Key Points
Small deletions in the RPS14 region of 5q must be considered in atypical 5q− syndrome and nonclassical Diamond Blackfan anemia.
Abstract
Classical 5q− syndrome is an acquired macrocytic anemia of the elderly. Similar to Diamond Blackfan anemia (DBA), an inherited red cell aplasia, the bone marrow is characterized by a paucity of erythroid precursors. RPS14 deletions in combination with other deletions in the region have been implicated as causative of the 5q− syndrome phenotype. We asked whether smaller, less easily detectable deletions could account for a syndrome with a modified phenotype. We employed single-nucleotide polymorphism array genotyping to identify small deletions in patients diagnosed with DBA and other anemias lacking molecular diagnoses. Diminutive mosaic deletions involving RPS14 were identified in a 5-year-old patient with nonclassical DBA and in a 17-year-old patient with myelodysplastic syndrome. Patients with nonclassical DBA and other hypoproliferative anemias may have somatically acquired 5q deletions with RPS14 haploinsufficiency not identified by fluorescence in situ hybridization or cytogenetic testing, thus refining the spectrum of disorders with 5q− deletions.
Introduction
Diamond Blackfan anemia (DBA) is a rare, inherited1,2 red cell aplasia and cancer predisposition syndrome3 presenting at a median age of 2 months with macrocytic anemia, marrow erythroid hypoplasia, and reticulocytopenia.4 It is associated with elevated erythrocyte adenosine deaminase (eADA) activity in 80% of patients.5 Approximately 65% of patients have a mutation or deletion in 1 of 10 genes encoding ribosomal proteins (RP), resulting in RP haploinsufficiency.1,6-9 In contrast to DBA, the 5q− syndrome, a subtype of myelodysplastic syndrome (MDS), is an acquired red cell failure disorder associated with interstitial deletions of chromosome 5q, predominantly diagnosed in elderly women.10 Ebert et al showed that the erythroid defect in 5q− syndrome is caused by the monoallelic loss of RPS14,11 establishing most DBA and the 5q− syndrome as ribosomopathies.
Here we describe 2 patients, 1 diagnosed with DBA and 1 with myelodysplastic syndrome, who were found to have acquired, clonal abnormalities with smaller than previously reported deletions of 5q, which led to a successful treatment intervention in one of them.
Study design
SNP array genotyping
Single-nucleotide polymorphism (SNP) array genotyping was performed on whole-blood and lineage-specific DNA from 105 patients from the DBA Registry and 1 patient referred with an atypical red cell aplasia, as previously described.7
Estimation of mosaic monosomy
Regions of mosaic monosomy were identified by visual inspection of minor allele frequency plots. A normal continuous distribution function (CDF) of the heterozygous minor alleles was averaged from 5 dizygous regions for each array analyzed. The normal CDF was fitted to the empirical CDF of the mosaic region to estimate fractional mosaicism, as previously described.12
Accession numbers
Array data were deposited in the National Center for Biotechnology Information Gene Expression Omnibus Database under accession number GSE42570.
All clinical investigations were conducted according to Declaration of Helsinki principles. These studies were approved by The Feinstein Institute for Medical Research, North Shore-LIJ Health System institutional review board, and written informed consent was received from participants before their inclusion in the study.
