeficient mice enables us to apply a mass spectrometry–based approach that is usually challenging with primary samples due to the limited sample size. This methodology provides direct evidence for the localization of proteins at the leukemia cell surface, information that cannot be derived reliably from transcriptomic studies or obtained at a larger scale with currently available antibodies.
Our proteomic data include cell surface proteins that share potentially important functions for the maintenance and propagation of the disease. Different strategies can be used to select candidate genes for future functional investigation. For instance, a comparison with expression data from other tissue could identify ectopic cell surface marker expression in leukemia. Indeed, we identified different plexin receptors, which are typically expressed in neuronal tissues, on ALL cells. The plexin ligands semaphorins, including Sema4D (CD100), which is known to sustain proliferation and survival of leukemic cells,28 were also detected. Short lists can be derived from the surfaceome data by intersection with surface marker information that reflects relevant biological or disease states (eg, stem and progenitor cells and disease progression). We detected different ephrins that had been linked to cancerogenesis and metastasis.29 Categorization by functional annotation is another way to prioritize candidates for further investigation. We identified members of the tetraspanins (Table1), a type of receptor with potential death receptor function on normal and malignant B cells,30 and modulators of the cellular immune response including CD59 and members of the signal regulatory protein family that could have implications for cellular antileukemic responses.31 Also striking is the predominance of proteins from the major facilitator superfamily, including different solute carrier proteins, which regulate the intracellular flux of metabolic substrates, which are essential for cell survival and for metabolic interactions with the microenvironment.32 Surfaceome data could therefore be used to generate libraries of practicable size for functional genomic approaches using in vitro and in vivo leukemia xenograft models in order to identify critical interactions of leukemia cells with the microenvironment to sustain the disease.
The comparison between malignant and normal hematopoietic subpopulations can be used to identify new features in both normal and malignant hematopoiesis. We used a gene expression data set from FCM-sorted normal hematopoietic subpopulations to preselect candidate leukemia markers. Because gene expression data do not always correlate with protein levels at the cell surface,33-35 and because subcellular location is not reliably predicted based on transcript information, there is an added value to determine membrane location by CSC. Coustan-Smith et al22 used gene expression profiling to identify genes that were preferentially expressed in ALL cells compared with sorted normal CD10+ CD19+ lymphoid progenitors. The ALL surfaceome data include 21 out of the 26 membrane proteins that were reported in this gene expression profiling study with a threefold higher transcript level in leukemia cells. From the markers that were validated by FCM in the Coustan-Smith study, we retrieved 15 (CD44, CD49F, CD69, CD72, CD73, CD79B, CD86, CD97, CD99, CD102, CD120A, CD123, CD130, CD200, and CD300A), but most of our candidates did not overlap with the genes reported in this study. As a proof of concept, we identified CD58, a marker commonly used for detection of MRD for ALL.6,23,36 The bottleneck for further validation studies is the availability of appropriate antibodies. We tested 9 antibodies and found informative differences between leukemic and normal lymphoid cells in patient samples for all of them, with most predominant differences for CD63, CD97, and CD157, as well as informative antigenic shifts during treatment, which can be helpful for the identification of leukemia cells.3,24,37,38 Taken together, we provide initial evidence that this approach will contribute to the identification of new surface markers.
There is still a need for better discrimination between normal and leukemia cells by FCM, especially for disease monitoring.6,39,40 With the development of multiparametric FCM, we postulate that better discrimination between cellular subpopulations will be possible by increasing the number of different surface markers and analysis in a multidimensional space.41 The clinical significance of risk stratification by the monitoring of MRD is well established for ALL.25,42-44 The availability of more sensitive FCM assays would have several advantages including speed and cost reduction for MRD monitoring. In this study, we did not identify markers that per se will improve discrimination of leukemia cells significantly, but we show that the addition of new markers to the conventional backbone of diagnostic markers can improve the separation of malignant and normal cell subpopulations using a bidimensional PCA.
Our discovery-driven LC-MS approach for the analysis of the cell surface proteome of ALL cells allows for the identification of new or unknown cell surface markers. The methodology could be used for biomarker discovery in defined subsets, such as early thymic precursor ALL45 or translocation t(17;19)-positive ALL.25 The CSC technology used here has limitations. It is nonquantitative, directed against a specific subset of the surface proteome, and cannot ensure a complete cell surface proteome mapping yet. Also, we cannot make a statement about the spatial and temporal co-occurrence of all identified proteins. It is important to emphasize the potential of the part of the detected surfaceome that is left unexplored due to lack of appropriate affinity probes. Further progress can be expected with the development of new antibodies and alternative quantitative proteomic technologies such as targeted mass spectrometry techniques46,47 or mass spectrometry–based FCM (cytometry by time-to-flight), which rapidly increases the numbers of parameters that can be detected at the same time at the single-cell level.48,49 Our work constitutes the basis for prospective validation of new leukemia markers and a valuable resource to build and to characterize the leukemia surfaceome, including information about cell surface–exposed proteins, their N-glycosite occupancy, and tools for their targeted mass-spectrometric assessment. This knowledge will contribute to accelerate the clinical validation of useful markers for multidimensional visualization and distinction of normal and malignant hematopoietic subpopulations.
This article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Rosaria Diano, Claudia Möller, Severine Gilloz, Fabienne Ulmer, and Daniela Morf for their excellent help with FCM analysis.
This work was supported by grants from National Center of Competence in Research Neural Plasticity and Repair (B.W.); the Mach-Gaensslen Foundation (B.W. and J.-P.B.); the Swiss Cancer League (J.-P.B.); the Empiris Foundation (J.-P.B.); the Novartis Research Foundation (J.-P.B.); the Swiss National Science Foundation (J.-P.B.); the Hanne-Liebermann Foundation and the Foundation Kind und Krebs (J.-P.B.); and the clinical research focus program “human hemato-lymphatic diseases” of the University of Zurich (J.-P.B.), GAUK 15710 (L. Slamova), P301/10/1877 (E.M. and L. Slamova), NT13462 (E.M.), and ERA-NET PRIOMEDCHILD (V.H.J.v.d.V. and T.S.).
Authorship
Contribution: B.W. and J.-P.B. designed the research; P.M., A.H., M. Schmitz, L. Slamova, and L. Sedek performed the research; P.M. and A.H. analyzed data; V.H.J.v.d.V. performed the PCA; P.M., A.H., B.C.B., B.W., and J.-P.B. wrote the manuscript; E.M., T.S., V.H.J.v.d.V., and J.-P.B. coordinated clinical diagnostics and acquisition of material; G.C., M. Stanulla, and M. Schrappe provided biobanked samples; and all authors discussed the results and implications and commented on the manuscript at all stages.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Bernd Wollscheid, Department of Biology, Institute of Molecular Systems Biology, Eidgenössische Technische Hochschule Zurich, Wolfgang-Pauli-Strasse 16, 8093 Zurich, Switzerland; e-mail: wbernd@ethz.ch; and Jean-Pierre Bourquin, Department of Oncology, University Children’s Hospital Zurich, August-Forel-Strasse 1, 8008 Zurich, Switzerland; e-mail: jean-pierre.bourquin@kispi.uzh.ch.
References
Author notes
P.M. and A.H. contributed equally to this study as senior authors.