Key Points
PBX3 is a critical co-factor of HOXA9 in AMLs, particularly those carrying MLL rearrangements.
Targeting HOXA9/PBX3 interaction holds a therapeutic potential to treat leukemia.
Abstract
Although PBX proteins are known to increase DNA-binding/transcriptional activity of HOX proteins through their direct binding, the functional importance of their interaction in leukemogenesis is unclear. We recently reported that overexpression of a 4-homeobox-gene signature (ie, PBX3/HOXA7/HOXA9/HOXA11) is an independent predictor of poor survival in patients with cytogenetically abnormal acute myeloid leukemia (CA-AML). Here we show that it is PBX3, but not PBX1 or PBX2, that is consistently coexpressed with HOXA9 in various subtypes of CA-AML, particularly MLL-rearranged AML, and thus appears as a potential pathologic cofactor of HOXA9 in CA-AML. We then show that depletion of endogenous Pbx3 expression by shRNA significantly inhibits MLL-fusion–mediated cell transformation, and coexpressed PBX3 exhibits a significantly synergistic effect with HOXA9 in promoting cell transformation in vitro and leukemogenesis in vivo. Furthermore, as a proof of concept, we show that a small peptide, namely HXR9, which was developed to specifically disrupt the interactions between HOX and PBX proteins, can selectively kill leukemic cells with overexpression of HOXA/PBX3 genes. Collectively, our data suggest that PBX3 is a critical cofactor of HOXA9 in leukemogenesis, and targeting their interaction is a feasible strategy to treat presently therapy resistant CA-AML (eg, MLL-rearranged leukemia) in which HOXA/PBX3 genes are overexpressed.
Introduction
Homeobox genes are highly conserved in mammals and are crucial in regulating cell differentiation and proliferation. There are 4 (ie, A, B, C, and D) clusters which include 39 individual homeotic or Hox genes in mammals.1,2 Hox proteins can form heterodimers or heterotrimers with members of the 3-amino-acid loop extension (TALE) family of cofactors including Pbx and Meis proteins to regulate the transcription of downstream target genes directly.1-5 Aberrant overexpression of a set of HOXA genes including HOXA2-A10 and their cofactors, such as MEIS1, has been frequently observed in acute myeloid leukemia (AML) with chromosomal translocations involving the MLL, NUP98, and MYST3 genes, and in a subset of AML with normal cytogenetics.1,2,6-9 Overexpression of individual HOXA genes can induce myeloproliferation and block differentiation.8,10 Coexpression of Meis1 and Hoxa9 is sufficient to transform normal hematopoietic progenitor cells and to induce a rapidly fatal leukemia in transplanted mice,8,11-13 and their aberrant overexpression is required for the induction and maintenance of MLL-rearranged leukemia.14-19
In contrast, although Pbx proteins are also well known for their interaction with Hox proteins that increase the DNA-binding affinity of Hox proteins and thereby enhances the transcription of the downstream target genes,1,3,20-22 the cooperation between Pbx proteins and Hox proteins (eg, Hoxa9) in cell transformation and leukemogenesis is unclear. As Pbx1 is the founding member of the Pbx family, previous studies focused mainly on Pbx1 and showed that Pbx1 could form heterodimers or heterotrimers with Hox and/or Meis proteins1,3,9,20,21 ; however, no synergistic effect between Hoxa9 and Pbx1 was observed in cell transformation or leukemogenesis.7,11,12
We recently showed that increased expression of a 4-homeobox-gene signature (composed of HOXA7, HOXA9, HOXA11, and PBX3) is an independent predictor of shorter overall survival (OS) in patients with cytogenetically abnormal AML (CA-AML).23 Unlike the functions of the HOXA genes (especially HOXA9) that have been extensively studied in AMLs and particularly, in MLL-rearranged leukemia,7,8,16,17,24-26 the role of PBX3 in leukemogenesis is poorly understood. The identification of this HOXA/PBX3 prognostic gene signature triggered our interest to investigate whether there is a synergistic effect between PBX3 and HOXA9 in cell transformation and leukemogenesis.
To this end, through checking expression profiles of 3 independent large-scale patient sets, we first showed that PBX3 is the only member of the PBX family that is consistently coexpressed with HOXA9 in various subtypes of CA-AML, particularly in MLL-rearranged AML; in contrast, both PBX1 and PBX2 tend to exhibit an inverse correlation of expression with HOXA9 in CA-AML. A similar pattern was observed in MLL fusion-mediated mouse leukemia models. Thus, our data suggest that it is PBX3, but not PBX1 or PBX2, that is a potential pathologic cofactor of HOXA9 in CA-AML. We then showed that depletion of expression of Pbx3 dramatically inhibited MLL-AF9–induced transformation/immortalization of mouse normal bone marrow (BM) progenitor cells. Furthermore, we demonstrated that forced expression of PBX3 exhibited a significantly synergistic effect with HOXA9 in promoting cell transformation/immortalization in vitro and leukemogenesis in vivo. Finally, we treated a group of leukemia cells with HXR9, a small, cell-permeable peptide that was designed and proven to specifically disrupt the formation of HOX/PBX heterodimers.27 We found that the cells with higher levels of HOXA/PBX3 expression are more sensitive to HXR9 treatment than those with lower levels. Thus, targeting the HOXA/PBX3 pathway may provide a new strategy to substantially improve outcomes of patients with nonfavorable CA-AML, such as MLL-rearranged leukemia.
Methods
Microarray data
The microarray data of the United States (USA) set23,28 (including 97 CA-AML cases and 10 normal control samples), Germany set29 (including 71 CA-AML cases), The Netherlands set30 (including 271 CA-AML cases), and the mouse bone marrow transplantation (BMT) set28 (including 9 MLL-AF9 mouse leukemic BM cell samples and 6 normal control BM cell samples collected from primary or secondary BMT recipient mice) were generated by use of Affymetrix GeneChip Human Exon 1.0 ST arrays (Affymetirx), Stanford cDNA arrays (manufactured by the Stanford Functional Genomics Facility), Affymetrix U133 Plus2.0 arrays, and Affymetrix GeneChip Mouse Gene 1.0 ST arrays, respectively. The normalization of these microarray data were previously described.23,28-30 The complete-microarray datasets are available at the Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo/); accession nos. GSE30285 and GSE34184 for the USA set, GSE425 for the Germany set, GSE14468 for The Netherlands set, and GSE34185 for the mouse BMT set.
Retroviral constructs
HOXA9 coding region sequence was PCR amplified from human normal BM mononuclear cells with primers, forward 5′-ATAGAATTCATGGCCACCACTGGGGC-3′, and reverse 5′-ACCCTCGAGTCACTCGTCTTTTGCTC-3′, was then cloned into MSCVneo (Clontech), and named as MSCVneo-HOXA9. MSCVneo-MLL-AF9 is a kind gift from Dr Scott Armstrong. The coding region of PBX3 was synthesized by GenScript USA, and then was cloned into MSCV-PIG vector (containing a PGK-puromycin-IRES-GFP [PIG] cassette, kindly provided by Drs Hannon, Hammond, and He),31 and named as MSCV-PIG-PBX3. The pGFP-V-RS-Pbx3 shRNA construct (ie, pGFP-V-RS-shPbx3) and a noneffective scrambled control shRNA vector (ie, pGFP-V-RS–scrambled) were purchased from OriGene. All inserts were confirmed by sequencing.
Cell culture
HEK293T and Rat1a cells were cultured in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin (Invitrogen). MONOMAC-6 cells were grown in RPMI 1640 supplemented with 10% FBS, 2mM l-glutamine, nonessential amino acid, 1mM sodium pyruvate, and 9 μg/mL human insulin. THP-1 cells were maintained in RPMI 1640 containing 1% penicillin-streptomycin, 0.05mM 2-mercaptoethanol and 10% FBS (Invitrogen). ML-2 and Jurkat cells were maintained in RPMI 1640 containing 1% penicillin-streptomycin and 10% FBS (Invitrogen). K562 cells were maintained in Iscove modified Dulbecco medium with 4mM l-glutamine adjusted to contain 1.5g/L sodium bicarbonate and 10% FBS (Invitrogen).
Retrovirus preparation
As previously described,32-34 retrovirus for each construct was produced in 293T cells by cotransfecting the retroviral construct and pCL-Eco packaging vector (IMGENEX) with Effectene Transfection Reagent (QIAGEN). Rat1a cells were used to determine the viral titer.
In vitro colony-forming and replating assay
Hematopoietic progenitor cells (ie, lineage negative, Lin−) were obtained from 6-week-old B6.SJL (CD45.1) mice 5 days after 5-FU treatment (150 mg/kg) using the Mouse Lineage Cell Depletion kit (Miltenyi Biotec), and were then cotransduced with murine stem cell virus (MSCV) vectors, respectively, through “spinoculation.”23,28,32-34 Then, 2 aliquots of 1 × 104 of the transfected cells were plated into 4 35 mm Nunc Petri dishes in 1.1 mL of Methocult M3230 methylcellulose medium (StemCell Technologies) containing 10 ng/mL each of murine recombinant IL-3, IL-6, and GM-CSF and 30 ng/mL of murine recombinant stem cell factor (R&D Systems), along with 1.0 mg/mL of G418 (Gibco BRL) and 2.5 μg/mL of puromycin (Sigma-Aldrich). Cultures were incubated at 37°C in a humidified atmosphere of 5% CO2 in air for 7 days. Then, colony cells were collected and replated in methylcellulose dishes every 7 days with 1 × 104 cells as input for up to 4 passages. Because of the drug selections, all the colony-forming cells are probably 100% double-transduction positive.
Mouse BMT
Normal BM cells of B6.SJL (CD45.1) mice were retrovirally transduced with MSCVneo+ MSCV-PIG (as control), MSCVneo+ MSCV-PIG-PBX3 (ie, PBX3), MSCVneo-HOXA9+ MSCV-PIG (ie, HOXA9), and MSCVneo-HOXA9+ MSCV-PIG-PBX3 (ie, PBX3+HOXA9), respectively, through 2 rounds of spinoculation.23,28,32-34 The transduced cells were plated in methylcellulose dishes to form colonies under the selection with G418 and puromycin as described above. Six days later, colony cells were collected and washed, and then injected by tail vein into lethally irradiated (960 rads) 8- to 10-week-old C57BL/6 (CD45.2) recipient mice with 1 × 106 donor cells plus a radioprotective dose of whole BM cells (0.5 × 106; freshly harvested from a C57BL/6 mouse) per recipient mouse. All the mice were maintained in our animal facility to monitor for leukemogenesis for up to 200 days. All experiments on mice in our research protocol were approved by Institutional Animal Care and Use Committee (IACUC) of the University of Chicago.
Cytospin
For cytospin preparation, 50 000 cells were washed twice and were diluted in 200 μL of cold PBS. Each sample was loaded into the appropriate wells of the cytospin, and then spun at 2000 rpm for 2 minutes. The slides were dried in a desiccation chamber for 20 minutes and were stained with Wright-Giemsa.
Peptide treatment and cell proliferation, viability, and apoptosis assays
As previously described,27,35,36 HXR9 is an 18-amino-acid peptide consisting of the previously identified hexapeptide sequence that can bind to PBX and 9 C-terminal arginine residues (R9) that facilitate cell entry. CXR9 is a control peptide that lacks a functional hexapeptide sequence but includes the R9 sequence. Both peptides were synthesized using conventional column-based chemistry and purified to at least 80% (Biosynthesis). Cells were plated in 96-well plates in 200 μL of medium in triplicate and allowed to recover for 20 hours. Then the cells were treated with the active peptide HXR9 or the control peptide CXR9 with the indicated concentration for 6, 24, or 48 hours. Cell number was counted at the indicated time point for cell proliferation assay. To assess cell viability, cells were processed in a similar manner; after exposure to HXR9 or CXR9 as indicated concentration for 24 hours. Assessment of cell viability was performed using the MTS assay (Promega) according to the manufacturer's instructions. In the apoptosis assay, cells were plated in a 6 well plate in 2 mL of medium and allowed to recover for 20 hours. Then the cells were treated with HXR9 or CXR9 for 4 hours. Apoptotic cells were identified using flow cytometry and the annexin-V–APC apoptosis detection kit (BD Pharmingen) as described by the manufacturer's protocol.
RNA extraction and quantitative RT-PCR
Total RNA was extracted with the miRNeasy extraction kit (QIAGEN) and was used as template to synthesize cDNA for quantitative RT-PCR (qPCR) analysis in a 7900HT real-time PCR system (Applied Biosystems). qPCR with SYBR Green dye (QIAGEN) was used to determine expression of genes. GAPDH or PGK1 were used as endogenous controls. Each sample was run in triplicate.
Results
It is PBX3, but not PBX1 or PBX2, that is a potential pathologic cofactor of HOXA9 in human CA-AML
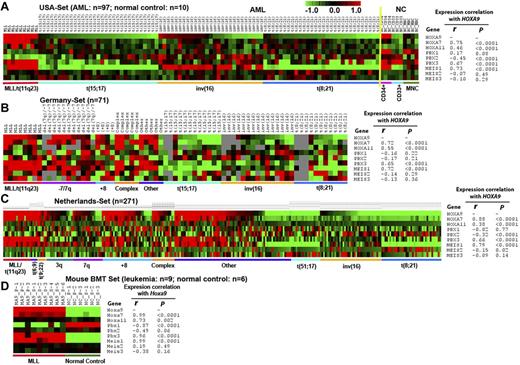
To determine whether PBX1, PBX2, and PBX3 are coexpressed with HOXA9 in CA-AML, we performed Pearson correlation analysis of 3 independent AML datasets. In the USA set (including 97 CA-AML patient samples and 10 normal BM control cell samples), we found that only PBX3, but not PBX1 or PBX2, exhibited a significantly positive correlation of expression (r = 0.67; P < .0001) with HOXA9 across the 107 samples; actually, PBX2 even showed a significantly negative correlation of expression (r = -0.45; P < .0001) with HOXA9 (Figure 1A). In the MEIS family, only MEIS1, but not MEIS2 or MEIS3, exhibited a significantly positive correlation of expression (r = 0.73; P < .0001) with HOXA9 across the samples (Figure 1A). Similar patterns were observed in the Germany set (including 71 CA-AML patient samples; Figure 1B) and The Netherlands set (including 271 CA-AML patient samples; Figure 1C). In all 3 sample sets, both PBX3 and MEIS1 were highly coexpressed with HOXA9 in MLL-rearranged AML (Figure 1A-C). These 3 genes were expressed at a high level also in some nonfavorable prognosis AML cases with 3q, 7q, +8, or complex karyotypes but were expressed at a low level in favorable subtypes of AML, such as those carrying t(15;17), inv(16), and t(8;21) (see Figure 1A-C and supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Besides PBX3 and MEIS1, most of the HOXA cluster genes also exhibited a significantly positive correlation of expression with HOXA9 across the samples in the 3 datasets (supplemental Figures 1-3). Thus, it is clear that PBX3 is the only member of the PBX family that is coexpressed with the HOXA cluster genes (eg, HOXA9). Therefore, PBX3, but not PBX1 or PBX2, that is probably a pathologic cofactor of HOXA9 in CA-AML, particularly those with nonfavorable prognosis.
Expression patterns of HOXA7, HOXA9, HOXA11, PBX1, PBX2, PBX3, MEIS1, MEIS2, and MEIS3, and the expression correlations between HOXA9 and other genes. (A) The USA set23,28 (including 9 MLL-rearranged, 31 t(15;17), 27 inv(16), 30 t(8;21) AML cases and 10 normal control samples); (B) the Germany set29 (including 8 MLL-rearranged, 11 (−7)/7q, 4 (+8), 5 complex, 12 t(15;17), 15 inv(16), 11 t(8;21) AML, and 5 other CA-AML cases); (C) The Netherlands set30 (including 16 MLL-rearranged, 5 t(6;9); 4 t(9;22), 18 (−3q), 18 (−7q), 31 (+8), 14 complex, 19 t(15;17), 38 inv(16), 37 t(8;21) AML, and 71 other CA-AML cases); and (D) the mouse bone marrow transplantation (BMT) set28 (including 9 MLL-AF9 mouse leukemic BM cell samples and 6 normal control BM cell samples collected from primary or secondary BMT recipient mice), respectively. Pearson correlation was applied to analyze the correlation. Expression data were mean centered and the relative value for each sample is represented by a color, with red representing a high expression and green representing a low expression (scale shown in the top right).
Expression patterns of HOXA7, HOXA9, HOXA11, PBX1, PBX2, PBX3, MEIS1, MEIS2, and MEIS3, and the expression correlations between HOXA9 and other genes. (A) The USA set23,28 (including 9 MLL-rearranged, 31 t(15;17), 27 inv(16), 30 t(8;21) AML cases and 10 normal control samples); (B) the Germany set29 (including 8 MLL-rearranged, 11 (−7)/7q, 4 (+8), 5 complex, 12 t(15;17), 15 inv(16), 11 t(8;21) AML, and 5 other CA-AML cases); (C) The Netherlands set30 (including 16 MLL-rearranged, 5 t(6;9); 4 t(9;22), 18 (−3q), 18 (−7q), 31 (+8), 14 complex, 19 t(15;17), 38 inv(16), 37 t(8;21) AML, and 71 other CA-AML cases); and (D) the mouse bone marrow transplantation (BMT) set28 (including 9 MLL-AF9 mouse leukemic BM cell samples and 6 normal control BM cell samples collected from primary or secondary BMT recipient mice), respectively. Pearson correlation was applied to analyze the correlation. Expression data were mean centered and the relative value for each sample is represented by a color, with red representing a high expression and green representing a low expression (scale shown in the top right).
It is Pbx3, but not Pbx1 or Pbx2, that is co-overexpressed with Hoxa9 in MLL-fusion–mediated murine leukemia
As the MLL-rearranged AML is the only subtype of CA-AML in which PBX3 is consistently co-overexpressed with HOXA9 in almost all patient cases tested (see Figure 1A-C, supplemental Table 2), we next investigated the coexpression pattern between Pbx3 and Hoxa9 in murine genetic models of MLL-rearranged leukemia. As previously described,28 we retrovirally transduced MLL-AF9 into BM progenitor cells of donor mice and transplanted them into congenic recipient mice to induce leukemia, and leukemic BM cells from primary recipient mice were collected and transplanted into secondary recipient mice to develop leukemia again. Then, we performed Affymetrix GeneChip array assays of 9 mouse leukemic BM samples (3 from primary and 6 from secondary BMT recipient mice), along with 6 normal control BM samples (3 each from primary and secondary BMT recipient mice with retrovirally transduced control vector).28 Consistent with what we observed in human AML carrying MLL rearrangements (Figure 1A-C), we also found that Pbx3 is the only member of the Pbx family that is co–up-regulated with Hoxa9 (as well as Meis1 and other Hoxa genes) by MLL fusion proteins in mouse leukemic cells (Figure 1D, supplemental Figure 4).
Expression of Pbx3 is required for cell transformation mediated by MLL fusions
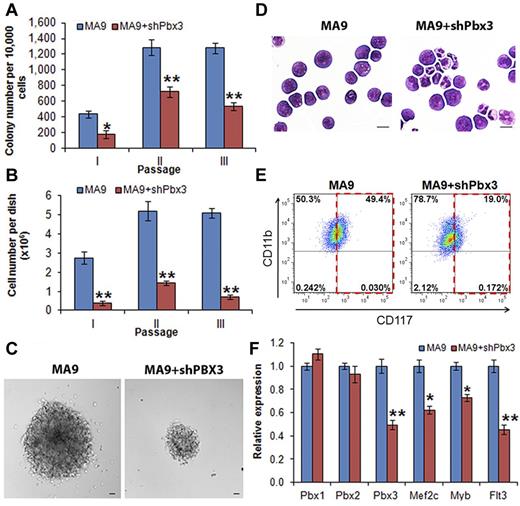
To investigate whether expression of PBX3 is required for cell transformation mediated by MLL fusion proteins, we conducted in vitro colony-forming/replating assays. We cotransduced shRNA of Pbx3 or scrambled shRNA (as a negative control) with MLL-AF9 into mouse normal BM progenitor (ie, lineage negative, Lin−) cells that were plated onto methylcellulose medium with G418 and puromycin selection. Cells from the colonies were replated every 7 days for up to 3 passages. As shown in Figure 2A, cells transduced with MLL-AF9 plus scrambled shRNA (labeled as MA9) could form more than 1000 colonies after 2 passages of plating, whereas cells transduced with MLL-AF9 plus Pbx3 shRNA (ie, MA9+shPbx3) formed significantly (P < .001, t test) fewer colonies after replating. Moreover, the cell number per dish was also significantly lower (P < .001, t test) in the cells transduced with MA9+shPbx3 than in those with MA9 alone (Figure 2B). We also observed that the size of individual colonies was much smaller (Figure 2C) in MA9+shPbx3 colonies than in MA9 colonies. Furthermore, MA9 colony cells displayed a very immature morphology (basophilic cytoplasm, large nucleus, evident nucleolus, lacy chromatin), whereas colony cells with MA9+shPbx3 were partially differentiated with some granulocytes (Figure 2D). Flow cytometric analysis showed that MA9+shPbx3 cells had a much lower proportion of c-Kit+ blast cells (19% versus 49%; Figure 2E). To eliminate the possibility that our Pbx3 shRNA has an off-target effect on repressing expression of Pbx1 or Pbx2, we performed qPCR and showed that only expression of Pbx3, but not that of Pbx1 or Pbx2, was significantly down-regulated (P < .001) by Pbx3 shRNA (Figure 2F). As a functional consequence, the depletion of Pbx3 expression resulted in a significant down-regulation of expression of downstream targets of the Hox/Pbx/Meis axis, including Mef2c, Myb, and Flt316,37-39 (Figure 2F). These 3 downstream target genes have been shown to play critical oncogenic role in the development and maintenance of MLL-rearranged leukemia.37,38,40,41 Thus, our data suggest that the expression of Pbx3 is probably required for cell transformation (ie, immortalization) mediated by MLL fusion proteins.
Knockdown expression of Pbx3 by shRNA inhibits cell transformation mediated by MLL-AF9. Colony-forming and replating assays of mouse normal BM progenitor cells transduced with MSCVneo-MLL-AF9+pGFP-V-RS-scrambled (ie, MA9) or MSCVneo-MLL-AF9+ pGFP-V-RS-Pbx3 shRNA (ie, MA9+shPbx3). Duplicates were plated for each combination with 1 × 104 cells per dish, and every 7 days the cells were replated for up to 3 passages. Mean values and standard deviations (mean ± SD) from 2 independent experiments are shown (*P < .01; **P < .001, 2-tailed t test). (A) Numbers of colonies per dish (only colonies with ≥ 50 cells/colony were counted; 1 × 104 input cells) in each passage are shown. (B) Average numbers of colony cells per dish are shown. (C) Morphology of colonies of secondary passage. Scale bars represent 100 μm. (D) Morphology of cells of secondary passage. Cells were stained with Wright-Giemsa. Scale bars represent 10 μm. (E) Flow cytometric analysis of colony-forming cells (secondary passage) with APC-labeled anti-CD117 (ie, c-Kit) antibody and eFluor 450–labeled anti-CD11b (ie, Mac-1) antibody. (F) qPCR assay of the endogenous expression of Pbx1, Pbx2, Pbx3, Mef2c, Myb, and Flt3 in colony cells collected from the secondary passage.
Knockdown expression of Pbx3 by shRNA inhibits cell transformation mediated by MLL-AF9. Colony-forming and replating assays of mouse normal BM progenitor cells transduced with MSCVneo-MLL-AF9+pGFP-V-RS-scrambled (ie, MA9) or MSCVneo-MLL-AF9+ pGFP-V-RS-Pbx3 shRNA (ie, MA9+shPbx3). Duplicates were plated for each combination with 1 × 104 cells per dish, and every 7 days the cells were replated for up to 3 passages. Mean values and standard deviations (mean ± SD) from 2 independent experiments are shown (*P < .01; **P < .001, 2-tailed t test). (A) Numbers of colonies per dish (only colonies with ≥ 50 cells/colony were counted; 1 × 104 input cells) in each passage are shown. (B) Average numbers of colony cells per dish are shown. (C) Morphology of colonies of secondary passage. Scale bars represent 100 μm. (D) Morphology of cells of secondary passage. Cells were stained with Wright-Giemsa. Scale bars represent 10 μm. (E) Flow cytometric analysis of colony-forming cells (secondary passage) with APC-labeled anti-CD117 (ie, c-Kit) antibody and eFluor 450–labeled anti-CD11b (ie, Mac-1) antibody. (F) qPCR assay of the endogenous expression of Pbx1, Pbx2, Pbx3, Mef2c, Myb, and Flt3 in colony cells collected from the secondary passage.
Coexpression of PBX3 and HOXA9 recapitulates the phenotypes observed in MLL-AF9–mediated cell transformation
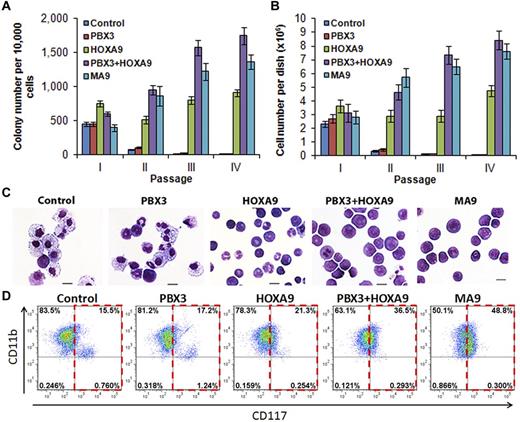
We next sought to determine whether there is a synergistic effect between PBX3 and HOXA9 in cell transformation. As shown in Figure 3A, cells transduced with PBX3 alone formed only a few colonies after a series of replating, in a manner similar to those transduced with control vector; in contrast, cells transduced with HOXA9 alone formed hundreds of colonies after replating. Remarkably, cotransduction of both PBX3 and HOXA9 (ie, PBX3+HOXA9) caused significantly (P < .001, t test) more colonies as well as more colony cells per dish than did HOXA9 alone, to a level similar to that of transduction of MLL-AF9 (Figure 3A-B). In addition, the cells from the PBX3+HOXA9 colonies were more immature compared with those from the HOXA9 only colonies, in a manner similar to cells from the MLL-AF9 colonies (Figure 3C-D). These results indicate that PBX3 has a synergistic effect with HOXA9 in transforming normal hematopoietic progenitor cells through promoting self-renewal and proliferation of the cells, and coexpression of PBX3 and HOXA9 can, at least in part, recapitulate the phenotype caused by ectopic expression of MLL fusion genes in cell transformation.
PBX3 enhances colony-forming/immortalization capacity of mouse normal bone marrow progenitor cells induced by HOXA9. Colony-forming and replating assays of mouse normal BM progenitor (ie, lineage negative) cells transduced with MSCV-PIG+MSCVneo (ie, control), MSCV-PIG-PBX3+MSCVneo (ie, PBX3), MSCV-PIG+MSCVneo-HOXA9 (ie, HOXA9), MSCV-PIG-PBX3+MSCVneo-HOXA9 (ie, PBX3+HOXA9), or MSCV-PIG+MSCVneo-MLL-AF9 (ie, MA9). Cells were plated for each transduction with 1 × 104 cells per dish duplicates, and every 7 days the cells were replated for up to 4 passages. Mean ± SD values from 2 independent experiments are shown. (A) Numbers of colonies per dish in each passage are shown. (B) Cell numbers per dish are shown. (C) Morphology of cells of secondary passage. Cells were stained with Wright-Giemsa. Scale bars represent 10 μm. (D) Flow cytometric analysis of colony-forming cells (secondary passage) with APC-labeled anti-CD117 (ie, c-Kit) antibody and eFluor 450–labeled anti-CD11b (ie, Mac-1) antibody.
PBX3 enhances colony-forming/immortalization capacity of mouse normal bone marrow progenitor cells induced by HOXA9. Colony-forming and replating assays of mouse normal BM progenitor (ie, lineage negative) cells transduced with MSCV-PIG+MSCVneo (ie, control), MSCV-PIG-PBX3+MSCVneo (ie, PBX3), MSCV-PIG+MSCVneo-HOXA9 (ie, HOXA9), MSCV-PIG-PBX3+MSCVneo-HOXA9 (ie, PBX3+HOXA9), or MSCV-PIG+MSCVneo-MLL-AF9 (ie, MA9). Cells were plated for each transduction with 1 × 104 cells per dish duplicates, and every 7 days the cells were replated for up to 4 passages. Mean ± SD values from 2 independent experiments are shown. (A) Numbers of colonies per dish in each passage are shown. (B) Cell numbers per dish are shown. (C) Morphology of cells of secondary passage. Cells were stained with Wright-Giemsa. Scale bars represent 10 μm. (D) Flow cytometric analysis of colony-forming cells (secondary passage) with APC-labeled anti-CD117 (ie, c-Kit) antibody and eFluor 450–labeled anti-CD11b (ie, Mac-1) antibody.
Coexpression of PBX3 and HOXA9 causes leukemia rapidly in mice
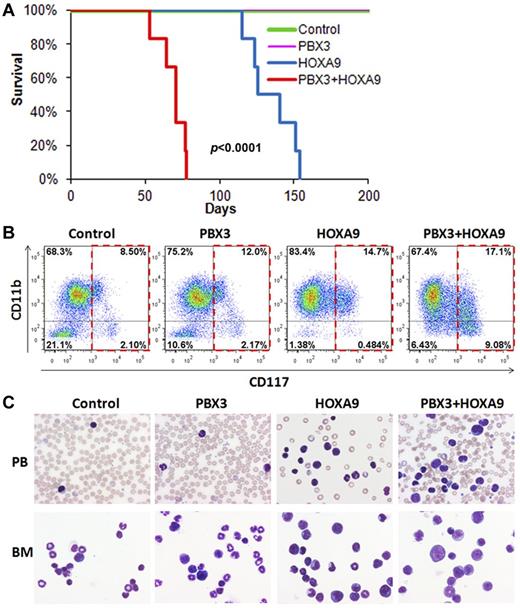
To assess the potential synergistic effect between PBX3 and HOXA9 in inducing leukemia in vivo, we performed mouse BMT assays. As shown in Figure 4A, all transplanted mice with co-overexpression of PBX3 and HOXA9 developed a fatal AML within 80 days, significantly faster than the mice with overexpression of HOXA9 alone (P < .0001, log-rank test), whereas PBX3 overexpression alone could not induce leukemia. Furthermore, coexpression of PBX3 and HOXA9 resulted in a higher proportion of leukemic blast cells (c-Kit+) than overexpression of HOXA9 or PBX3 alone (Figure 4B). Cytospin analysis of mouse peripheral blood (PB) and BM cells also showed that there is a remarkable synergistic effect between PBX3 and HOXA9 in inducing accumulation of leukemic blast cells in PB and BM (see Figure 4C). Thus, PBX3 exhibited a significantly synergistic effect with HOXA9 in promoting cell transformation and leukemogenesis.
PBX3 exhibited a synergistic effect with HOXA9 in inducing leukemia in transplanted mice. (A) The PBX3+HOXA9 (ie, MSCV-PIG-PBX3+MSCVneo-HOXA9; n = 6) group developed leukemia significantly faster (median overall survival, 71 days versus 140 days; P < .0001, log-rank test) than the HOXA9 alone (ie, MSCV-PIG+MSCVneo-HOXA9; n = 6) group, whereas the control group (ie, MSCV-PIG+MSCVneo; n = 6) and the PBX3 alone (ie, MSCV-PIG-PBX3+MSCVneo; n = 7) group did not develop leukemia. Kaplan-Meier curves are shown. (B) Flow cytometric analysis of BM cells of representative mice stained with APC-labeled anti-CD117 (ie, c-Kit) antibody and eFluor 450–labeled anti-CD11b (ie, Mac-1) antibody. (C) Comparison of cell morphology between the 4 mouse groups. Peripheral blood (PB) and BM cell cytospin via Wright-Giemsa staining are shown.
PBX3 exhibited a synergistic effect with HOXA9 in inducing leukemia in transplanted mice. (A) The PBX3+HOXA9 (ie, MSCV-PIG-PBX3+MSCVneo-HOXA9; n = 6) group developed leukemia significantly faster (median overall survival, 71 days versus 140 days; P < .0001, log-rank test) than the HOXA9 alone (ie, MSCV-PIG+MSCVneo-HOXA9; n = 6) group, whereas the control group (ie, MSCV-PIG+MSCVneo; n = 6) and the PBX3 alone (ie, MSCV-PIG-PBX3+MSCVneo; n = 7) group did not develop leukemia. Kaplan-Meier curves are shown. (B) Flow cytometric analysis of BM cells of representative mice stained with APC-labeled anti-CD117 (ie, c-Kit) antibody and eFluor 450–labeled anti-CD11b (ie, Mac-1) antibody. (C) Comparison of cell morphology between the 4 mouse groups. Peripheral blood (PB) and BM cell cytospin via Wright-Giemsa staining are shown.
HXR9 selectively inhibits cell proliferation and promotes apoptosis in AML cells with a high level of expression of HOXA/PBX3 genes
Recently, a small, cell-permeable peptide (ie, HXR9) which carries the core motif of HOX proteins for binding with PBX proteins was designed by Morgan and colleagues and was proven to specifically disrupt the formation of HOX/PBX heterodimers.27 Their previous studies showed that HXR9 exhibited remarkable anti–tumor effects in vitro and in vivo in various types of aggressive solid tumors,27,36,42 with no deleterious side effects in HXR9-treated mice.27 Thus, it is important to investigate whether such antagonists can selectively kill leukemic cells with overexpression of the HOXA/PBX3 signature.
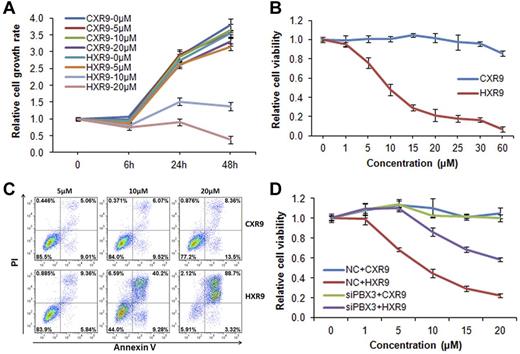
First, we treated MONOMAC-6/t(9;11) cell line with HXR9 or the negative control (ie, CXR9) at different concentrations (ie, 5μM, 10μM, and 20μM) for 6, 24, and 48 hours, respectively. As shown in Figure 5A, HXR9, compared with CXR9, dramatically inhibited cell growth at a concentration of 10μM or 20μM at all time points, particularly, at 48 hours after treatment. It also showed a dose-dependent effect of HXR9 on inhibition of MONOMAC-6 cell growth and the IC50 is 10.54 ± 0.14 μM (Figure 5B). In addition, there was much more late-phase apoptosis induced by HXR9 than the peptide control, CXR9, at the concentration of 10μM or 20μM (40.2% versus 6.07% or 88.7% versus 8.36%; Figure 5C). As shown in Figure 5D, the depletion of endogenous expression of PBX3 in MONOMAC-6 cells by siRNA oligos could significantly inhibit (P = .013; paired t test) the effect of HXR9, indicating that HXR9 exerts its effect through targeting the PBX3-associated pathway. Furthermore, we also treated colony-forming cells of PBX3+HOXA9, MLL-AF9, or control group (secondary passage; Figure 3) with HXR9 (CXR9 was used as a negative control) and showed that HXR9 exhibited a much significant inhibitory effect on the first 2 groups compared with the control group (supplemental Figure 5). Thus, our data suggest that the efficacy of HXR9 is highly probable to be dependent on the presence of HOXA9/PBX3.
HXR9 selectively inhibits proliferation and promotes apoptosis of MONOMAC-6 cells. (A) Cell proliferation assay after the treatment with CXR9 or HXR9 at the concentration of 0, 5, 10, and 20μM, respectively, for 6, 24, or 48 hours (h). Cells were counted at indicated time points and the cell number was normalized with that before peptide treatment. Mean ± SD values from 2 independent experiments are shown. (B) Cell viability assay after the treatment with CXR9 or HXR9 at the concentration of 0, 1, 5, 10, 20, 25, 30, and 60μM, respectively, for 24 hours. Cell viability was measured with MTS assay. Mean ± SD values from 3 independent experiments are shown. (C) Cell apoptosis assay after the treatment with CXR9 or HXR9 at the concentration of 5, 10, and 20μM, respectively, for 24 hours. Apoptosis was measured by flow cytometry analysis of MONOMAC-6 cells stained with FITC-labeled annexin V and counterstained with PI. Percentage of cells is shown in the corner of each quadrant. Results are representatives of 3 independent experiments. (D) Cells were transfected with PBX3 siRNA oligos (ie, siPBX3) or scrambled oligos (ie, NC) and then treated with CXR9 or HXR9 at the concentration of 0, 1, 5, 10, 15, and 20μM, respectively. Cell viability was measured with MTS assay 24 hours after treatment. Mean ± SD values from 2 independent experiments are shown.
HXR9 selectively inhibits proliferation and promotes apoptosis of MONOMAC-6 cells. (A) Cell proliferation assay after the treatment with CXR9 or HXR9 at the concentration of 0, 5, 10, and 20μM, respectively, for 6, 24, or 48 hours (h). Cells were counted at indicated time points and the cell number was normalized with that before peptide treatment. Mean ± SD values from 2 independent experiments are shown. (B) Cell viability assay after the treatment with CXR9 or HXR9 at the concentration of 0, 1, 5, 10, 20, 25, 30, and 60μM, respectively, for 24 hours. Cell viability was measured with MTS assay. Mean ± SD values from 3 independent experiments are shown. (C) Cell apoptosis assay after the treatment with CXR9 or HXR9 at the concentration of 5, 10, and 20μM, respectively, for 24 hours. Apoptosis was measured by flow cytometry analysis of MONOMAC-6 cells stained with FITC-labeled annexin V and counterstained with PI. Percentage of cells is shown in the corner of each quadrant. Results are representatives of 3 independent experiments. (D) Cells were transfected with PBX3 siRNA oligos (ie, siPBX3) or scrambled oligos (ie, NC) and then treated with CXR9 or HXR9 at the concentration of 0, 1, 5, 10, 15, and 20μM, respectively. Cell viability was measured with MTS assay 24 hours after treatment. Mean ± SD values from 2 independent experiments are shown.
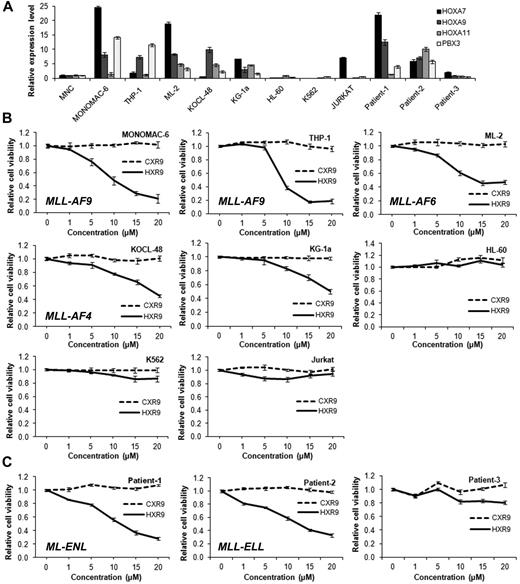
We next expanded the study to treat a panel of 8 leukemia cell lines with HXR9 or CXR9 at a concentration of 0, 1, 5, 10, 15, and 20μM, respectively for 24 hours, and then assessed cell viability. The 8 cell lines exhibited various levels of HOXA/PBX3 expression (Figure 6A). As shown in Figure 6B, HXR9 exhibited a robust inhibitory effect on cell viability of 3 MLL-rearranged leukemia cell lines including MONOMAC-6, THP-1/t(9;11) and ML-2/t(6;11), whereas it exhibited a relatively mild effect on KOCL-48/t(4,11) and KG-1a cells (a non–MLL-rearranged AML cell line), along with a very minor (or even no) effect on HL-60 (a non–MLL-rearranged AML cell line), K562 (a erythroid/megakaryocytic Ph-positive CML cell line) and Jurkat (a T-cell acute leukemia cell line). Remarkably, the effects of HXR9 on those cell lines are largely correlated with the endogenous expression levels of the HOXA/PBX3 genes (Figure 6A). Equally remarkably, the expression of downstream targets of the HOX/PBX/MEIS axis, including MEF2C, MYB, and FLT3,16,37-39 also changed coordinately after HXR9 treatment (supplemental Figure 6).
The inhibitory effect of HXR9 on cell viability is associated with the endogenous expression of the HOXA/PBX3 genes in corresponding leukemic cells. (A) qPCR analysis of HOXA7, HOXA9, HOXA11, and PBX3 expression in 8 leukemic cell lines and 3 primary AML patient leukemic BM cells. Expression level of each gene in each leukemia sample was normalized to that in human normal BM mononuclear cells (MNC). (B-C) Cell viability assay after the treatment with CXR9 and HXR9 on the 8 leukemic cell lines (B) and 3 primary AML samples (C) at the concentration of 1μM, 5μM, 10μM, 15μM, and 20μM for 24 hours. Cell viability was measured with MTS assay. Mean values and SD from 3 independent experiments are shown.
The inhibitory effect of HXR9 on cell viability is associated with the endogenous expression of the HOXA/PBX3 genes in corresponding leukemic cells. (A) qPCR analysis of HOXA7, HOXA9, HOXA11, and PBX3 expression in 8 leukemic cell lines and 3 primary AML patient leukemic BM cells. Expression level of each gene in each leukemia sample was normalized to that in human normal BM mononuclear cells (MNC). (B-C) Cell viability assay after the treatment with CXR9 and HXR9 on the 8 leukemic cell lines (B) and 3 primary AML samples (C) at the concentration of 1μM, 5μM, 10μM, 15μM, and 20μM for 24 hours. Cell viability was measured with MTS assay. Mean values and SD from 3 independent experiments are shown.
Finally, we also tested leukemic BM cell samples from 3 primary AML patients. We cultured cells for 3 to 5 days and then treated them with HXR9 or CXR9. As shown in Figure 6A, 2 patient samples (ie, patients 1 and 2) with MLL-rearrangements have high levels of expression of the HOXA/PBX3 genes, whereas the third patient sample (ie, patient 3) with a t(15;17) abnormality has a low level of HOXA/PBX3 expression. As expected, HXR9 exhibited a remarkable inhibitory effect on the 2 MLL-rearranged AML samples but not on the third patient sample (Figure 6C).
Collectively, our data demonstrate that HXR9 selectively decreases cell proliferation and promotes apoptosis in cells with a high level of expression of the HOXA/PBX3 genes, such as MLL-rearranged leukemic cells, probably resulting from the specific disruption of the interaction between HOXA and PBX3 proteins, followed by the consequent inhibition on expression of critical downstream target genes of the HOXA/PBX3 axis that are important for cell proliferation and anti-apoptosis.
Discussion
A previous study suggested that the induction and maintenance of MLL-fusion–mediated cell transformation is dependent on the redundant contributions of Pbx2 and Pbx3; however, knockout of each alone did not affect the transformation.14 In contrast, in this study, we showed that knockdown of Pbx3 alone by shRNA could significantly inhibit MLL-fusion–mediated cell transformation (Figure 2). Interestingly, similar controversial observations were reported previously with regard to whether Hoxa9 is required for MLL-rearranged leukemia. Two studies used Hoxa9 knockout mouse models and showed that Hoxa9 is not required for cell transformation and leukemogenesis mediated by MLL-GAS7 or Mll-AF9,43,44 whereas other studies used Hoxa9 knockout or shRNA knockdown models and showed that Hoxa9 is required for MLL-ENL, MLL-AF9, or MLL-AF4–mediated cell transformation and leukemogenesis.16,17,19 The discrepancy between those studies was thought to be because of quantitative differences in Hoxa9 dependency because of different compositions of stem/progenitor populations being used for the different experiments.43 Similarly, our result is not necessarily in contradiction with that of Wong et al, as we used different models.14 Indeed, the discrepancy between these 2 studies might be attributed to different types of donor cells being used, as Wong et al used fetal liver cells from Pbx3−/− mice, whereas we used BM progenitor (ie, lineage negative) cells from 6-week-old B6.SJL mice.14 It is possible that their fetal liver cells had a higher level of endogenous Pbx2 expression than the BM progenitor cells we used, and thus in their model they had to deplete expression of both Pbx3 and Pbx2 to show a significant inhibitory effect, whereas in our model it was not necessary to further knockdown expression of Pbx2 as its endogenous expression level was already low. Actually, as shown in Figure 1, PBX2/Pbx2 expression is probably further down-regulated by MLL fusions in both human and murine MLL-rearranged leukemia relative to normal BM control cells. As shown in Figure 2F, only expression of Pbx3, but not that of Pbx1 or Pbx2, was significantly down-regulated (P < .001) by Pbx3 shRNA, indicating that the effects of Pbx3 shRNA we observed (Figure 2A-E) are not because of any off-target effect of Pbx3 shRNA on expression of Pbx1 or Pbx2.
Previous studies indicated that there was a significantly synergistic effect between Hoxa9 and Meis1, but not between Hoxa9 and Pbx1, in transforming cells and inducing leukemia.7,11,12 Here we showed that, in fact, it is PBX3, but not PBX1 or PBX2, that exhibited a significantly positive correlation (r > 0.6, P < .0001, Pearson correlation) of expression with HOXA9 in CA-AML, particularly in MLL-rearranged AML cells, and a similar pattern was observed in MLL-AF9–mediated murine leukemic cells (Figure 1, supplemental Figures 1-4). This pattern implies that there might be a synergistic effect between PBX3 and HOXA9 in leukemogenesis. Indeed, we showed that they exhibited a synergistic effect in transforming normal hematopoietic progenitor cells through promoting self-renewal and proliferation of the cells, and their coexpression probably recapitulates the phenotypes caused by ectopic expression of MLL fusion genes (Figure 3). Furthermore, we showed that although PBX3 alone could not cause leukemia, coexpression of PBX3 and HOXA9 could cause leukemia in mice very rapidly and all mice died of AML within 80 days, significantly faster than HOXA9 alone mice (Figure 4A). Our flow cytometry and cytospin analyses showed that the PB and BM cells of the PBX3+HOXA9 leukemic mice had a greater proportion of immature blast cells than those of the HOXA9 or PBX3 alone mice (Figure 4B-C). Our data together with those reported previously7,8,16,17,23,24,26 suggest that both PBX3 and HOXA genes are required for the development and maintenance of some subtypes of CA-AML (eg, MLL-rearranged AML) and they have synergistic effects in transforming hematopoietic cells and causing leukemia.
The HOXA genes and PBX3 are aberrantly overexpressed in nonfavorable CA-AML and they play their synergistic functions probably through forming heterodimers; antagonists that specifically disrupt the formation of HOX/PBX heterodimers would be useful drugs to treat nonfavorable CA-AML. Interestingly, Morgan and colleagues developed such an antagonist, namely HXR9, and have shown its promising anti–tumor effects both in vitro and in vivo in a group of aggressive solid tumors.27,36,42 Remarkably, as a proof of our concept, we show here that HXR9 can selectively inhibit cell proliferation, decrease cell viability, and promote apoptosis in leukemia cells with a high level of expression of the HOXA/PBX3 genes, for example, those with MLL rearrangements (Figures 5–6, supplemental Figure 5); the effects of HXR9 are correlated with endogenous expression levels of the HOXA/PBX3 gene signature and the degrees of the down-regulation of downstream target genes of this gene signature (supplemental Figure 6).
Taken together, we show here that it is PBX3, but not PBX1 or PBX2, that is consistently coexpressed with HOXA9 in CA-AML, particularly in MLL-rearranged AML cells, and thus it is PBX3 that is a potential pathologic cofactor of HOXA9 in leukemogenesis. We also show that PBX3 is required for MLL-fusion–mediated cell transformation, and there is a synergistic effect between PBX3 and HOXA9 in cell transformation and leukemogenesis. Furthermore, we show that the HOXA/PBX3 gene signature is also an attractive druggable target by use of antagonists, such as HXR9 or its derivatives. As it was previously shown that HXR9 did not cause evident side effect/toxicity in mice,27 HXR9 or its derivatives hold a great potential to be clinically applied in the treatment of nonfavorable CA-AML (eg, MLL-rearranged leukemia) alone, or, in combination with other effective therapeutic drug(s), to substantially improve the survival of the leukemia patients. In addition, mutations of the nucleophosmin (NPM1) gene occur in around one-half of patients with cytogenetically normal AML (ie, CN-AML, which accounts for around 40%-50% of the entire set of AML) and are associated with increased expression of multiple homeobox genes including HOXA7, HOXA9, and PBX3.45-48 Thus, HXR9 and its derivatives are probably also applicable to treat CN-AML with NPM1 mutations. Therefore, therapeutically targeting the interaction between the PBX3 and HOXA proteins may have a broad application in treating more than 30% to 40% of patients with AML.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Drs Michelle Le Beau and Richard Larson for patient information and samples used in this project, and Drs Gregory Hannon, Scott Hammond, Lin He, and Scott Armstrong for providing retroviral vectors.
This work was supported in part by the Leukemia & Lymphoma Society (L.L.S.) Translational Research Grant (J.D.R. and J.C.), the National Institutes of Health (NIH) R01 grant CA127277 (J.C.), American Cancer Society (ACS) Research Scholar grant (J.C.), the G. Harold and Leila Y. Mathers Charitable Foundation (J.C.), LLS Special Fellowship (Z.L.), Gabrielle's Angel Foundation for Cancer Research (J.C., Z.L., H.H., and X.J.), The Spastic Paralysis Foundation of the Illinois, Eastern Iowa Branch of Kiwanis International (J.D.R.), The Fidelity Foundation (J.D.R. and J.C.), The University of Chicago Committee on Cancer Biology (CCB) Fellowship Program (X.J.), NIH/NCCAM P01 AT004418 (C.S.Y.), The Intramural Research Program of National Human Genome Research Institute, NIH (A.E. and P.P.L.), The Deutsche José Carreras Leukämie Stiftung (DJCLS) grant R 06/41v (L.B.), and The Deutsche Forschungsgemeinschaft (DFG) Heisenberg-Stipendium BU 1339/3-1 (L.B).
National Institutes of Health
Authorship
Contribution: Z.L. and J.C. conceived and designed the study; Z.L., H.H., X.J., A.G.E., L.B., P.P.L., J.D.R., C.-S.Y., and J.C. provided financial support; C.-Z.W., A.G.E., L.B., P.J.M.V., R.D., B.L., P.P.L., R.M., J.D.R., C.-S.Y., and J.C. provided study materials or reagents; all authors collected and assembled data; Z.L., Z.Z., Y.L., S.A., C.H., and J.C. conducted data analysis and/or interpretation; Z.L., Z.Z., Y.L., S.A., P.C., H.H., X.J., G.-M.H., R.B.K., H.R., and J.C. conducted experiments; Z.L. and J.C. prepared the paper; and all authors helped to revise and approved the final paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jianjun Chen or Zejuan Li, Dept of Medicine, University of Chicago, 900 E 57th St, KCBD Rm 7134, Chicago, IL 60637; e-mail: jchen@medicine.bsd.uchicago.edu or zjli@uchicago.edu.
References
Author notes
Z.L, Z.Z., Y.L., and S.A. contributed equally to this work.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal