Key Points
Simvastatin and tBHQ suppress KLF1 and BCL11 gene expression and additively increase fetal hemoglobin in primary human erythroid cells.
Because both drugs are FDA-approved, these findings could lead to clinical trials in the relatively near future.
Abstract
Although increased fetal hemoglobin (HbF) levels have proven benefit for people with β-hemoglobinopathies, all current HbF-inducing agents have limitations. We previously reported that drugs that activate the NRF2 antioxidant response signaling pathway increase HbF in primary human erythroid cells. In an attempt to increase HbF levels achieved with NRF2 activators, in the present study, we investigated potential complementary activity between these agents and HMG-CoA reductase inhibitors (statins) based on their ability to induce KLF2 protein levels. Experiments in K562 cells showed that simvastatin increased KLF2 mRNA and protein and KLF2 binding to HS2 of the β-globin locus control region and enhanced γ-globin mRNA production by the NRF2 activator Tert-butylhydroquinone (tBHQ). When tested in differentiating primary human erythroid cells, simvastatin induced HbF alone and additively with tBHQ, but it did not increase KLF2 mRNA or locus control region binding above levels seen with normal differentiation. Investigating alternative mechanisms of action, we found that both simvastatin and tBHQ suppress β-globin mRNA and KLF1 and BCL11A mRNA and protein, similar to what is seen in people with an HPFH phenotype because of KLF1 haploinsufficiency. These findings identify statins as a potential class of HbF-inducing agents and suggest a novel mechanism of action based on pharmacologic suppression of KLF1 and BCL11A gene expression.
Introduction
The β-hemoglobinopathies are inherited disorders caused by mutations that alter β-globin protein structure (eg, sickle cell disease, SCD) or gene expression (eg, β-thalassemia, β-thal). In contrast to most genetic diseases, humans have 2 potential replacement genes, the γ-globin genes, which are expressed during fetal development but then silenced soon after birth. When these genes are expressed in adults with β-hemoglobinopathies as a result of mutations, the clinical manifestations of SCD and β-thal are greatly improved because of the ability of the γ-globin protein to inhibit sickle Hb polymerization and to reduce the globin chain imbalance of β-thal.1,2 This has led to a 3-decade-long search for pharmacologic agents that can reactivate γ-globin gene expression and fetal hemoglobin (HbF) production. Proof-of-principle studies using DNA methyltransferase inhibitors in small numbers of near end-stage β-thal and SCD patients have shown that significant clinical benefits can be achieved.3,4 However, toxicity associated with these agents and other issues has prevented their widespread use. Although hydroxyurea has been approved by the Food and Drug Administration (FDA) as an HbF-inducing agent, its use has been limited because it is effective in only approximately half of SCD patients,5 it is less effective in β-thal,6 and it has a low therapeutic index because of suppression of blood counts, making close monitoring a necessity. Because of these and other concerns, hydroxyurea is significantly under used in both the United States7 and Africa.8
A major goal of hemoglobinopathy research is to develop improved pharmacologic inducers of γ-globin gene expression and HbF production that are safe, effective, and have the ease of use and affordability that would make them applicable to the great majority of patients who lack access to modern medical care. We recently reported that pharmacologic activation of the nuclear factor erythroid 2-related factor 2 (NRF2) antioxidant response signaling pathway also directly activates γ-globin gene expression and increases HbF levels in primary human erythroid cells.9 Drugs that activate this pathway are prime candidates for further development because several are already in clinical trials for the prevention and treatment of cancer and cardiovascular and neurodegenerative diseases, they can be taken orally, and they generally have a favorable side effect profile.10 This discovery has prompted us to investigate other drug classes that have the potential to augment the HbF induction achieved with NRF2-activating agents. One such class of drugs is the statins.
Statins (3-hydroxy-3-methyl-glutaryl-CoA reductase inhibitors) are among the most widely prescribed drugs because of their ability to reduce the risk of cardiovascular disease (CVD). It has been shown recently that these agents are able to reduce CVD risk well beyond what had been expected based solely on their ability to lower serum cholesterol levels.11 Studies investigating this activity have shown that the statins activate genes that are involved in the shear stress response in vascular endothelial cells.12 These genes include endothelial nitric oxide synthase and thrombomodulin.13,14 The products of these genes mediate vascular dilation and inhibit thrombus formation and local inflammation, thereby helping prevent acute cardiovascular events. Research into the mechanisms by which statins affect the expression of these and other relevant genes has identified Kruppel-like factor 2 (KLF2) as a key mediator of this response.15 Other studies have shown that the physiologic shear stress response in vascular endothelial cells is dependent on NRF2.16 Tying these observations together, KLF2 has been shown to synergistically enhance the effects of NRF2 on target gene induction in endothelial cells.17 These results led us to hypothesize that the statins could enhance the ability of NRF2-inducing agents to stimulate γ-globin gene expression and HbF production in erythroid cells. In support of this hypothesis, KLF2 is essential for primitive murine erythropoiesis18 and it and the erythroid-specific Kruppel-like factor KLF1 have compensatory roles in murine β-globin expression.19 If our hypothesis is correct, it could lead to trials of statins alone or in combination with NRF2-activating agents in people with β-hemoglobinopathies.
Methods
Cells, reagents, and flow cytometry
K562 cells were cultured in RPMI 1640 medium with l-glutamine (Mediatech) with 10% FBS and 1% Pen/Strep. G-CSF–primed peripheral blood CD34+ cells were obtained from the hematopoietic cell processing core at the University of Washington using an institutional review board–approved protocol in accordance with the Declaration of Helsinki. We used vitro differentiation system of Sankaran et al.20 Tert-butylhydroquinone (tBHQ) and 5-azacytadine were from Sigma-Aldrich and simvastatin sodium salt was from EMD Biosciences. 5-Azacytidine (5-Aza) was dissolved in PBS. tBHQ and statin were both dissolved in DMSO. The following Abs from BD Biosciences were used for flow cytometry: CD235a-FITC (559943), anti–CD71-APC (M-A712), and matching isotype control Ab FITC Mouse IgG2Bk (555742).
RNA and hemoglobin analysis
Total RNA was isolated from cells with RNeasy columns (QIAGEN). cDNAs were generated from equal amounts of RNA using the iScript cDNA Synthesis Kit (Bio-Rad). Quantitative real-time PCR (qPCR) was performed with iQ SYBR Green Super Mix (Bio-Rad) with 2 μL of diluted cDNA. mRNA levels were calculated by the method of Larionov et al21 relative to GAPDH mRNA levels. Real-time PCR primers for mRNA quantification were: KLF2 5′-AGACCTACACCAAGAGTTCGCATC-3′ (F), 5′-ATCGCACAGATGGCACTGGAATG-3′ (R); KLF1 5′-TCAGTACCAAGGGCACTTC-3′ (F), 5′-GTTGCGGCAAGAGCTACACC-3′ (R); BCL11A XL 5′-ATGCGAGCTGTGCAACTATG-3′ (F), 5′-GTAAACGTCCTTCCCCACCT-3′ (R); Primer sequences for GAPDH, β-actin, γ-globin, β-globin, and NQO1 have been published previously.9 Hb HPLC analysis was performed as described in Ou and Rogenard22 using a PolyCAT-A cation exchange column (The Nest Group).
siRNA transfection
K562 cells were transfected with 10nM siGENOME SMART siRNA for KLF2 (M-006928-05; Dharmacon) or 10nM nontargeting control siRNA (siGENOME D-001210-02-05; Dharmacon) for 48-72 hours. HiPerfect transfection reagent (QIAGEN) was used to transfect cells according to the manufacturer's instructions.