Results and discussion
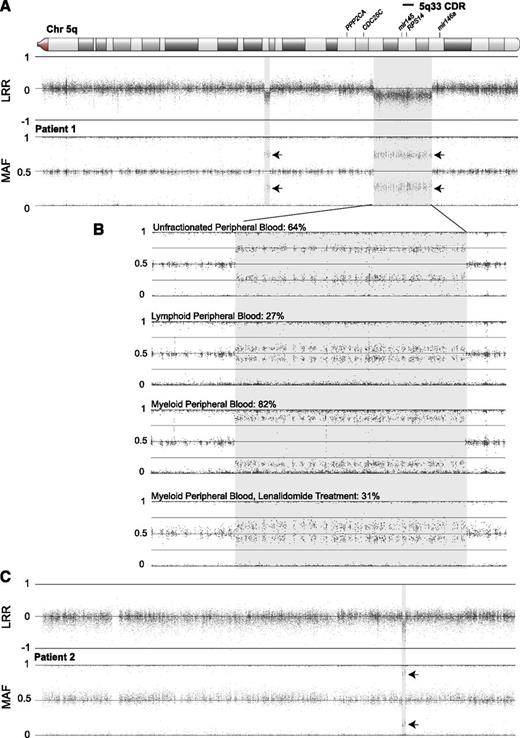
Patient 1 presented in 1991 at 5 years of age with macrocytic anemia (hemoglobin [Hb], 8.4 g/dL; mean corpuscular volume [MCV], 108.2 fL; reticulocyte count, 0.4%) and normal white blood cell (5300/µL) and platelet (279 000/µL) counts. Her bone marrow (BM) evaluation showed decreased cellularity with a marked selective decrease in red blood cell precursors, as well as normal myeloid and megakaryocyte morphology. BM karyotype done initially and repeated in 1996 was normal 46,XX. The eADA activity was 0.61 IU/gm Hb (normal, 0.33-0.96 IU/gm Hb). There were no physical anomalies or significant family history. A diagnosis of nonclassical DBA was made on the basis of the patient’s atypical presenting age. She did not respond to corticosteroids and was maintained on monthly red blood cell transfusions for approximately 20 years. Commercial mutation analysis for the known DBA-associated RP genes was negative. SNP array genotyping of peripheral blood DNA identified mosaic monosomy in 2 regions on 5q: a 16.1-Mb region and a smaller, more centromeric 1.5-Mb region (Figure 1A). The larger region includes the entire 5q− syndrome commonly deleted region (CDR) at 5q33, including RPS14.13 The monosomic fraction was 64% in whole-blood DNA, 27% in lymphoid populations, and 82% in myeloid populations (Figure 1B). This abnormality was not found in the patient’s fibroblasts or in either parent (not shown).
SNP array genotyping demonstrates mosaic deletions of chromosome 5q that overlap the 5q33 CDR in 2 patients. Regions of copy loss (shaded) are indicated by decreased log R signal ratio (LRR). Splitting of the heterozygous minor allele frequencies (MAFs) around the expected 0.5 level in regions of reduced copy number indicates mosaic monosomy/disomy (arrows). The displayed region of chromosome 5q is boxed in the ideogram. (A) Patient 1 has 2 apparently discrete regions of mosaic monosomy in 64% of peripheral blood DNA: a 16.1-Mb deletion extending from chr5:141 108 260 to 157 224 755 (involving PCDH1 to CLINT1) that includes all of the 5q33 CDR, as well as a more centromeric, 1.5-Mb deletion at chr5:110 887 600 to 112 390 739 (involving STARD4 to MCC). Similarities in LRR and MAF signal values at both regions in this and subsequent experiments (Figure 2) suggest the possibility of an intrachromosomal rearrangement joining the 2 regions. (B) Analysis of patient 1 mosaicism indicates a nonuniform distribution of copy loss in circulating cell fractions and improvement with lenalidomide treatment. Mosaicism was estimated to involve 64% of whole-blood DNA at the larger deletion region. Lymphoid-enriched (CD3+ or CD19+) and myeloid-enriched (CD3− and CD19−) DNA prepared from magnetic bead-separated peripheral blood populations showed substantial disparity, with monosomy estimated in only 27% of the lymphoid but 82% of the myeloid population. Similar analysis of the myeloid fraction from blood obtained after 3 months of treatment with lenalidomide showed marked diminution of the monosomic fraction to an estimated 31% of myeloid DNA. Similar changes were also observed in the smaller deletion region (not shown). (C) Patient 2 has an 897-Kb deletion extending from chr5:149 496 080 to 150 393 600 (involving PDGFRB to TNIP1), with monosomy in 77% of the peripheral blood DNA sample. This deletion is approximately half the size and is entirely contained within the current 5q33 CDR. The 5q33 CDR and genes with potential relevance to the 5q syndrome are indicated above the chromosome. Coordinates are expressed relative to National Center for Biotechnology Information Build 36.1.
SNP array genotyping demonstrates mosaic deletions of chromosome 5q that overlap the 5q33 CDR in 2 patients. Regions of copy loss (shaded) are indicated by decreased log R signal ratio (LRR). Splitting of the heterozygous minor allele frequencies (MAFs) around the expected 0.5 level in regions of reduced copy number indicates mosaic monosomy/disomy (arrows). The displayed region of chromosome 5q is boxed in the ideogram. (A) Patient 1 has 2 apparently discrete regions of mosaic monosomy in 64% of peripheral blood DNA: a 16.1-Mb deletion extending from chr5:141 108 260 to 157 224 755 (involving PCDH1 to CLINT1) that includes all of the 5q33 CDR, as well as a more centromeric, 1.5-Mb deletion at chr5:110 887 600 to 112 390 739 (involving STARD4 to MCC). Similarities in LRR and MAF signal values at both regions in this and subsequent experiments (Figure 2) suggest the possibility of an intrachromosomal rearrangement joining the 2 regions. (B) Analysis of patient 1 mosaicism indicates a nonuniform distribution of copy loss in circulating cell fractions and improvement with lenalidomide treatment. Mosaicism was estimated to involve 64% of whole-blood DNA at the larger deletion region. Lymphoid-enriched (CD3+ or CD19+) and myeloid-enriched (CD3− and CD19−) DNA prepared from magnetic bead-separated peripheral blood populations showed substantial disparity, with monosomy estimated in only 27% of the lymphoid but 82% of the myeloid population. Similar analysis of the myeloid fraction from blood obtained after 3 months of treatment with lenalidomide showed marked diminution of the monosomic fraction to an estimated 31% of myeloid DNA. Similar changes were also observed in the smaller deletion region (not shown). (C) Patient 2 has an 897-Kb deletion extending from chr5:149 496 080 to 150 393 600 (involving PDGFRB to TNIP1), with monosomy in 77% of the peripheral blood DNA sample. This deletion is approximately half the size and is entirely contained within the current 5q33 CDR. The 5q33 CDR and genes with potential relevance to the 5q syndrome are indicated above the chromosome. Coordinates are expressed relative to National Center for Biotechnology Information Build 36.1.
Prompted by these SNP array results, in 2010, a BM evaluation at 24 years of age revealed reduced cellularity (30%-60%) with a marked decrease in the erythroid series; karyotype analysis showed an interstitial 5q deletion in 12 of 20 cells [46,XX,der(5)del(5)(q15q22)del(5)(q32q33)], with normal MDS fluorescence in situ hybridization panel. Karyotype of cultured skin fibroblasts was normal, consistent with somatic copy loss. No DNA was available from previous BM evaluations for SNP array analysis.
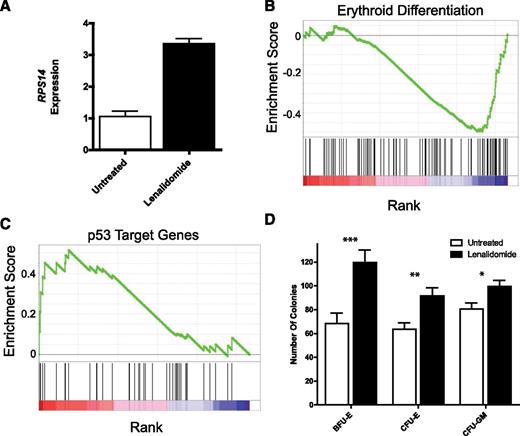
With the diagnosis revised from DBA to 5q− syndrome, patient 1 was successfully treated with lenalidomide.14 She had a complete, sustained clinical response and remains transfusion-independent at the time of this report (Hb, 13.5 g/dL; MCV, 99.1 fL; white blood cell count, 5700/µL; platelet count, 177 000/µL). SNP array on peripheral blood 3 months after initiation of lenalidomide treatment showed reduction of the abnormal clone, going from 82% to 31% in circulating myeloid cells (Figure 1B), and repeat BM karyotype after 6 months of lenalidomide therapy revealed the 5q− deletion in 5 of 20 cells, with the remaining cells showing normal 46,XX. This reduction was associated with increased expression of RPS14 mRNA in bone marrow cells, increased expression of mRNAs associated with an erythroid differentiation program, decreased expression of p53 target genes, and global improvement in the colony-forming potential of marrow-derived CD34+ cells in comparison with marrow obtained before treatment (Figure 2).
Lenalidomide treatment normalizes bone marrow mRNA expression and colony-forming potential. (A) Bone marrow expression of RPS14 as measured by quantitative real-time PCR is markedly increased with lenalidomide treatment and the concomitant reductions in 5q mosaic monosomy. Gene set enrichment analysis comparing pre- and post-lenalidomide-treatment bone marrow mRNA expression demonstrates significant increases in (B) erythroid differentiation program (False Discovery Rate < 0.05) and (C) a decrease in expression of p53 target genes, a putative mediator of ribosomal protein haploinsufficiency. (D) The in vitro colony-forming potential for BFU-E, CFU-E, and to a lesser extent, CFU-GM was also significantly improved with lenalidomide therapy. *P value of Student t test < .05; **P value of student t test < .005; ***P value of student t test < .001; error bars denote SEM.
Lenalidomide treatment normalizes bone marrow mRNA expression and colony-forming potential. (A) Bone marrow expression of RPS14 as measured by quantitative real-time PCR is markedly increased with lenalidomide treatment and the concomitant reductions in 5q mosaic monosomy. Gene set enrichment analysis comparing pre- and post-lenalidomide-treatment bone marrow mRNA expression demonstrates significant increases in (B) erythroid differentiation program (False Discovery Rate < 0.05) and (C) a decrease in expression of p53 target genes, a putative mediator of ribosomal protein haploinsufficiency. (D) The in vitro colony-forming potential for BFU-E, CFU-E, and to a lesser extent, CFU-GM was also significantly improved with lenalidomide therapy. *P value of Student t test < .05; **P value of student t test < .005; ***P value of student t test < .001; error bars denote SEM.
Patient 2 presented in 2004 at 17 years of age with macrocytic anemia (Hb, 9.2 g/dL; MCV, 112.4 fL; reticulocyte count, 2.1%) and slightly elevated white blood cell (16 420/µL) and platelet (478 000/µL) counts. She had no physical anomalies or significant family history. She subsequently required intermittent red cell transfusions, and BM evaluation revealed hypercellularity (95%), marked erythroid hypoplasia, increased megakaryocyte number with mild atypical forms, and rare hypolobulated nuclei. Cytogenetic evaluation, eADA activity (0.54 IU/gm Hb), and HbF (1.8%) were normal. DBA gene mutation analysis was negative. The patient was unresponsive to erythropoietin, corticosteroids, and cyclosporine. A diagnosis of an atypical chronic myeloproliferative disorder was made, and she underwent successful hematopoietic stem cell transplantation.
SNP array on DNA from a pretransplant blood sample from patient 2 showed a small, mosaic deletion on a region of 5q containing RPS14 (77% monosomy over 897 Kb; Figure 1C), establishing the diagnosis of acquired 5q− syndrome. This deletion, undetected by karyotype analysis and routine fluorescence in situ hybridization, is completely contained within the 5q33 CDR.13 This is the smallest deletion reported in association with 5q− syndrome. A retrospective diagnosis of refractory cytopenias with multilineage dysplasia with a microdeletion within 5q was made. The patient was lost to follow-up, and therefore no germline DNA was available for further study.
We describe the identification of unrecognized somatic 5q deletions in 2 young patients. Both patients shared a presentation atypical for DBA.4 These patients are also atypical for 5q− syndrome. The deletions in our patients are small in comparison with previously reported deletions, which often span most of 5q and are characteristic of 5q− syndrome. Furthermore, the 2 deletions include neither the megakaryocyte regulatory miRNAs, mir145 (patient 2) or mir146a (both patients), nor the 2 regulatory phosphatases, CDC25c and PP2A (both patients). The megakaryocytic changes in 5q− syndrome have been attributed to loss of these regulatory miRNAs.15 The absence of such changes in patient 1 and minimal changes in patient 2 suggest the distinctive hematological features in 5q− syndrome are, in large part, a result of the effects of other specific genes (not RPS14) and are further supportive of their role in the platelet phenotype of the disorder. Haploinsufficiency of CDC25c and PP2A have been implicated in the lenalidomide response.16 Although we did not exclude possible additional mutations in 1 or both of these genes, patient 1, who is diploid at these loci, demonstrated a sustained hematological response to lenalidomide. Additional studies beyond the scope of this report, such as sequence analysis of these genes, would be necessary to clarify the role of these phosphatases in the lenalidomide response in 5q− syndrome.
We have demonstrated 1 clearly somatic and 1 most likely somatic acquired deletion of RPS14 that cannot be detected by routine fluorescence in situ hybridization probing. Although 5q− syndrome or refractory cytopenias with multilineage dysplasia rarely enter the pediatric hematologist’s differential diagnosis, our results suggest small deletions in the RPS14 region of 5q must be considered in nonclassical cases of DBA; that is, those patients with normal eADA activity, no congenital anomalies, and/or more than 1 year of age at presentation, and for patients who do not initially respond to corticosteroid therapy, lacking a mutation in a known DBA-associated RP gene. Further, some “non-5q− syndrome” MDS patients who respond to lenalidomide may have unidentified small 5q deletions involving RPS14. Careful use of fluorescence in situ hybridization probes that span the critical genes, or the application of more sensitive alternative techniques, such as SNP array genotyping, may allow identification of previously unclassifiable patients. These findings suggest such studies should be considered in patients of any age with unexplained red cell aplasia.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
The data reported in this article have been deposited in the Gene Expression Omnibus database (accession number GSE42570).
Acknowledgments
The authors especially thank the DBA patients, their families, and their physicians, who tirelessly provide the Diamond Blackfan Anemia Registry with data, as well as the Diamond Blackfan Anemia Foundation and the Daniella Maria Arturi Foundation. The authors also acknowledge Fatima Al-Shahrour for the gene expression analysis.
This work was supported in part by grants from the Diamond Blackfan Anemia Foundation (H.T.G., D.M.B., A.V., E.M.), the Pediatric Cancer Foundation (J.M. Lipton), the Arkansas Biosciences Institute (J.E.F.), The Feinstein Institute for Medical Research General Clinical Research Center (M01 RR018535; A.V., J.M. Lipton), the Centers for Disease Control-sponsored Diamond Blackfan Anemia Surveillance and Awareness Program (A.V., E.M.), and the National Institutes of Health (R01-HL079571 [A.V., E.A., J.M. Lipton], R01-HL107558 [H.T.G.], R109-MOHLKE [A.V., E.A., J.M.Lipton], K02-HL111156 [H.T.G.], K08-HL092224 [J.E.F.], and National Human Genome Research Institute Intramural funds [D.M.B.]), St. Baldrick’s Foundation Scholar Career Development Award (S.A.S.), and St. Baldrick's Consortium on Pediatric Myelodysplastic Syndrome (J.M. Liu).
Authorship
Contribution: A.V. and J.M. Lipton designed research, analyzed and interpreted data, and drafted the manuscript; J.E.F., A.N., T.C.M., S.A.S., M.L., H.T.G., J.M. Liu, S.R.E., B.L.E., and D.M.B. designed and performed research, analyzed and interpreted data, and edited the manuscript; E.A. and E.M. contributed, analyzed, and interpreted data and edited the manuscript; L.B. interpreted data and edited the manuscript; and R.J.A. interpreted and analyzed data, and edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for R.J.A. is Phoenix Children’s Hospital, University of Arizona College of Medicine, Translational Genomics Research Institute, Phoenix, AZ.
Correspondence: Adrianna Vlachos, Steven and Alexandra Cohen Children's Medical Center of New York, Hematology/Oncology and Stem Cell Transplantation, 269-01 76th Ave, New Hyde Park, NY 11040; e-mail: avlachos@nshs.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal