Key Points
miR-30c is a direct target of C/EBPα and upregulated by C/EBPα-p42.
NOTCH1 is a direct target of miR-30c and regulated by C/EBPα and miR-30c.
Abstract
The transcription factor CCAAT enhancer binding protein α (C/EBPα) is a master regulator in granulopoiesis and is frequently disrupted in acute myeloid leukemia (AML). We have previously shown that C/EBPα exerts its effects by regulating microRNAs (miRs) such as miR-223 and miR-34a. Here, we confirm miR-30c as a novel important target of C/EBPα during granulopoiesis. Thus, wild-type C/EBPα-p42 directly upregulates miR-30c expression, whereas C/EBPα-p30, found in AML, does not. miR-30c is downregulated in AML, especially in normal karyotype AML patients with CEBPA mutations. An induced C/EBPα knockout in mice leads to a significant downregulation of miR-30c expression in bone marrow cells. We identified NOTCH1 as a direct target of miR-30c. Finally, a block of miR-30c prevents C/EBPα-induced downregulation of Notch1 protein and leads to a reduced CD11b expression in myeloid differentiation. Our study presents the first evidence that C/EBPα, miR-30c, and Notch1 together play a critical role in granulocytic differentiation and AML, and particularly in AML with CEBPA mutations. These data reveal the importance of deregulated miRNA expression in leukemia and may provide novel biomarkers and therapeutic targets in AML.
Introduction
CCAAT enhancer binding protein α (C/EBPα) functions as a key regulator of granulocytopoiesis.1 C/EBPα is activated in early myeloid precursors and directs them to granulocytic maturation.2,3 Loss of C/EBPα functions has been linked to leukemogenesis, suggesting an important role for C/EBPα as a tumor suppressor.4,5 In acute myeloid leukemia (AML), C/EBPα is deregulated by various mechanisms including its own mutations.6 Mutations in the CEBPA gene are reported for approximately 10% of all AMLs.7 The mutations of CEBPA are point mutations in the C-terminal basic region–leucine zipper domain and frame shift mutations in the N-terminal domain. The CEBPA N-terminal mutation results in a shorter form of C/EBPα, C/EBPα-p30, which fails to induce differentiation and exhibits a dominant-negative function over C/EBPα-p42.4,5 A conditional silencing of C/EBPα in mice shows a selective block in the differentiation of granulocytes.8
MicroRNAs (miRNAs) are a group of gene regulators that play important roles in biologic processes such as cell proliferation, differentiation, and apoptosis, all of which are frequently affected in cancer. A growing number of studies demonstrate that the deregulation of miRNAs is associated with the development of cancer, including leukemia.9,10 Former studies have demonstrated the regulation of specific miRNAs by C/EBPα. Fazi et al first identified miR-223 as a direct target of C/EBPα. The C/EBPα-induced upregulation of miR-223 leads to granulopoiesis.11 Moreover, recent studies from our group show a direct regulation of miR-223 and miR-34a by C/EBPα in normal granulopoiesis and emphasize that C/EBPα acts as a tumor suppressor gene via transactivation of these miRNAs.12,13 Furthermore, in AML, where the expression of C/EBPα is deregulated, the transactivation of both miRNAs is inhibited and myeloid differentiation is blocked.11-13
For granulocytes, past studies have reported a high expression of miRNA-30c.14,15 We found that upon overexpression of C/EBPα, miR-30c was upregulated. Some other studies describe a function of miR-30c in several cancers such as bladder16 tumor, breast cancer,17 and endometrial cancer.18 In breast cancer and AML with NPM1 mutations, miR-30c functions like a tumor suppressor.19,20 There has been no report that shows any specific function of miR-30c in granulopoiesis and AML independent of NPM1 mutations.
In the present study, we first investigated the role of miR-30c in C/EBPα-induced granulocytic differentiation and AML, in particular, in normal karyotype AML with CEBPA mutations. We identified the miR-30c gene as a new direct C/EBPα target. A in silico analysis identified NOTCH1 as a putative target of miR-30c, and by using a luciferase assay we could show that NOTCH1 is directly regulated by miR-30c. Here we show that an induction of C/EBPα-p42, but not of C/EBPα-p30, leads to an increased miR-30c expression and, consequently, downregulation of the direct target gene NOTCH1. A block of miR-30c by locked nucleic acids (LNAs) prevents the C/EBPα-induced repression of Notch1 protein expression; thus, miR-30c is necessary for C/EBPα to block Notch1. Furthermore, a block of miR-30c leads to reduced CD11b expression during granulopoiesis.
Our study supports a new molecular network involving miR-30c, C/EBPα, and Notch1 as major components of the granulocyte differentiation program.
Methods
Human cell samples from AML patients
AML patient samples were obtained from University Hospital Münster (Münster, Germany), University Hospital Leipzig (Leipzig, Germany), and Munich Leukemia Laboratory ( Munich, Germany). The study protocols used for AML patient sample collection were approved by the ethics committees of the participating centers. All patients provided written informed consent in accordance with the Declaration of Helsinki. All samples were analyzed by cytogenetic and molecular genetic analysis.
Hematopoietic CD34+ cells were isolated from leukapherisates using a CD34+ selection kit (Miltenyi Biotec). The percentage of cells positive for the CD34 antigen was analyzed by fluorescence-activated cell sorting (FACS) analysis using PE-conjugated mouse anti-human CD34 antibody (BD Bioscience) and was found to be around 93%.
Cell cultures
293T cells for lentivirus production were cultivated in Dulbecco’s modified Eagle’s medium complemented with 10% heat-inactivated fetal bovine serum (FBS) and 1% penicillin-streptomycin. K562-C/EBPα-p42-ER, K562-C/EBPα-p30-ER, and K562-ER cells were maintained in RPMI 1640 without phenol red, supplemented with 10% charcoal-treated FBS, 1% penicillin-streptomycin, and 1 μg/mL puromycin.21 K562 and U937 cells were cultured in RPMI 1640 supplemented with 10% FBS and 1% penicillin-streptomycin. For differentiation of K562-C/EBPα-ER cell lines, cells (1 × 106) were induced with 5 μM β-estradiol (Sigma) dissolved in ethanol. U937 cells (1 × 106) were induced by the addition of 1 µM of retinoic acid (Sigma-Aldrich) dissolved in dimethyl sulfoxide. MV4-11 cells were maintained in Iscove’s modified Dulbecco’s medium (IMDM) supplemented with 10% FBS and 1% penicillin-streptomycin. The primary human CD34+ progenitor cells were cultivated at 2.5 × 105/mL in IMDM supplemented with 10% FBS, 50 ng/µL stem cell factor, 50 ng/µL FLT3-Ligand, and 50 ng/µL IL-6. IL-6 was replaced with 50 ng/µL IL-3 after the first 3 days of culture. Granulocytic differentiation was induced with 100 ng/µL granulocytic colony-stimulating factor.22 Treatment of the cells was done every third day of the experiment process.
Inducible C/EBPα knockout mice
The adult C/EBPaflox/flox;Mx-Cre conditional knockout mice8 were treated with polyinosinic-polycytidylic acid (poly I:C) to induce Mx-Cre recombinase–mediated excision of C/EBPα in hematopoietic cells. Briefly, 600 µg Poly I:C was given by intraperitoneal injection every alternate day for a total of 5 injections, and mice were sacrificed 2 weeks after the last injection. Bone marrow was harvested and stained for myeloid precursors to confirm the block on myeloid differentiation. We sorted for LSK (Lin– cKit+ Sca1+), common myeloid precursors (CMP), granulocytic-macrophage precursors, megakaryocyte-erythrocyte precursors, and granulocytes using FACS (ARIA).
miRNA and mRNA detection by quantitative real-time PCR
Total RNA from cells was extracted using TRIzol reagent (Invitrogen). The miRNA quantification was performed as previously described12 by using RNUB6 expression for normalization. Corresponding reverse transcription (RT) and polymerase chain reaction (PCR) primers for RNUB6, snoRNA-202, snoRNA-135, miRNA-30c, and miR-223 were obtained from Applied Biosystems. mRNA amplification was performed as previously described12 by using glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression for normalization. All PCR reactions were performed in triplicate in a Rotor-Gene RG-3000 cycler (Corbett Research Australia). Primer sequences are provided in supplemental Table 1 (available on the Blood Web site).
Immunoblot analyses
Immunoblot analyses were performed as previously described.12 For Notch1 protein detection, a rabbit monoclonal antibody anti-Notch1 (Epitomics), and for Trib2 protein detection, the mouse monoclonal antibody anti-Trib2 (sc-100878; Santa Cruz Biotechnology) were used. The polyclonal rabbit anti-GAPDH (sc-25778) antibody was used for normalization. The immunoreactivity was determined using an enhanced chemiluminescence method (Amersham Biosciences) per the manufacturer’s instructions. The band intensities were quantified using ImageJ software (National Institutes of Health, Bethesda, MD).
DNA constructs and cloning
The C/EBPα-expressing lentiviral vector was prepared by digesting pBluescript-C/EBPα with BamHI and XhoI, repairing ends with DNA polymerase I (Klenow fragment), and ligating into the XbaI-digested pCDH1-MCS1-EF1-copGFP plasmid (System Biosciences).
The wild-type C/EBPα-pcDNA3 construct, as well as the control vector pcDNA3, has been published previously.4
For the NOTCH1 3′ untranslated region (UTR) luciferase vector, the UTR sequence was amplified from human genomic DNA and inserted into the unique XbaI restriction site 3′ to the luciferase gene in the pGL3-control plasmid (Promega). The primer sequences have been reported before.23 The pGL3 mutant NOTCH1 3′UTR vector was generated using the QuikChange II XL Site-Directed Mutagenesis kit (Stratagene) according to the manufacturer’s instructions.
For the miR-30c expression vector, the mature miR-30c sequence was cloned into the pcDNA 6.2-GW/EmGFP-miR plasmid using the BLOCK-iT Pol II miR RNAi Expression Vector Kit (Invitrogen).
All primer sequences are provided in supplemental Table 1. The correct assembly of the vectors was verified by sequencing.
Lentiviral infection
293T cells were cotransfected using calcium-phosphate transfection with either an empty vector or C/EBPα-pCDH1-MCS1-EF1-copGFP, along with the packaging plasmids pVSVG, pRSV-Rev, and pMDL g/pRRE. Virus-containing supernatants were collected at 24 and 48 hours after transfection, filtered through a 0.45-µm filter and centrifuged at 35 000 rpm for 2 hours at 4°C in a Sorvall WX ultracentrifuge using a TH-641 rotor. Pellets were resuspended in phosphate-buffered saline and stored at −80°C. Lentiviral transduction of K562 cells was performed in 6-well culture dishes for 2 consecutive days.
Chromatin immunoprecipitation
The chromatin immunoprecipitation assay was performed using ChIP-IT Express Enzymatic (Active Motif) according to the manufacturer’s instructions. Primer sequences are provided in supplemental Table 1.
Transfections
Transfection of C/EBPα-expressing construct (C/EBPα-pcDNA3) in K562 cells was performed with the Nucleofector kit (Lonza). 5 μg of DNA plasmid was used for each transfection. Locked nucleic acid (LNA) oligonucleotides were obtained from Exiqon. LNAs (200 nM) were individually transfected in K562 and K562-C/EBPα-p42-ER cells using the Nucleofector kit (Lonza). The pcDNA6.2-GW/EmGFP-miR-30c and pGL3-3′UTR-Notch1 were transfected in MV4-11 cells using Lipofectamine LTX (Invitrogen).
Luciferase reporter assay
To test whether miR-30c directly targets Notch1, MV4-11 cells were transiently transfected with 0.7 µg of each reporter construct (wild-type or mutant NOTCH1 3′UTR), 0.1 µg of Renilla construct, and 1.4 µg pcDNA6.2-GW/EmGFP-miR-30c or control plasmid by lipofectamine LTX (Invitrogen) as described by the manufacturer. Firefly luciferase activities from the promoter constructs and Renilla luciferase activity from the internal control plasmid were determined 24 hours after transfection using the Dual-Luciferase Reporter Assay System (Promega). Values were normalized by using the Renilla luciferase.
Cell differentiation
Cell differentiation was assessed by FACS analysis using PE-conjugated mouse anti-human CD11b antibody (BD Biosciences) as previously described.24
Statistical analysis
We used Student t test to determine the statistical significance of experimental results. A P value ≤ .05 was considered significant. The results were represented as the mean ± SD from 3 independent experiments.
Results
C/EBPα-p42 upregulates miR-30c during granulopoiesis
Past studies have shown a high expression of miR-30c in granulocytes.14,15 To assess the role of miR-30c during granulopoiesis, we analyzed its expression upon lentiviral transduction of C/EBPα in K562 cells. With a quantitative real-time polymerase chain reaction (Q-RT-PCR) in K562, we observed a high expression level of miR-30c (Figure 1A). We also analyzed the expression levels of miR-223, a miRNA that has been reported to be upregulated during granulopoiesis.11,12 As expected, miR-223 expression is increased upon C/EBPα overexpression (Figure 1A). To further understand the regulation of miR-30c by C/EBPα proteins, we used an inducible cell line model K562-C/EBPα-ER. These cells do not express endogenous C/EBPα and have been reported as a model for granulopoiesis in the context of C/EBPα proteins.21 K562-C/EBPα-ER cell lines were established by stably transfecting K562 cells with a plasmid encoding an estrogen-inducible C/EBPα (p42 form as well as mutant p30) estrogen receptor fusion protein.21 After treatment with β-estradiol, the K562-C/EBPα-p42ER cells undergo myeloid differentiation.13 Our data show that activation of C/EBPα leads to an upregulation of miR-30c expression in K562- C/EBPα-p42ER cells (Figure 1B). The expression of miR-30c was found to be high up to 12 hours after β-estradiol treatment. D’Alo et al could show that C/EBPα is able to differentiate K562-C/EBPα-p42ER cells during a very short time interval.21 CEBPA N-terminal frame-shift mutation is responsible for premature termination of the full-length C/EBPα-p42 isoform, while keeping the truncated C/EBPα-p30 protein intact,4 and is associated with AML in mice.19 Therefore, we investigated whether the p30 mutant form mediates miR-30c expression. We observed that treatment of K562-C/EBPα-p30ER cells with β-estradiol fails to show any increase in miR-30c expression (Figure 1C). We did not observe any change in miR-30c expression in the control cell line K562-ER (Figure 1D). These data suggest that upregulation of miR-30c during granulocytic differentiation is a specific function of the C/EBPα-p42 isoform. To further understand the regulation of miR-30c in granulocytic differentiation, we analyzed the expression during retinoic acid (RA)-induced differentiation of U937 cells. We observed that RA is able to induce the expression of miR-30c (Figure 1E). To assess the role of miR-30c during granulopoiesis, we analyzed its expression during differentiation of primary human CD34+ hematopoietic progenitor cells obtained from leukapheresis. CD34+ cells were cultured in the presence of a sequential cytokine cocktail that has been shown to induce granulopoiesis.22 We observed granulocytic differentiation of CD34+ cells as evaluated by expression of granulocytic markers.13 miR-30c expression was analyzed by RT-PCR 7 and 14 days after induction of differentiation. We observed a gradual increase of miR-30c expression during granulocytic differentiation (Figure 1F).
C/EBPα-p42 regulates miR-30c during granulopoiesis. (A) Lentiviral overexpression of C/EBPα in K562 cells. Total RNA was isolated at day 7 and analyzed by Q-RT-PCR with oligos for miR-30c and miR-223. Values were normalized to U6. Data are represented as mean ± SD from 3 independent experiments. *P ≤ .05. (B) K562-C/EBPα-p42ER, (C) K562-C/EBPα-p30ER, and (D) K562-ER cells were induced with β-estradiol (5 µM) for respective time points. Total RNA was analyzed by Q-RT-PCR with oligos for miR-30c. Values were normalized to U6. Data are represented as mean ± SD from 3 independent experiments. *P ≤ .05. (E) U937 cells were induced with retinoic acid (1 µM) for respective time points. Total RNA was analyzed by Q-RT-PCR with oligos for miR-30c. Values were normalized to U6. Data are represented as mean ± SD from 3 independent experiments. *P ≤ .05. (F) Hematopoietic CD34+ cells were cultured as discussed in Methods. Total RNA was isolated at different time points and analyzed by Q-RT-PCR with oligos for miR-30c. Values were normalized to U6. Data are represented as mean ± SD from 2 independent experiments.
C/EBPα-p42 regulates miR-30c during granulopoiesis. (A) Lentiviral overexpression of C/EBPα in K562 cells. Total RNA was isolated at day 7 and analyzed by Q-RT-PCR with oligos for miR-30c and miR-223. Values were normalized to U6. Data are represented as mean ± SD from 3 independent experiments. *P ≤ .05. (B) K562-C/EBPα-p42ER, (C) K562-C/EBPα-p30ER, and (D) K562-ER cells were induced with β-estradiol (5 µM) for respective time points. Total RNA was analyzed by Q-RT-PCR with oligos for miR-30c. Values were normalized to U6. Data are represented as mean ± SD from 3 independent experiments. *P ≤ .05. (E) U937 cells were induced with retinoic acid (1 µM) for respective time points. Total RNA was analyzed by Q-RT-PCR with oligos for miR-30c. Values were normalized to U6. Data are represented as mean ± SD from 3 independent experiments. *P ≤ .05. (F) Hematopoietic CD34+ cells were cultured as discussed in Methods. Total RNA was isolated at different time points and analyzed by Q-RT-PCR with oligos for miR-30c. Values were normalized to U6. Data are represented as mean ± SD from 2 independent experiments.
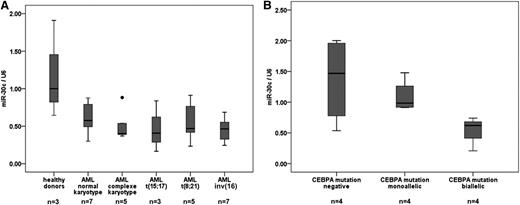
miR-30c is downregulated in normal karyotype AML patient samples with CEBPA mutations
Because miR-30c is regulated by the C/EBPα-p42 isoform and the mutant C/EBPα-p30 fails to regulate it, we hypothesized that this miRNA could be downregulated in AML patient samples, especially in normal karyotype AML with CEBPA mutations. Recent studies found CEBPA mutations predominantly in cytogenetically normal AML,4,5 having been reported in 13% to 19% of young AML patients.4,25,26 To understand its regulation in AML, the expression levels of miR-30c were quantified in diagnostic samples of AML patients. In general, miR-30c levels were substantially repressed in AML samples compared with healthy donors (Figure 2A). In particular, we found that miR-30c is slightly lower when expressed in normal karyotype AML patient samples with monoallelic CEBPA mutations, and is strikingly low when expressed in AML patient samples with biallelic CEBPA mutations in comparison with normal karyotype AML patient samples without CEBPA mutations (Figure 2B). Genetic and morphologic features of AML samples used are shown in supplemental Tables 2 and 3.
miR-30c is downregulated in normal karyotype AML patient samples with CEBPA mutations. (A) Q-RT-PCR for miR-30c was carried out using bone marrow cells derived from AML patient samples (n = 30) and healthy donors (n = 3). Values were normalized to U6 and further to the expression level of 3 healthy donors. (B) Q-RT-PCR for miR-30c was carried out using bone marrow cells derived from AML patient samples with monoallelic (n = 4), biallelic (n = 4), and without CEBPA mutations (n = 4). Values were normalized to U6 and further to the expression level of CEBPA mutations negative.
miR-30c is downregulated in normal karyotype AML patient samples with CEBPA mutations. (A) Q-RT-PCR for miR-30c was carried out using bone marrow cells derived from AML patient samples (n = 30) and healthy donors (n = 3). Values were normalized to U6 and further to the expression level of 3 healthy donors. (B) Q-RT-PCR for miR-30c was carried out using bone marrow cells derived from AML patient samples with monoallelic (n = 4), biallelic (n = 4), and without CEBPA mutations (n = 4). Values were normalized to U6 and further to the expression level of CEBPA mutations negative.
miR-30c is downregulated in inducible C/EBPα-knockout mice bone marrow cells
To assess the role of the miR-30c in vivo and to analyze the expression during myelopoiesis, we performed Q-RT-PCR on sorted precursor populations and mature granulocytes from mouse bone marrow. We observed a significantly increased expression of miR-30c in granulocytes (Figure 3A).
Regulation of miR-30c in inducible C/EBPα-knockout mice. (A) Q-RT-PCR for miR-30c in sorted mouse bone marrow cell populations. Values were normalized with snoRNA-135. Total data are represented as mean ± SD from 3 independent mice. *P ≤ .05. LSK (Lin– cKit+ Sca1− contains hematopoietic cells, both LTHSCs and STHSCs), CMP (common myeloid progenitors), GMP (granulocyte-macrophage precursors), MEP (megakaryocyte-erythrocyte precursors), and granulocytes. (B-C) Q-RT-PCR for C/EBPα (B) and miR-30c (C) was carried out using bone marrow cells derived from poly(I:C)-injected C/EBPαfl/fl;Mx-Cre (C/EBPαΔ/Δ) and C/EBPαfl/fl control. Values were normalized with GAPDH and snoRNA-202, respectively, and further to the expression of C/EBPαfl/fl control. Total data are represented as mean ± SD from 5 independent mice. **P ≤ .005.
Regulation of miR-30c in inducible C/EBPα-knockout mice. (A) Q-RT-PCR for miR-30c in sorted mouse bone marrow cell populations. Values were normalized with snoRNA-135. Total data are represented as mean ± SD from 3 independent mice. *P ≤ .05. LSK (Lin– cKit+ Sca1− contains hematopoietic cells, both LTHSCs and STHSCs), CMP (common myeloid progenitors), GMP (granulocyte-macrophage precursors), MEP (megakaryocyte-erythrocyte precursors), and granulocytes. (B-C) Q-RT-PCR for C/EBPα (B) and miR-30c (C) was carried out using bone marrow cells derived from poly(I:C)-injected C/EBPαfl/fl;Mx-Cre (C/EBPαΔ/Δ) and C/EBPαfl/fl control. Values were normalized with GAPDH and snoRNA-202, respectively, and further to the expression of C/EBPαfl/fl control. Total data are represented as mean ± SD from 5 independent mice. **P ≤ .005.
miR-30c is a direct target of C/EBPα
Next we investigated whether C/EBPα-p42 directly binds to miR-30c. We found several putative C/EBPα binding sites in the minimal promoter regions upstream of miR-30c-1, miR-30c-1 host gene NFYC, and miR-30c-2. Therefore, we performed chromatin immunoprecipitation experiments in K562-C/EBPα-p42ER cells. The cells were treated for with β-estradiol for 4 hours, and the chromatin fragments were immunoprecipitated with an anti-C/EBPα antibody. The DNA fragments were analyzed with specific primers for the indicated regions of the miR-30c regulatory region (Figure 4A). We could observe an enrichment of DNA from all predicted C/EBPα binding sites (Figure 4B) compared with the immunoglobulin G (IgG) control. Furthermore, we could see a changing in the binding affinity of chromatin after C/EBPα induction in K562 cells compared with the control, as measured by Q-RT-PCR (Figure 4C). These data suggest that C/EBPα-p42 interacts with the miR-30c regulatory region.
miR-30c is a direct target of C/EBPα. (A) Schematic representation of examined putative C/EBPα binding sites (α) in the promoter regions of miR-30c-1, the host gene NFYC, and miR-30c-2 (primer #1-#4). (B) Chromatin derived from K562-C/EBPα-p42-ER cells induced for 4 hours with β-estradiol was immunoprecipitated with anti-C/EBPα or IgG antibodies. Recovered DNA was PCR amplified with different primers specific for C/EBPα-binding amplificon (primers #1-#4). (C) Q-RT-PCR with primers for indicated regions with the samples from (B). Shown is the fold change in binding affinity of the anti-C/EBPα antibody normalized to IgG from β-estradiol (active C/EBPα) –treated vs control vehicle–treated cells. Total data are represented as mean ± SD from 2 independent experiments.
miR-30c is a direct target of C/EBPα. (A) Schematic representation of examined putative C/EBPα binding sites (α) in the promoter regions of miR-30c-1, the host gene NFYC, and miR-30c-2 (primer #1-#4). (B) Chromatin derived from K562-C/EBPα-p42-ER cells induced for 4 hours with β-estradiol was immunoprecipitated with anti-C/EBPα or IgG antibodies. Recovered DNA was PCR amplified with different primers specific for C/EBPα-binding amplificon (primers #1-#4). (C) Q-RT-PCR with primers for indicated regions with the samples from (B). Shown is the fold change in binding affinity of the anti-C/EBPα antibody normalized to IgG from β-estradiol (active C/EBPα) –treated vs control vehicle–treated cells. Total data are represented as mean ± SD from 2 independent experiments.
NOTCH1 is a direct target of miR-30c
Using the TargetScanHuman 6.0 (www.targetscan.org) prediction algorithm, we identified a putative binding site of miR-30c in the 3′UTR region of NOTCH1, which is highly conserved in human, mice, rat, and dog (Figure 5A-B). We hypothesized that NOTCH1 is the key target of miR-30c.
NOTCH1 is a direct target of miR-30c. (A) Schematic representation of miR-30c site in the human NOTCH1 3′UTR. The numbers (+1520 - +1526) represent the nucleotides relative to the termination codon of human NOTCH1. (B) Conservation of the miR-30c binding site in the NOTCH1 3′UTR of human, mice, rat, and dog. (C) K562-C/EBPα-p42ER and (D) K562-C/EBPα-p30ER cells were induced with β-estradiol (5 µM) for respective time points. Total protein was analyzed by western blot analysis with the anti-Notch1 antibody. Values below the gel image indicate the Notch1 protein levels normalized to GAPDH. Total RNA was analyzed by Q-RT-PCR with oligonucleotides for Notch1. Data are represented as mean ± SD from 3 independent experiments. (E) K562-C/EBPα-p42ER cells were induced with β-estradiol (5 µM) for 6 hours. Total protein was analyzed by western blot analysis with anti-Trib2 antibody. Values below the gel image indicate the Trib2 protein levels normalized to GAPDH. (F) K562 cells were transfected with control and pcDNA6.2-GW/EmGFP-miR-30c vectors and cultivated for 48 hours. Total protein was analyzed by western blot analysis with the anti-Notch1 antibody. Values below the gel image indicate the Notch1 protein levels normalized to GAPDH. (G) Hematopoietic CD34+ cells were cultured as discussed in “Methods.” Total protein was analyzed by western blot analysis with the anti-Notch1 antibody. Values below the gel image indicate the Notch1 protein levels normalized to GAPDH. (H) Schematic representation of predicted and mutated miR-30c binding site in the luciferase construct used for reporter assays. (I) Luciferase assays in MV4-11 cells cotransfected with Notch1 3′UTR constructs (wild-type and mutant) and pcDNA6.2-GW/EmGFP-miR-30c. The bars represent luciferase activity for the corresponding vectors. Data are represented as mean ± SD from 3 independent experiments. **P ≤ .01.
NOTCH1 is a direct target of miR-30c. (A) Schematic representation of miR-30c site in the human NOTCH1 3′UTR. The numbers (+1520 - +1526) represent the nucleotides relative to the termination codon of human NOTCH1. (B) Conservation of the miR-30c binding site in the NOTCH1 3′UTR of human, mice, rat, and dog. (C) K562-C/EBPα-p42ER and (D) K562-C/EBPα-p30ER cells were induced with β-estradiol (5 µM) for respective time points. Total protein was analyzed by western blot analysis with the anti-Notch1 antibody. Values below the gel image indicate the Notch1 protein levels normalized to GAPDH. Total RNA was analyzed by Q-RT-PCR with oligonucleotides for Notch1. Data are represented as mean ± SD from 3 independent experiments. (E) K562-C/EBPα-p42ER cells were induced with β-estradiol (5 µM) for 6 hours. Total protein was analyzed by western blot analysis with anti-Trib2 antibody. Values below the gel image indicate the Trib2 protein levels normalized to GAPDH. (F) K562 cells were transfected with control and pcDNA6.2-GW/EmGFP-miR-30c vectors and cultivated for 48 hours. Total protein was analyzed by western blot analysis with the anti-Notch1 antibody. Values below the gel image indicate the Notch1 protein levels normalized to GAPDH. (G) Hematopoietic CD34+ cells were cultured as discussed in “Methods.” Total protein was analyzed by western blot analysis with the anti-Notch1 antibody. Values below the gel image indicate the Notch1 protein levels normalized to GAPDH. (H) Schematic representation of predicted and mutated miR-30c binding site in the luciferase construct used for reporter assays. (I) Luciferase assays in MV4-11 cells cotransfected with Notch1 3′UTR constructs (wild-type and mutant) and pcDNA6.2-GW/EmGFP-miR-30c. The bars represent luciferase activity for the corresponding vectors. Data are represented as mean ± SD from 3 independent experiments. **P ≤ .01.
Because C/EBPα-p42 directly regulates miR-30c (Figures 1B and 4) and NOTCH1 is a putative target of miR-30c (Figure 5A), we analyzed the potential of the C/EBPα-p42 protein in regulating the Notch1 protein. C/EBPα-p42 function in K562 cells is able to cause suppression of the Notch1 protein, although its transcript levels were unaltered (Figure 5C) after C/EBPα induction. We found an inverse correlation of miR-30c expression (Figure 1B) and Notch1 protein level (Figure 5C). The mutant C/EBPα (C/EBPα-p30) fails to downregulate the Notch1 protein level (Figure 5D).
Recent works have shown that TRIB2 is a transcriptional target of NOTCH1,27 which directly inhibits the function of C/EBPα-p42 and causes AML in mice.28,29 Therefore, we examined the protein expression of Trib2 upon C/EBPα induction in K562-C/EBPα-p42ER cells. We observed a decrease in Trib2 protein after 6 hours. The decrease of Trib2 is temporally displaced to the downregulation of Notch1, which indicates TRIB2 as a transcriptional target of NOTCH1 (Figure 5E). These data hint that C/EBPα-p42 plays a critical role in the regulation of the Notch1 protein level during granulocytic differentiation. To validate NOTCH1 as a target of miR-30c, we analyzed Notch1 protein levels after overexpression of miR-30c in K562 cells and found reduced Notch1 protein levels (Figure 5F). Furthermore, we detected a decrease of the Notch1 protein expression in human CD34+ cells during granulocytic differentiation (Figure 5G).
To test whether NOTCH1 is a direct target of miR-30c, we conducted luciferase reporter assays in MV4-11 cells, which do not express miR-30c,14 with a construct containing 3′UTR of the NOTCH1 gene with the putative binding site (Notch1 3′UTR WT; Figure 5H). We also generated a control vector containing a mutated miR-30c binding site (Notch1 3′UTR mut; Figure 5H). These reporter constructs were cotransfected with either pcDNA6.2-GW/EmGFP-miR-30c or a control vector. Relative luciferase activity was significantly decreased 24 hours after miR-30c transfection compared with transfection with the control vector pcDNA6.2-GW/EmGFP-miR-30c, indicating that miR-30c targets Notch1 mRNA via a direct interaction with the 3′UTR. The vector containing the mutated miR-30c binding site shows no reduced luciferase activity (Figure 5I), indicating a miR-30c-specific inhibition of Notch1.
miR-30c is necessary for C/EBPα to block Notch1 and to induce myeloid differentiation
Because miR-30c is directly regulated by C/EBPα-p42 (Figures 1B and 4) and directly targets Notch1 (Figure 5), we hypothesized that C/EBPα blocks Notch1 protein expression through upregulation of miR-30c and that this is necessary for granulocytic differentiation. Blocking of miR-30c with LNA oligonucleotides in K562 cells led to an increase of Notch1 protein expression. Moreover, the suppression of Notch1 caused by ectopic expression of C/EBPα was substantially alleviated by blocking miR-30c (Figure 6A). Collectively, these data suggest that miR-30c is necessary for C/EBPα to block Notch1 protein expression during granulocytic differentiation.
miR-30c is necessary for C/EBPα to block Notch1 and to induce myeloid differentiation. (A) K562 cells were cotransfected with pcDNA3-C/EBPα or pcDNA3-empty vector along with LNA oligonucleotides for miR-30c or scramble (control) for 48 hours. Total protein was analyzed by western blot analysis with anti-Notch1 antibody. Values below the gel image indicate the Notch1 protein levels normalized to GAPDH. (B) FACS analysis of CD11b expression during blocking of miR-30c with LNA oligonucleotides in β-estradiol (5 µM)–treated K562-C/EBPα-p42ER cells by flow cytometry. Representative FACS data are from K562-C/EBPα-p42ER cells transfected for 24 hours. Total data are represented as mean ± SD from 2 representative experiments. *P ≤ .005.
miR-30c is necessary for C/EBPα to block Notch1 and to induce myeloid differentiation. (A) K562 cells were cotransfected with pcDNA3-C/EBPα or pcDNA3-empty vector along with LNA oligonucleotides for miR-30c or scramble (control) for 48 hours. Total protein was analyzed by western blot analysis with anti-Notch1 antibody. Values below the gel image indicate the Notch1 protein levels normalized to GAPDH. (B) FACS analysis of CD11b expression during blocking of miR-30c with LNA oligonucleotides in β-estradiol (5 µM)–treated K562-C/EBPα-p42ER cells by flow cytometry. Representative FACS data are from K562-C/EBPα-p42ER cells transfected for 24 hours. Total data are represented as mean ± SD from 2 representative experiments. *P ≤ .005.
A block of miR-30c by LNA oligonucleotides in K562-C/EBPα-p42ER cells induced with β-estradiol to undergo myeloid differentiation leads to a reduced number of CD11b+ cells (Figure 6B). These data illustrate that miR-30c is necessary for C/EBPα to induce myeloid differentiation.
Discussion
The transcription factor C/EBPα is a master regulator in normal hematopoiesis.30 Previous studies have shown that C/EBPα acts via regulation of specific miRNAs. Fazi et al first identified miR-223 as a direct target of C/EBPα.11 The C/EBPα-mediated upregulation of miR-223 leads to granulocytic differentiation.11,12 Recent studies from our group show a direct regulation of miR-223 and miR-34a by C/EBPα in normal granulopoiesis. C/EBPα transactivates these miRNAs and acts as a tumor suppressor gene in this way.12,13 Moreover, in AML in which C/EBPα is deregulated, the transactivation of miR-34a and miR-223 is inhibited, and myeloid differentiation is blocked.11-13
In the present study, we identified the miR-30c gene as a novel direct target of C/EBPα-p42 in granulopoiesis (Figure 4). Although high expression of miR-30c in granulocytes has been indicated,14,15 specific regulation and function of this miRNA has not been addressed. Therefore, we first examined the role of miR-30c in C/EBPα-induced granulocytic differentiation and AML. The 2 major mutations reported for C/EBPα are distributed in the N- and C-terminal domains, signifying the relevance of these domains of C/EBPα in leukemogenesis. Both mutations are predicted to confer loss of function in the C/EBPα protein as the resulting mutant forms fail to induce granulopoiesis, are defective in inhibiting myeloid cell proliferation,21 and induce AML.31 The N-terminal frame-shift mutation of C/EBPα is responsible for premature termination of the full-length C/EBPα-p42 isoform, while keeping the truncated C/EBPα-p30 protein intact.4 To examine the regulation of miR-30c by 2 C/EBPα isoforms, we used the K562-C/EBPα-ER cell lines stably transfected with plasmid encoding an estrogen-inducible C/EBPα (either p42 form or the mutant p30 form) estrogen-receptor fusion protein.21 Our data show that only C/EBPα-p42 regulates miR-30c during granulocytic differentiation (Figure 1B), whereas the shorter-form C/EBPα-p30 fails to induce the expression of miR-30c (Figure 1C). Mutations in the CEBPA gene are reported for ∼10% of all AMLs7 and are predominantly found in cytogenetically normal AML,4,5 having been reported in 13% to 19% of young AML patients.4,25,26 Our analyses show that miR-30c is suppressed in different subtypes of AML (Figure 2A). Direct comparison of expression levels in normal karyotype AML clearly indicates reduced levels of miR-30c in samples with especially biallelic CEBPA mutations (Figure 2B). These observations suggest that miR-30c is a key mediator of tumor suppressor function of C/EBPα in AML; likewise, other studies in breast cancer and AML with NPM1 mutations showed a tumor suppressor–like function of miR-30c.19,20 An in vivo investigation of the whole-mouse bone marrow cells showed an increased expression of miR-30c in granulocytes (Figure 3A). A disruption of the C/EBPα gene in adult mice selectively blocks granulocytic development.8 Induction of Cre recombinase activity in C/EBPαfl/fl Mx-Cre mice resulted in hematopoietic-specific knockout of C/EBPα as evident by the arrest of myeloid differentiation at the CMP stage. We observed a significant downregulation of miR-30c in the bone marrow lacking C/EBPα compared with control (Figure 3B-C).
In the current study, we first showed that NOTCH1 is a crucial direct-target gene of miR-30c. We identified a conserved binding site of miR-30c in the 3′UTR of Notch1 using a luciferase reporter assay in MV4-11 cells (Figure 5). NOTCH1 encodes a membrane receptor and transcriptional regulator, which plays a critical role in T-cell development.32 Recent studies have shown that an activation of NOTCH1 signaling either by treatment with Notch ligands or by enforced expression of the constitutively activated NOTCH1 gene resulted in inhibition of granulocytic differentiation and preservation of a more immature phenotype in the myeloid cell lines 32D and HL-60, and in primary murine and human myeloid progenitor cells.33-36 Notch signaling can play both oncogenic and tumor-suppressor roles in solid tumors; in the hematopoietic system, it is exclusively oncogenic, notably in T-cell acute lymphocytic leukemia, a disease characterized by Notch1-activating mutations.37 Our data show first that an induction of C/EBPα-p42 is able to downregulate the protein but not the mRNA level of Notch1 (Figure 5C). miR-30c expression (Figure 1B) and Notch1 protein (Figure 5C) show an inverse correlation. C/EBPα-p30 fails to downregulate the Notch1 protein and mRNA level (Figure 5D). A specific subset of human AMLs associated with impaired C/EBPα function also shows a high Trib2 expression.27,28 Because Wouters et al identified TRIB2 as a direct transcriptional target of oncogenic NOTCH1,27 which functionally inactivates C/EBPα by causing its degradation,28,29 we also analyzed the Trib2 protein level by C/EBPα-p42 overexpression and observed a decrease (Figure 5F). Our data suggest that C/EBPα-p42 plays a critical role in the regulation of the Notch1 protein level during granulocytic differentiation. An inhibition of miR-30c with LNA oligonucleotides in K562 cells leads to an increase in the Notch1 protein level (Figure 6A) and to a reduced number of CD11b+ cells during myeloid differentiation (Figure 6B). Collectively, our study indicates that miR-30c is necessary for C/EBPα to block Notch1 protein and induce myeloid differentiation. A recent study shows that a block of miR-30 by antisense oligonucleotides enhances self-renewal, tumor regeneration, and metastasis in differentiated breast cancer cells.19 Even though miR-30c has been shown to be involved in AML with NPM1 mutations19 and its high expression levels in granulocytes,14,15 little is known about the regulation of this miRNA in granulopoiesis. Our data reveal novel insights into the function of miR-30c in granulocytic differentiation.
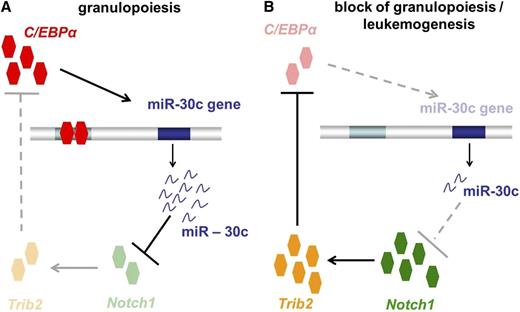
In summary, our study demonstrates that miR-30c plays a critical role in regulating the myeloid differentiation induced by C/EBPα (Figure 7A). There is strong evidence that miR-30c acts via repression of Notch1 and Trib2 to promote granulocytic differentiation. In AML, especially in AML with CEBPA mutations, loss of function of C/EBPα results in a block of miR-30c expression, which in turn results in a lack of Notch1 and Trib2 inhibition (Figure 7B). Our study makes evident that C/EBPα, miR-30c, Notch1, and Trib2 together play a critical role in granulocytic differentiation and AML, particularly AML with CEBPA mutations. This new molecular network may provide miR-30c as a novel biomarker and therapeutic target in AML.
Model of how transcription factor C/EBPα-induced microRNA miR-30c inactivates Notch1 during granulopoiesis and is downregulated in AML. (A) During granulopoiesis, C/EBPα transactivates miR-30c, which in turn leads to Notch1 and Trib2 repression, resulting in myeloid differentiation. (B) In AML, C/EBPα fails to transactivate miR-30c, which results in a lack of Notch1 inhibition.
Model of how transcription factor C/EBPα-induced microRNA miR-30c inactivates Notch1 during granulopoiesis and is downregulated in AML. (A) During granulopoiesis, C/EBPα transactivates miR-30c, which in turn leads to Notch1 and Trib2 repression, resulting in myeloid differentiation. (B) In AML, C/EBPα fails to transactivate miR-30c, which results in a lack of Notch1 inhibition.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This study was supported by grants from DFG (German Research Foundation), Deutsche Krebshilfe, Wilhelm-Sander-Stiftung (G.B.), German José Carreras Foundation (G.B., C.K.), and the National Institutes of Health (CA118316) (D.G.T.).
Authorship
Contribution: C.K. and G.B. designed the research, analyzed the data, and wrote the paper; G.B. supervised the work; C.K., V.M., A.A.W., D.G., and J.-U.H. performed experiments; C.M.-T. provided AML patient samples; S.S. provided AML patient samples with CEBPA mutations; D.G.T. provided K562-ER cell lines and inducible C/EBPα knockout mice; and V.M., D.G., A.A.W., D.B.-H., J.-U.H., S.S., D.G.T., and D.N. commented on the paper.
Conflict-of-interest disclosure: S.S. declares equity ownership in the MLL Munich Leukemia Laboratory. The remaining authors declare no competing financial interests.
Correspondence: Prof Dr med Gerhard Behre, Division of Hematology and Oncology, Leipzig University Hospital, Johannisallee 32A, 04103 Leipzig, Germany; e-mail: gerhard.behre@medizin.uni-leipzig.de.






This feature is available to Subscribers Only
Sign In or Create an Account Close Modal