Western blotting
For total cell lysates, pellets were lysed in cell lysis buffer (9803; Cell Signaling Technology) and combined with equal volumes of 2× Laemmli buffer supplemented with 100mM DTT, protease (11836170001; Roche), and phosphatase inhibitors (P2580; Sigma-Aldrich). Nuclear extracts were prepared, electrophoresed, and blotted as described previously.9 The following primary Abs were used: KLF2 (sc-28675; Santa Cruz Biotechnology), TATA-binding protein (ab818; Abcam), BCL11A (ab19487; Abcam), GAPDH (sc-47724; Santa Cruz Biotechnology). KLF1 Ab was a generous gift from Dr Merlin Crossley.
ChIP analysis
ChIP assays were performed in triplicate using 2 × 106 cells per experiment. The NRF2 ChIP was described previously.9 The KLF2 ChIP protocol followed the NRF2 ChIP protocol with the following changes: before cross-linking, cells were washed in PBS and resuspended at 5 × 105 cells/mL in PBS. Ethylene glycol bissuccinimidylsuccinate in dimethylformamide was added to a final concentration of 1.5mM and cells were shaken at room temperature (RT) for 30 minutes. Formaldehyde was then added to a final concentration of 1% and shaken at RT for 10 minutes. The cross-linking reaction was quenched with glycine at a final concentration of 20mM and shaken for 15 minutes at RT. Immunoprecipitations were performed with 5 μg of anti-NRF2 (sc-13032; Santa Cruz Biotechnology), 5 μg of anti-KLF2 (sc-28675; Santa Cruz Biotechnology) or 5 μg of AffiniPure rabbit anti–rat IgG (Jackson ImmunoResearch Laboratories). The immunoprecipitated DNA was quantified in triplicate by quantitative real-time PCR. Primers are in Table 1.
Quantitative real-time PCR primers for ChIP
| Gene . | Forward . | Reverse . |
|---|---|---|
| β-globin | GAGACGCAGGAAGAGATCCATCTACA | CTCTGCCCTGACTTTTATGCCCAGCC |
| γ-globin | AACGGTCCCTGGCTAAACTC | GCTGAAGGGTGCTTCCTTTT |
| HS2 | GACCTCCCATAGTCCAAGCA | CATTCTGTCTCAGGCATCCA |
| HS3 | ATAGACCATGAGTAGAGGGCAGAC | TGATCCTGAAAACATAGGAGTCAA |
| HS4 | GCCCTTTTCTCTCTCCCACTCAGC A | AGTATGACATGGAAATGATCAGGCTT |
| HS5 | GGGAAGAAGATACCTGTGGAGCTAAT | AGGGTAAGAGACAGGGAAGGAGGTG |
| NQO1 | CAGTGGCATGCACCCAGGGAA | GCATGCCCCTTTTAGCCTTGGCA |
| Necdin | GTGTTATGTGCGTGCAAACC | CTCTTCCCGGGTTTCTTCTC |
| Gene . | Forward . | Reverse . |
|---|---|---|
| β-globin | GAGACGCAGGAAGAGATCCATCTACA | CTCTGCCCTGACTTTTATGCCCAGCC |
| γ-globin | AACGGTCCCTGGCTAAACTC | GCTGAAGGGTGCTTCCTTTT |
| HS2 | GACCTCCCATAGTCCAAGCA | CATTCTGTCTCAGGCATCCA |
| HS3 | ATAGACCATGAGTAGAGGGCAGAC | TGATCCTGAAAACATAGGAGTCAA |
| HS4 | GCCCTTTTCTCTCTCCCACTCAGC A | AGTATGACATGGAAATGATCAGGCTT |
| HS5 | GGGAAGAAGATACCTGTGGAGCTAAT | AGGGTAAGAGACAGGGAAGGAGGTG |
| NQO1 | CAGTGGCATGCACCCAGGGAA | GCATGCCCCTTTTAGCCTTGGCA |
| Necdin | GTGTTATGTGCGTGCAAACC | CTCTTCCCGGGTTTCTTCTC |
Stable transfection of murine klf2 plasmid
The construct pBK-CMV/klf2 was a generous gift from Dr Jerry Lingrel.23 The control vector, pBK-CMV, was created by removing the klf2 gene using the flanking BAMHI and HINDIII sites. One × 107 K562 cells were electroporated with 10 mg of linearized plasmid and allowed to recover for 48 hours. Cells were then placed in selective medium containing 1 mg/mL of G418 for a minimum of 2 weeks before colonies were isolated. Pools of approximately 30 clones were selected and klf2 expression was confirmed by real-time PCR using primers for murine klf2.24
Statistical analysis
Results
Simvastatin induces KLF2 mRNA and protein expression in K562 cells
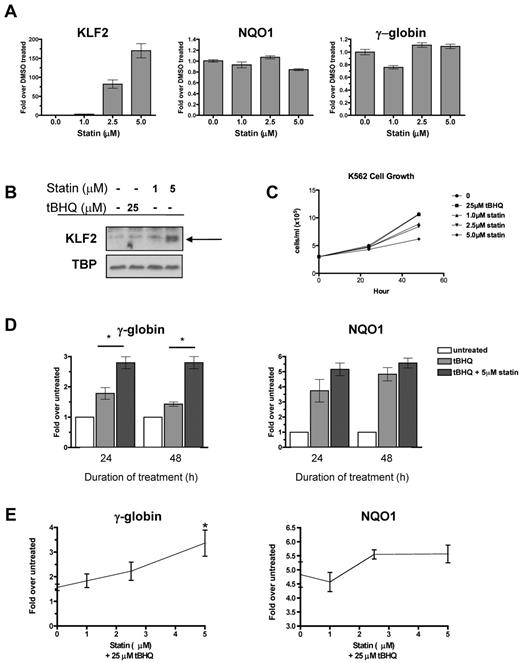
Multiple statins, including simvastatin, lovastatin, mevastatin, and cerivastatin, have been shown to induce KLF2 mRNA in endothelial cells.15,25 To begin testing our hypothesis, we first determined whether statins induce KLF2 expression in the K562 erythroleukemia cell line. We applied increasing doses of simvastatin for 24 hours and then determined the steady-state KLF2 mRNA and nuclear protein levels. Simvastatin increased KLF2 mRNA levels in a dose-dependent fashion, with the highest dose (5μM) increasing KLF2 mRNA 170-fold over the DMSO control (Figure 1A). Simvastatin had no effect on expression of the NRF2 target gene NQO1 or γ-globin mRNA levels (Figure 1A). Simvastatin also induced KLF2 nuclear protein levels, whereas treatment with the NRF2 inducer tBHQ had no effect on KLF2 protein levels (Figure 1B). K562 cell growth was not affected at 24 hours, but was suppressed by simvastatin at 48 hours (Figure 1C).
Statin increases KLF2 expression and enhances tBHQ-induced γ-globin gene expression. (A) Effect of statin on KLF2, NQO1, and γ-globin steady-state mRNA levels. K562 cells were treated with increasing doses of statin for 24 hours before RNA isolation. (B) Effect of statin on KLF2 nuclear protein levels. K562 cells were treated with 25μM tBHQ or 1 or 5μM of statin for 24 hours before nuclear protein harvest. (C) Effect of tBHQ and statin on K562 cell growth. 3.0 × 105 cells were treated with 25μM tBHQ or 1, 2.5, or 5μM statin and counted at 24 and 48 hours. (D) Effect of tBHQ and combination of tBHQ and statin treatment on γ-globin and NQO1 steady-state mRNA levels at 24 and 48 hours. Results expressed as the fold increase over untreated controls. (E) Statin dose response enhances tBHQ-induced γ-globin and NQO1 steady-state mRNA levels. K562 cells were treated with 25μM tBHQ or 25μM tBHQ and statin (1μM-5μM) for 48 hours before RNA isolation. Real-time PCR results are expressed as the fold increase over untreated controls. Error bars represent ± SEM. *P < .05 compared with tBHQ alone.
Statin increases KLF2 expression and enhances tBHQ-induced γ-globin gene expression. (A) Effect of statin on KLF2, NQO1, and γ-globin steady-state mRNA levels. K562 cells were treated with increasing doses of statin for 24 hours before RNA isolation. (B) Effect of statin on KLF2 nuclear protein levels. K562 cells were treated with 25μM tBHQ or 1 or 5μM of statin for 24 hours before nuclear protein harvest. (C) Effect of tBHQ and statin on K562 cell growth. 3.0 × 105 cells were treated with 25μM tBHQ or 1, 2.5, or 5μM statin and counted at 24 and 48 hours. (D) Effect of tBHQ and combination of tBHQ and statin treatment on γ-globin and NQO1 steady-state mRNA levels at 24 and 48 hours. Results expressed as the fold increase over untreated controls. (E) Statin dose response enhances tBHQ-induced γ-globin and NQO1 steady-state mRNA levels. K562 cells were treated with 25μM tBHQ or 25μM tBHQ and statin (1μM-5μM) for 48 hours before RNA isolation. Real-time PCR results are expressed as the fold increase over untreated controls. Error bars represent ± SEM. *P < .05 compared with tBHQ alone.
Simvastatin enhances induction of γ-globin gene expression by tBHQ
tBHQ is a well-studied activator of NRF2 signaling. We have shown previously that tBHQ increases γ-globin gene expression in K562 and primary human erythroid cells.9 As a next step in testing our hypothesis that pharmacologic induction of KLF2 will enhance NRF2-mediated induction of γ-globin gene expression, we treated K562 cells with 25μM tBHQ and with a combination of tBHQ and 5μM simvastatin (Figure 1D). These doses were chosen because both result in consistent induction of target genes NQO1 (tBHQ) and KLF2 (simvastatin). When used alone, tBHQ produced a 1.8-fold induction of γ-globin mRNA at 24 hours. This declined to 1.4-fold over the untreated control at 48 hours. The addition of simvastatin to tBHQ produced a statistically significant increase (P < .05 compared with tBHQ alone) in γ-globin mRNA to 2.8-fold over control, which was maintained for 48 hours. We also examined the ability of simvastatin cotreatment to enhance induction of the NRF2 target gene NQO1 (Figure 1D). Simvastatin treatment also increased NQO1 mRNA, although to a lesser extent than was seen with γ-globin. Dose-response experiments (Figure 1E) demonstrated that the ability of simvastatin to enhance tBHQ induction of both γ-globin and NQO1 is dose dependent.
KLF2 is required for and substantially contributes to simvastatin enhancement of tBHQ-induced γ-globin expression
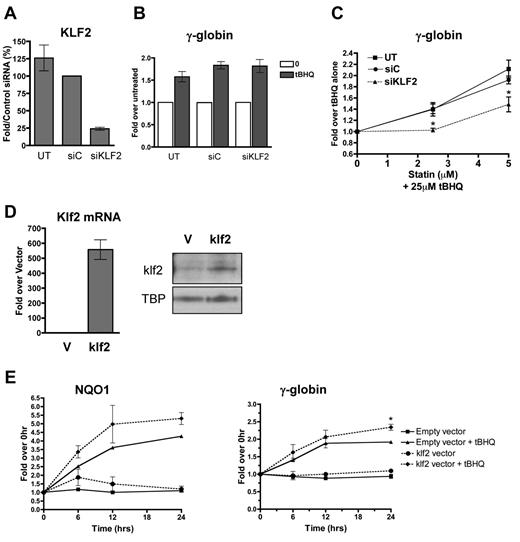
To determine whether the enhancement of NRF2 induction of γ-globin gene expression by simvastatin was because of its ability to induce KLF2 expression, we first used siRNA to reduce the levels of KLF2 in K562 cells. Cells were transfected with control siRNA or siRNA specific for KLF2 mRNA. Twenty-four hours after transfection, cells were treated with tBHQ alone or in combination with increasing concentrations of simvastatin. KLF2 siRNA reduced KLF2 mRNA levels to approximately 25% of those seen with the control siRNA (Figure 2A). This reduction in KLF2 mRNA had no effect on tBHQ induction of γ-globin mRNA (Figure 2B). However, the KLF2 knockdown significantly (P < .05) decreased the ability of simvastatin to enhance tBHQ-induced γ-globin expression at both the 2.5 and 5μM simvastatin doses (Figure 2C). The data in this figure are expressed as the fold increase over tBHQ treatment to emphasize the effect that simvastatin has on tBHQ-induced γ-globin gene expression.
KLF2 is essential for and contributes significantly to statin enhancement of tBHQ-induced γ-globin gene expression. K562 cells were transiently transfected with siRNA specific for KLF2. Twenty-four hours later, cells were treated with tBHQ or tBHQ and simvastatin (statin) for 48 hours before RNA isolation. Mock transfection (UT) and control siRNA (siC) were used as controls. (A) qPCR results measuring KLF2 steady-state mRNA levels 24 hours after siRNA transfection presented as the percentage relative to siC. (B) Effect of KLF2 siRNA on 25μM tBHQ-induced γ-globin mRNA levels. Results are expressed as the fold increase over untreated controls to emphasize that KLF2 knock-down had no effect on tBHQ induction of γ-globin. (C) Effect of KLF2 siRNA on statin-enhanced tBHQ-induced γ-globin mRNA levels. Results are expressed as the fold increase over tBHQ alone to emphasize the fold induction-dependent on the addition of statin. Error bars represent ± SEM. *P < .05 compared with siC at 2.5 or 5μM statin (n = 3). (D-E) klf2 overexpression significantly enhances tBHQ-induced γ-globin gene expression. (D) Real-time analysis of mRNA levels and Western blot analysis of nuclear extracts of empty vector (V) used as control and cells stably overexpressing murine klf2. TBP was used as a nuclear loading control. (E) Effect of overexpressing klf2 on tBHQ induction of V and γ-globin mRNA. Cells overexpressing V or klf2 were treated with tBHQ and harvested for RNA at 6, 12, and 24 hours after treatment. Results of real-time PCR are presented as the relative mRNA expression compared with 0-hour controls. Error bars represent ± SEM. *P < .05 (n = 2).
KLF2 is essential for and contributes significantly to statin enhancement of tBHQ-induced γ-globin gene expression. K562 cells were transiently transfected with siRNA specific for KLF2. Twenty-four hours later, cells were treated with tBHQ or tBHQ and simvastatin (statin) for 48 hours before RNA isolation. Mock transfection (UT) and control siRNA (siC) were used as controls. (A) qPCR results measuring KLF2 steady-state mRNA levels 24 hours after siRNA transfection presented as the percentage relative to siC. (B) Effect of KLF2 siRNA on 25μM tBHQ-induced γ-globin mRNA levels. Results are expressed as the fold increase over untreated controls to emphasize that KLF2 knock-down had no effect on tBHQ induction of γ-globin. (C) Effect of KLF2 siRNA on statin-enhanced tBHQ-induced γ-globin mRNA levels. Results are expressed as the fold increase over tBHQ alone to emphasize the fold induction-dependent on the addition of statin. Error bars represent ± SEM. *P < .05 compared with siC at 2.5 or 5μM statin (n = 3). (D-E) klf2 overexpression significantly enhances tBHQ-induced γ-globin gene expression. (D) Real-time analysis of mRNA levels and Western blot analysis of nuclear extracts of empty vector (V) used as control and cells stably overexpressing murine klf2. TBP was used as a nuclear loading control. (E) Effect of overexpressing klf2 on tBHQ induction of V and γ-globin mRNA. Cells overexpressing V or klf2 were treated with tBHQ and harvested for RNA at 6, 12, and 24 hours after treatment. Results of real-time PCR are presented as the relative mRNA expression compared with 0-hour controls. Error bars represent ± SEM. *P < .05 (n = 2).
To further investigate the role of KLF2 in simvastatin enhancement of tBHQ-induced γ-globin gene expression, we next created a K562 cell line that stably overexpresses murine Klf2. Cells transfected with an empty vector served as the negative control. As shown in Figure 2D, murine Klf2 was overexpressed at both the mRNA and protein levels. Treating the Klf2-overexpressing cells with 25μM tBHQ increased both NQO1 and γ-globin mRNA levels compared with control cells stably transfected with the empty vector (Figure 2E). For γ-globin, overexpression of Klf2 resulted in a 2.4-fold enhancement of tBHQ induction compared with 2.0-fold induction with the control cell line at 24 hours (P < .05) (Figure 2E). These results support our model that simvastatin is able to enhance tBHQ-mediated NRF2 induction of γ-globin gene expression by increasing KLF2 levels in erythroid cells.
tBHQ and simvastatin treatments induce binding of NRF2 and KLF2 to HS2 of the β-globin LCR
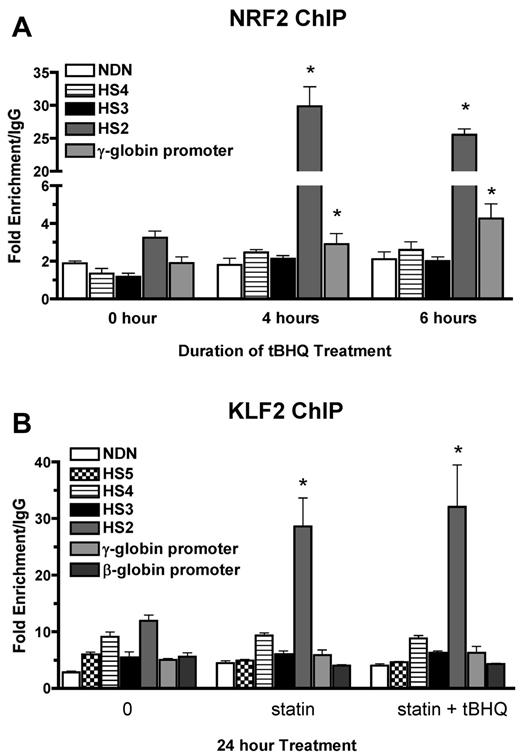
We have shown previously that tBHQ induces binding of NRF2 to a 7/7-bp antioxidant response element consensus sequence in the proximal γ-globin gene promoter in K562 and differentiating human primary erythroid cells.9 The NRF2 gene was originally cloned based on in vitro binding to the core enhancer region of HS2 of the β-globin locus control region (LCR) in K562 cells.26 To determine whether tBHQ also increases NRF2 binding to HS2 in K562 cells, we performed the ChIP assay after treatment with 25μM tBHQ. This treatment increased NRF2 binding to HS2 of the LCR from 3-fold (untreated) to 30-fold over the nonspecific IgG control after 4 hours of tBHQ treatment (Figure 3A). Immunoprecipitation of the negative control necdin (NDN) gene promoter was not increased. We also observed the expected enhanced binding to the γ-globin promoter. NRF2 binding to HS3 and HS4 was not increased with tBHQ treatment.
NRF2 and KLF2 bind HS2 of β-globin LCR in K562 cells. (A) NRF2 ChIP analysis of K562 cells treated with 25μM tBHQ for 0, 4, and 6 hours. (B) KLF2 ChIP analysis of K562 cells treated with 5μM statin or 5μM statin + 25μM tBHQ for 24 hours. qPCR was performed on immunoprecipitated DNA using primers that amplify HS5, HS4, HS3, HS2, γ-globin, and the β-globin promoter. The necdin promoter was used as a negative control and the NQO1 promoter was used as a positive control for the NRF2 ChIP. Results are expressed as the fold enrichment over IgG. Error bars represent ± SEM. *P < .05 compared with 0 hours or 0μM (n = 2).
NRF2 and KLF2 bind HS2 of β-globin LCR in K562 cells. (A) NRF2 ChIP analysis of K562 cells treated with 25μM tBHQ for 0, 4, and 6 hours. (B) KLF2 ChIP analysis of K562 cells treated with 5μM statin or 5μM statin + 25μM tBHQ for 24 hours. qPCR was performed on immunoprecipitated DNA using primers that amplify HS5, HS4, HS3, HS2, γ-globin, and the β-globin promoter. The necdin promoter was used as a negative control and the NQO1 promoter was used as a positive control for the NRF2 ChIP. Results are expressed as the fold enrichment over IgG. Error bars represent ± SEM. *P < .05 compared with 0 hours or 0μM (n = 2).
KLF2 has been shown to bind to HS2 and HS3 of the human β-globin LCR, as well as to the γ-globin promoters in mouse embryonic blood cells.27 To determine whether the increase in KLF2 levels by simvastatin treatment led to increased KLF2 binding to any of these regions in K562 cells, we performed ChIP assays after treatment with simvastatin alone or simvastatin plus tBHQ for 24 hours. Under basal conditions, KLF2 is enriched by approximately 10-fold compared with IgG at HS2 (Figure 3B). Treatment with simvastatin alone or in combination with tBHQ increased KLF2 enrichment by approximately 3-fold over baseline. Simvastatin treatment did not increase KLF2 binding to the other LCR HS's, the γ-globin promoter, the β-globin promoter, or the promoter of the negative control necdin gene. These results demonstrate that NRF2 and KLF2 both bind to HS2 of the β-globin LCR in K562 cells and that this binding is significantly increased after pharmacologic induction of the 2 proteins, suggesting a role for HS2 in the induction of γ-globin gene expression by both simvastatin and tBHQ in K562 cells.
Simvastatin and tBHQ additively increase HbF in differentiating primary human erythroid cells
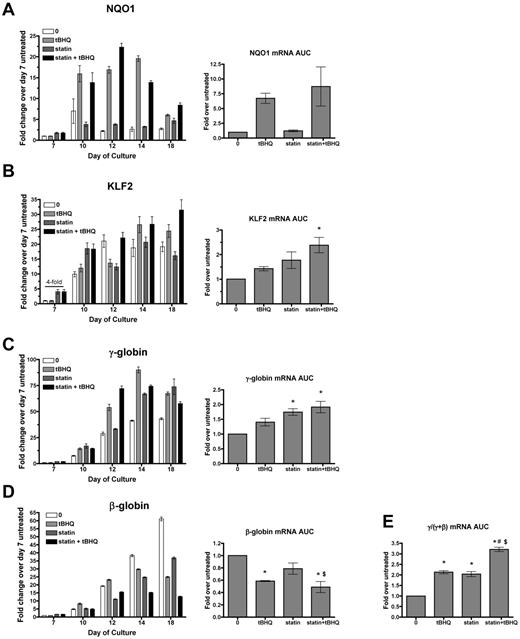
To determine whether simvastatin can augment the ability of tBHQ to increase γ-globin mRNA production and HbF levels in primary human erythroid cells, we used a previously described 2-phase in vitro erythroid differentiation system.20 G-CSF–mobilized CD34+ peripheral blood cells from normal donors were expanded for 7 days in medium containing FLT-3 ligand, SCF, IL-3, and IL-6. On day 7, cytokines were changed to erythropoietin, IL-3, SCF, and β-estradiol for erythroid differentiation through day 20. As previously reported, tBHQ was added every other day starting on day 9.9 We determined that the simvastatin half-life in PBS at 37°C is approximately 9 days. For the experiments discussed below, the drug was therefore added only once during each experiment on day 5. Preliminary experiments showed that adding simvastatin on this day produced maximal γ-globin induction and that adding the drug twice during differentiation did not change the results (data not shown). For each gene studied, data are first presented from a single representative experiment at multiple time points during the erythroid phase of differentiation so that temporal patterns of gene expression can be seen (Figure 4). Expression is also presented as the average area under the curve (AUC) for each gene from 3 independent experiments. Rather than comparing mRNA levels at a single time point for each experimental condition, this allows quantification of the total mRNA from each gene available for translation into protein over the full course of erythroid differentiation.
Simvastatin and tBHQ increase γ-globin mRNA and decrease β-globin mRNA in primary human erythroid cells. Human CD34+ peripheral blood cells were treated with simvastatin (statin) and tBHQ during in vitro erythroid differentiation. The effects of 5μM tBHQ, 5μM statin, and a combination of statin and tBHQ on gene expression were determined for NQO1 (A), KLF2 (B), γ-globin (C), and β-globin (D). For each gene studied, data are first presented from a single representative experiment at multiple time points during the erythroid phase of differentiation relative to untreated sample on day 7. Data from 3 independent experiments are also presented as the mean AUC for each gene. This data representation allows quantification of the total mRNA from each gene available for translation into protein over the full course of erythroid differentiation. (E) AUC for γ/(γ + β) mRNA. Error bars represent ± SEM. *P < .05 compared with untreated; #P < .05 compared with tBHQ; $P < .05 compared with statin (n = 3).
Simvastatin and tBHQ increase γ-globin mRNA and decrease β-globin mRNA in primary human erythroid cells. Human CD34+ peripheral blood cells were treated with simvastatin (statin) and tBHQ during in vitro erythroid differentiation. The effects of 5μM tBHQ, 5μM statin, and a combination of statin and tBHQ on gene expression were determined for NQO1 (A), KLF2 (B), γ-globin (C), and β-globin (D). For each gene studied, data are first presented from a single representative experiment at multiple time points during the erythroid phase of differentiation relative to untreated sample on day 7. Data from 3 independent experiments are also presented as the mean AUC for each gene. This data representation allows quantification of the total mRNA from each gene available for translation into protein over the full course of erythroid differentiation. (E) AUC for γ/(γ + β) mRNA. Error bars represent ± SEM. *P < .05 compared with untreated; #P < .05 compared with tBHQ; $P < .05 compared with statin (n = 3).
We first examined mRNA levels of the NRF2 target gene NQO1 to assess the activation of NRF2 antioxidant response signaling during differentiation. In the untreated controls, NQO1 mRNA levels remained low throughout erythroid differentiation. As expected, tBHQ alone or in combination with simvastatin increased NQO1 mRNA, whereas simvastatin alone did not. We next examined the effects of drug treatments on KLF2 mRNA. In untreated cells, KLF2 mRNA was initially low (day 7), but increased by approximately 20-fold during erythroid differentiation and reached a peak on days 12-18 (Figure 4B). With simvastatin alone or in combination with tBHQ, we found that KLF2 mRNA was increased on day 7 (4-fold over untreated control). However, over the full course of differentiation, there was only a nonsignificant trend of increased KLF2 mRNA AUC with either drug alone and a 2-fold increase when the drugs were combined. This contrasts with the 170-fold induction of KLF2 mRNA levels observed with simvastatin treatment of K562 cells (Figure 1A). We next determined the effects of drug treatments on γ-globin and β-globin mRNA levels throughout differentiation (Figure 4C-D). As expected from our previous results, over the course of differentiation, γ-globin mRNA was increased with tBHQ alone (1.4-fold) and with the combination of tBHQ + simvastatin (1.9-fold). However, again in contrast to results from K562 cells, γ-globin mRNA was significantly increased by simvastatin treatment alone (1.7-fold, P < .05). We also observed that all drug treatments decreased β-globin mRNA levels, with simvastatin lowering expression to 79% of untreated control, tBHQ to 51% of control and with tBHQ + simvastatin leading to the largest suppression to 41% of control. The greatest effect of the treatments was on the ratio of γ-globin mRNA to γ-globin + β-globin mRNA during differentiation. Both tBHQ and simvastatin increased the ratio to a similar degree (a 2.1- and 2.0-fold increase, respectively), whereas the combination of the 2 drugs increased the γ/(γ + β) ratio to higher levels than either drug alone at 3.2-fold over control (P < .001 vs untreated control and P < .01 vs tBHQ and simvastatin alone).
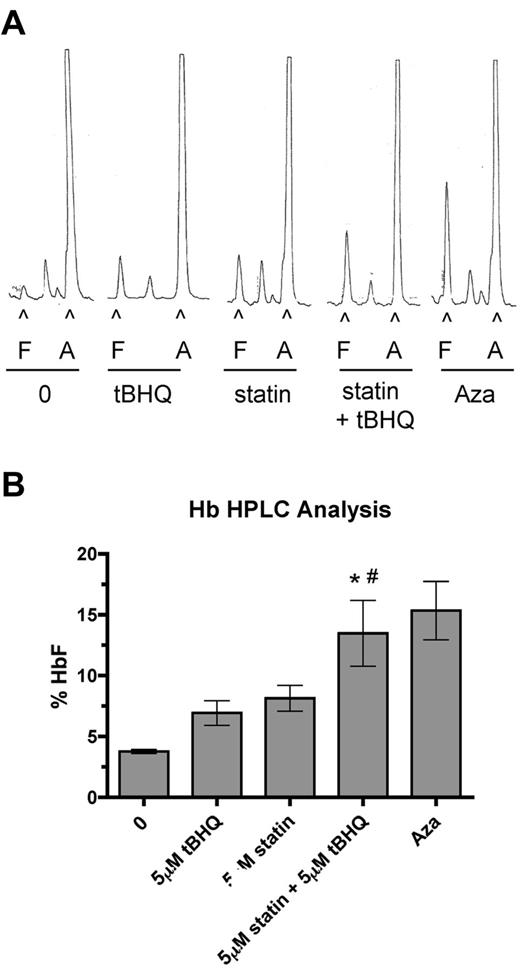
To determine the effect of drug treatments on HbF levels in primary human erythroid cells, we performed HPLC analysis on cell lysates at day 20 of differentiation. HPLC traces from a representative experiment are shown in Figure 5A. In these experiments 5-Aza was included as a positive control for HbF induction. Figure 5B shows the mean HbF levels from 3 separate experiments. The percentage of HbF was increased from a mean of 3.6% in untreated cells to 6.9% with tBHQ alone (3.3% > control), 8.1% with simvastatin alone (4.5% > control), and to 13.5% (9.9% > control) when the drugs were used together. This level of HbF induction was equivalent to that seen with 5-Aza, and the increase in the percentage of HbF with the 2 drugs combined was greater than the sum of the individual treatments. Among experiments using cells from 3 different donors, we observed a range of HbF concentrations for the combination drug treatment of 10.2%-18.6%.
Combination of simvastatin and tBHQ increases the proportion of HbF in primary human erythroid cells more than either drug alone. Human CD34+ peripheral blood cells were treated with 5μM tBHQ, 5μM statin, and a combination of statin + tBHQ during in vitro erythroid differentiation. At the end of differentiation on day 20, cells were lysed and the proportions of HbA (A) and HbF (F) were determined by ion-exchange HPLC. (A) Representative HPLC traces. (B) Quantitation of HPLC traces from 3 independent experiments (2 different donors). Error bars represent ± SEM. *P < .05 compared with untreated; #P < .05 compared with tBHQ.
Combination of simvastatin and tBHQ increases the proportion of HbF in primary human erythroid cells more than either drug alone. Human CD34+ peripheral blood cells were treated with 5μM tBHQ, 5μM statin, and a combination of statin + tBHQ during in vitro erythroid differentiation. At the end of differentiation on day 20, cells were lysed and the proportions of HbA (A) and HbF (F) were determined by ion-exchange HPLC. (A) Representative HPLC traces. (B) Quantitation of HPLC traces from 3 independent experiments (2 different donors). Error bars represent ± SEM. *P < .05 compared with untreated; #P < .05 compared with tBHQ.
Analysis of NRF2 and KLF2 binding to β-globin locus regulatory elements
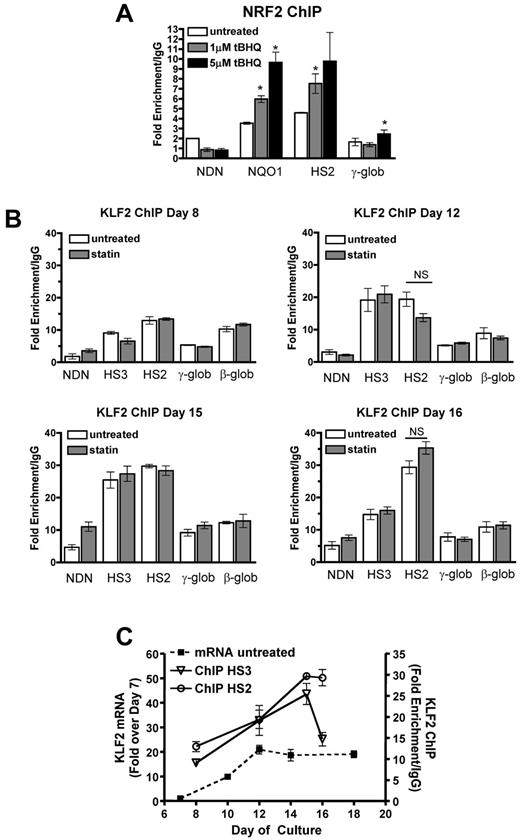
To further investigate the mechanism of tBHQ and simvastatin induction of HbF, we determined whether drug treatments increased NRF2 and KLF2 binding to HS2 of the β-globin LCR, as we had observed in K562 cells. ChIP assays using differentiating primary human erythroid cells showed that both 1 and 5μM tBHQ treatments strongly increased NRF2 binding to the positive control NQO1 promoter and to LCR HS2 (Figure 6A). We next investigated whether KLF2 binding to HS2 increased with simvastatin treatment (Figure 6B). For these experiments, we performed ChIP assays at several time points during erythroid differentiation. In the untreated control cells, binding of KLF2 was detectable at HS2, HS3, and the β-globin promoter as early as day 8, with the highest levels at HS2. The necdin gene promoter served as a negative control. On days 12 and 15, a further increase in KLF2 binding was seen at HS3 and HS2. On day 16, KLF2 binding to HS3 had declined, whereas it remained high at HS2. Again, in contrast to our results with K562 cells, on none of the days and at none of the sites we evaluated was KLF2 binding significantly increased by 5μM simvastatin, the dose that produced maximal induction of HbF. Results for untreated control cells are summarized in Figure 6C, where it can be seen that KLF2 mRNA levels and binding of KLF2 to HS2 and HS3 increased in parallel during erythroid differentiation. To our knowledge, this is the first characterization of KLF2 gene expression and DNA binding during differentiation of primary human erythroid cells.
NRF2 and KLF2 bind the β-globin LCR in differentiating primary human erythroid cells. (A) NRF2 ChIP analysis of day 13 CD34+ cells during erythroid differentiation. Cells were treated with 1 and 5μM tBHQ on days 9, 11, and 13. (B) KLF2 ChIP of CD34+ cells treated with 5μM statin on day 5. Cells were harvested for cross-linking and ChIP analysis on days 8, 12, 15, and 16 of differentiation. qPCR was performed on immunoprecipitated DNA using primers that amplify HS3, HS2, γ-globin, and the β-globin promoter. The necdin promoter was used as a negative control and the NQO1 promoter was used as a positive control for the NRF2 ChIP. Results are expressed as the fold enrichment over IgG. (C) Comparison of KLF2 mRNA levels and KLF2 protein bound to HS3 and HS2. The data are presented on the same graph to illustrate KLF2 binding at specific points during differentiation compared with KLF2 gene expression. Error bars represent ± SEM. *P < .05 compared with 0μM concentration of drug. NS indicates not significant by t test (P < .05, n = 2).
NRF2 and KLF2 bind the β-globin LCR in differentiating primary human erythroid cells. (A) NRF2 ChIP analysis of day 13 CD34+ cells during erythroid differentiation. Cells were treated with 1 and 5μM tBHQ on days 9, 11, and 13. (B) KLF2 ChIP of CD34+ cells treated with 5μM statin on day 5. Cells were harvested for cross-linking and ChIP analysis on days 8, 12, 15, and 16 of differentiation. qPCR was performed on immunoprecipitated DNA using primers that amplify HS3, HS2, γ-globin, and the β-globin promoter. The necdin promoter was used as a negative control and the NQO1 promoter was used as a positive control for the NRF2 ChIP. Results are expressed as the fold enrichment over IgG. (C) Comparison of KLF2 mRNA levels and KLF2 protein bound to HS3 and HS2. The data are presented on the same graph to illustrate KLF2 binding at specific points during differentiation compared with KLF2 gene expression. Error bars represent ± SEM. *P < .05 compared with 0μM concentration of drug. NS indicates not significant by t test (P < .05, n = 2).
tBHQ and simvastatin suppress KLF1 and BCL11A gene expression and protein levels.
Our primary cell experiments revealed both similarities to and important differences from our results with K562 cells and from our proposed model for simvastatin induction of HbF. tBHQ treatment of primary cells induced NRF2 signaling (demonstrated by induction of NQO1 gene expression), increased γ-globin mRNA levels, increased the percentage of HbF, and strongly enhanced binding of NRF2 to HS2 of the LCR. These results are consistent with our K562 cell data and our model for NRF2-activating agents. In addition, cotreatment of primary cells with tBHQ + simvastatin did increase HbF over what is seen with tBHQ alone. However, in primary erythroid cells, simvastatin did not significantly increase KLF2 gene expression or binding to globin locus regulatory elements but did increase HbF to levels equivalent to those seen with tBHQ when used alone. Another unexpected finding that was not predicted by either of our models was that both tBHQ and simvastatin strongly suppressed β-globin mRNA levels during erythroid differentiation. These results suggested that an alternative mechanism of HbF induction by simvastatin and an additional mechanism of action for tBHQ may be working in primary erythroid cells.
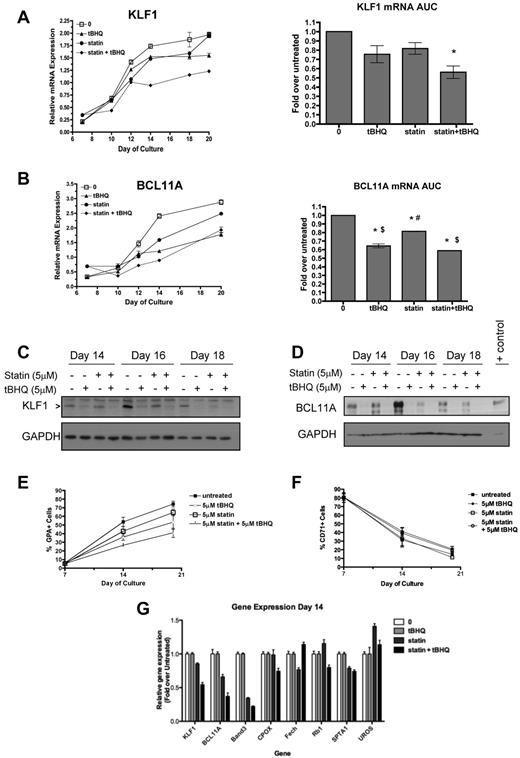
Our observations that both drugs suppress β-globin mRNA levels and increase HbF in primary cells led us to consider similar findings that have been reported recently for people who are haploinsufficient for KLF1.28 In these subjects, decreased KLF1 levels led directly to decreased β-globin and BCL11A gene expression. Decreased BCL11A protein levels in turn resulted in partial derepression of γ-globin gene expression. These changes produced a hereditary persistence of HbF (HPFH) phenotype.29 To determine whether tBHQ and simvastatin might be working through a similar mechanism, we evaluated the effects of tBHQ and simvastatin, alone or in combination, on KLF1 and BCL11A gene expression in our differentiation model. KLF1 mRNA, which normally increases during differentiation, was suppressed by either drug alone and to a greater extent by the combination of tBHQ and simvastatin (Figure 7A). AUC data showed that the combination of simvastatin and tBHQ resulted in a decrease in KLF1 mRNA to 56% of the level in untreated control cells over the course of differentiation (P < .05). Similar results were seen for BCL11A expression, as the drugs in combination suppressed mRNA levels to 59% of the untreated control level (P < .001; Figure 7B). Western blotting was performed to determine whether drug treatments were also associated with decreased levels of KLF1 and BCL11A protein. As shown in Figure 7C and D, all drug treatments reduced KLF1 and BCL11A protein on days 14, 16, and 18 compared with the untreated control.
Statin and tBHQ decrease KLF1 and BCL11A in primary human erythroid cells. Human CD34+ peripheral blood cells were treated with statin and tBHQ during in vitro erythroid differentiation. For each drug treatment, gene expression on days 7-20 and mRNA expressed as AUC are shown for each gene (combined data from 2 independent experiments and 2 different donors). (A) KLF1 gene expression and (B) BCL11a gene expression. (C-D) Effect of tBHQ, simvastatin, and the combination on KLF1 and BCL11A protein expression on days 14, 16, and 18 of differentiation. KLF1 and BCL11A were separated using a 15% and 10% SDS-polyacrylamide gel, respectively. Cell lysate from an adult CLL patient was used as a positive control for BCL11A XL. The percentage of GPA+ cells (E) and CD71+ cells (F) throughout erythroid differentiation are shown. (G) Effect of drug treatment on expression of genes that normally increase or decrease during erythroid differentiation. Error bars represent ± SEM. *P < .05 compared with untreated; #P < .05 compared with tBHQ; $P < .05 compared with statin.
Statin and tBHQ decrease KLF1 and BCL11A in primary human erythroid cells. Human CD34+ peripheral blood cells were treated with statin and tBHQ during in vitro erythroid differentiation. For each drug treatment, gene expression on days 7-20 and mRNA expressed as AUC are shown for each gene (combined data from 2 independent experiments and 2 different donors). (A) KLF1 gene expression and (B) BCL11a gene expression. (C-D) Effect of tBHQ, simvastatin, and the combination on KLF1 and BCL11A protein expression on days 14, 16, and 18 of differentiation. KLF1 and BCL11A were separated using a 15% and 10% SDS-polyacrylamide gel, respectively. Cell lysate from an adult CLL patient was used as a positive control for BCL11A XL. The percentage of GPA+ cells (E) and CD71+ cells (F) throughout erythroid differentiation are shown. (G) Effect of drug treatment on expression of genes that normally increase or decrease during erythroid differentiation. Error bars represent ± SEM. *P < .05 compared with untreated; #P < .05 compared with tBHQ; $P < .05 compared with statin.
Because KLF1 is not only a key regulator of γ- and β-globin gene expression, but is also required for normal erythroid differentiation and the expression of a wide array of erythroid-specific genes,30 we determined the effect of our drug treatments on differentiation as judged by cell-surface expression of glycophorin A (GPA), a target gene of KLF1. As shown in Figure 7E, GPA levels are decreased by drug treatments. This finding suggested 2 possible mechanisms as to how simvastatin and tBHQ cause suppression of KLF1 and BCL11A and changes in γ- and β-globin gene expression. First, the drugs might cause a generalized slowing of differentiation such that cells spend a longer period of time in an earlier phase of differentiation when expression of KLF1, BCL11A, and β-globin genes is lower and γ-globin mRNA is higher. The second possibility is that the drugs have a more specific effect on KLF1 gene expression that alters expression of its direct and indirect downstream target genes (including BCL11A and γ- and β-globin genes). In the first case, any genes or proteins that are up- or down-regulated during erythroid differentiation, regardless of whether they are downstream targets of KLF1, should be affected by our drug treatments. In the second case, genes or proteins that are downstream of KLF1 should be preferentially affected by the treatments, whereas genes in which the expression changes with differentiation but is not dependent on KLF1 should not. To test these models, we first determined the effects of drug treatments on cell-surface expression of the transferrin receptor CD71, which decreases during normal erythroid differentiation. As shown in Figure 7F, and in contrast to GPA, there was minimal change in CD71 levels with the drug treatments. Next, we examined expression of a panel of genes in which mRNA levels normally increase during erythroid differentiation31 but that are not affected by KLF1 knockout. These genes included CPOX,32 FECH,33 RB1,34 SPTA1,35 and UROS.33 Measurements were made on day 14 of differentiation. The erythrocyte Band 3 gene (SLC4A1) was included as a positive control gene that is directly regulated by KLF1.35 As shown in Figure 7G, KLF1, BCL11A, and Band 3 mRNA levels were all decreased by statin alone and even more so by statin + tBHQ. In contrast, mRNA levels for CPOX, FECH, RB1, SPTA1, and UROS were not consistently affected by drug treatments. These results suggest that a generalized slowing of differentiation is not the major mechanism by which tBHQ and simvastatin induce HbF.
Discussion
An estimated 99% of people born with hemoglobinopathies reside in nonindustrialized countries36 where few services exist for the diagnosis, management, or treatment of these diseases.37 Based on the success of programs for treating HIV and tuberculosis infections in the same or similar populations, our major goal has been to develop novel strategies for the pharmacologic induction of HbF that are effective, affordable, and do not require sophisticated medical services for their delivery. These considerations have led us to focus on drugs that activate the NRF2 antioxidant response and, in the present study, the potential use of statins to enhance HbF levels achieved by NRF2 activators. Both classes of drugs include members that are already FDA-approved or are in clinical trials for other indications.
Recent reports from the CVD literature show that NRF2 activity can be enhanced by KLF2, and that these 2 transcription factors synergistically induce genes that protect against cardiovascular disease.17 Therefore, we hypothesized that when combined with NRF2 activators, induction of KLF2 by statins would increase γ-globin gene expression and HbF production in erythroid cells. Our results from K562 cell experiments support this model, because simvastatin increased KLF2 mRNA and protein levels and KLF2 binding to the LCR but did not increase γ-globin gene expression. When used in combination with tBHQ, simvastatin enhanced tBHQ-induced γ-globin mRNA levels. However, our results with primary cells yielded several different results. In contrast to K562 cell experiments and our proposed model for statin action, in primary erythroid cells, simvastatin did not significantly increase KLF2 gene expression or binding to globin locus regulatory elements. However, simvastatin alone did increase HbF to levels equivalent to those seen with tBHQ when used alone. Another finding not predicted by our model was that both tBHQ and simvastatin independently suppressed β-globin mRNA levels during erythroid differentiation.
A potential explanation for the differences observed between K562 and primary cells is that K562 cells have low basal KLF2 mRNA levels (as seen in many cancer cell lines38 ), whereas KLF2 gene expression substantially increases during erythroid differentiation in primary cells. The only time we found increased KLF2 mRNA with statin treatment in primary cells was a 4-fold increase at the earliest time point, before KLF2 had begun the approximately 20-fold increase seen in untreated cells during differentiation. Simvastatin may be unable to increase KLF2 gene expression beyond that which occurs during normal differentiation in this setting.
Another major difference between K562 and primary cells was that simvastatin alone increased γ-globin mRNA and HbF production in primary cells but not in K562 cells. The facts that this induction occurred without increased KLF2, that it did not require tBHQ cotreatment, and that it was associated with decreased β-globin mRNA all suggested a different mechanism of action from our original model. These discrepancies emphasize that K562 cells may not be an appropriate model system for addressing some experimental questions.
Our observations that both simvastatin and tBHQ increased γ-globin mRNA, decreased β-globin mRNA, and increased HbF led us to test the alternative hypothesis that KLF1 and/or BCL11A gene expression might be suppressed by statin and tBHQ treatments in primary cells. Similar to what is seen in people haploinsufficient for KLF1, who also exhibit decreased β-globin mRNA and increased γ-globin mRNA and HbF,28 we found that drug treatments suppressed the levels of KLF1 and BCL11A mRNA and protein. These data suggest a model in which increased HbF is a result of increased γ-globin gene expression because of decreased BCL11A that, along with suppressed β-globin expression, is caused by decreased KLF1. An alternative explanation is that our observations could be the result of a generalized slowing of differentiation. To distinguish between these 2 models, we compared expression of genes known to be downstream of KLF1 (BCL11A, GPA, β-globin, γ-globin, and Band 3) with a panel of genes or proteins that decrease (CD71) or increase (CPOX, FECH, RB1, SPTA1, and UROS) during erythroid differentiation but are not affected by KLF1 knockout. Our results show little or no consistent effect of the drug treatments on KLF1 independent genes, suggesting a specific effect on the KLF1-BCL11A axis. However, even under this model it would not be surprising to see some effect of the drugs on erythroid differentiation, because Klf1-null mice exhibit altered erythroid differentiation based on decreased expression of the cell-cycle regulator E2f2, a direct KLF1 target gene.39
The discovery that the transcription factors BCL11A and KLF1 are master regulators of human fetal-to-adult β-like globin switching has given rise to speculation that they could be targets for drugs that induce HbF through inhibition of their activities.29,40 Our results suggest that tBHQ and simvastatin are potential lead compounds that can achieve partial reversal of the fetal-to-adult β-like globin gene switch by simultaneously suppressing KLF1 and BCL11A protein levels when used alone and to a greater extent in combination. Recently, simvastatin has been tested in a short-term study in SCD patients.41 The drug was well tolerated and improved vascular function, as evidenced by modulation of vascular biomarkers.41 Most importantly, this study has identified statins as a novel class of HbF-inducing agents and has shown that the combination of 2 FDA-approved compounds, tBHQ and simvastatin, produce an at least additive induction of HbF. Our results suggest that NRF2-activating agents and statins, alone or in combination, hold promise for increasing levels of HbF in people with β-thalassemia and SCD.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Drs Jerry Lingrel and Merlin Crossley for the generous gifts of reagents; Dr Lionel Lewis and Bernie Beaulieu for assistance with Hb HPLC; Katelyn Byrne for flow cytometry assistance; and Dr David Bodine and Cynthia Hahn for valuable discussions.
This work was supported by the National Institutes of Health (grant HL73442 to C.H.L.) and the Knights of the York Cross of Honour, a Masonic charitable organization. E.R.M. was supported by the National Cancer Institute (training grant T32 CA009658-19).
National Institutes of Health
Authorship
Contribution: E.R.M, E.K.S., and R.J.W. designed, performed, and analyzed the experiments and wrote the manuscript; and C.H.L. designed and oversaw the project, analyzed the experiments, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Christopher H. Lowrey, MD, Section of Hematology/Oncology, Dartmouth-Hitchcock Medical Center, One Medical Center Dr, Lebanon, NH 03756; e-mail: c.lowrey@dartmouth.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal