Key Points
EZH2 mutations occur in more than 25% of follicular lymphoma patients.
Mutations predominantly represent an early/clonal event in the pathogenesis of the disease.
Abstract
Gain of function mutations in the H3K27 methyltransferase EZH2 represent a promising therapeutic target in germinal center lymphomas. In this study, we assessed the frequency and distribution of EZH2 mutations in a large cohort of patients with follicular lymphoma (FL) (n = 366) and performed a longitudinal analysis of mutation during the disease progression from FL to transformed FL (tFL) (n = 33). Mutations were detected at 3 recurrent mutation hot spots (Y646, A682, and A692) in 27% of FL cases with variant allele frequencies (VAF) ranging from 2% to 61%. By comparing VAF of EZH2 with other mutation targets (CREBBP, MLL2, TNFRSF14, and MEF2B), we were able to distinguish patients harboring clonal EZH2 mutation from rarer cases with subclonal mutations. Overall, the high incidence of EZH2 mutations in FL and their stability during disease progression makes FL an appropriate disease to evaluate EZH2 targeted therapy.
Introduction
Next-generation sequencing (NGS) studies have shown frequent mutations in epigenetic regulators in almost all cases of follicular lymphoma (FL).1,2 These include EZH2, the catalytic subunit of PRC2, which catalyzes trimethylation of lysine 27 on histone H3 (H3K27me3), a repressive chromatin mark.3 Somatic gain-of-function mutations of EZH2 at codon Y646 (previously Y641) were identified in 7% to 22% of FLs and germinal center B-cell type diffuse large B-cell lymphomas leading to elevated H3K27 trimethylation4-8 with mutations at codons A682 and A692 described in isolated cases of diffuse large B-cell lymphomas.2,9-11 As highly selective EZH2 inhibitors have now been developed,12-14 we set out to assess EZH2 mutation status, the effect of mutations on global gene expression, and the clonal representation of EZH2 mutations as the disease progresses.
Study design
Patient samples
Genomic DNA from 181 diagnostic FL patients with accompanying clinical and gene expression data15 were obtained through the Lymphoma/Leukemia Molecular Profiling Project consortium. DNA from 185 additional FL patients (56 obtained at diagnosis and 129 at relapse) and 33 paired FL and transformed FL (tFL) samples were sourced from the tissue archive at the Barts Cancer Institute. The study was approved by the London Research Ethical Committee (05/Q0605/140) and was conducted in accordance with the Declaration of Helsinki.
Mutation analysis
NGS was performed on all 432 samples, with the entire coding region (n = 19 exons) of EZH2 screened in 46 FLs with the remaining 320 samples and the 33 paired FL-tFL cases restricted to exon 16 (Y646) and 18 (A682 and A692). The mutation analysis was performed by an NGS amplicon deep-sequencing assay using the Titanium amplicon chemistry (454 Life Sciences, Branford, CT)16,17 achieving at least a 200-fold coverage (sensitivity <5%). Exons 16 and 18 of EZH2 were also analyzed by bidirectional Sanger sequencing, as described previously (supplemental Table 1 on the Blood Web site).4,18 Subsequently, targeted resequencing of CREBBP, MLL2, TNFRSF14, and MEF2B was performed using the multiplex Access Array platform (Fluidigm) as per the manufacturer’s recommendations in selected FL cases with EZH2 mutation (variant allele frequencies [VAF] range: 3.1% to 49.1%). Corrected VAF of EZH2 for the sequential FL-tFL cases were determined using tumor cell content estimates calculated by the ASCAT algorithm19 using previously generated SNP6.0 array data.20
Gene expression data analysis
Existing gene expression profiling data15 from 181 FL samples from the Lymphoma/Leukemia Molecular Profiling Project FL cohort were analyzed as described in the supplemental Methods.
Results and discussion
Incidence of EZH2 mutations in FL is higher than previously reported
The incidence and distribution of EZH2 mutations were investigated in 366 FL patients (237 at diagnosis, 129 at relapse) using NGS and Sanger sequencing. Sequence analysis of the entire coding region of EZH2 in 46 FL cases confirmed recurrent mutations at codons Y646, A682, and A692, previously reported by Morin and colleagues,2,5 and the absence of additional mutational hotspots. We subsequently restricted our targeted resequencing to exons 16 and 18.
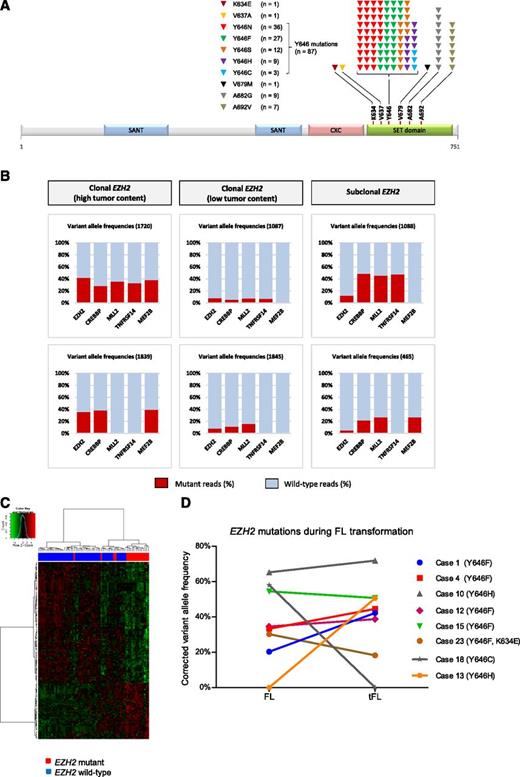
Sanger sequencing showed 63 EZH2 mutations in 62 patients (17%) (Table 1). Using the more sensitive NGS approach (≥200-fold coverage), EZH2 mutations were detected in 39 additional patients, increasing the total number of mutated patients to 101 (27.5%). Multiple mutations were observed in 4 patients; these were monoallelic (n = 2) or located to different reads (n = 2), suggesting either biallelic EZH2 mutation or the presence of mutations in different FL clones (supplemental Table 2). In total, 106 mutations were detected in 101 patients, which included 87 Y646, 9 A682G, and 7 A692 mutations at a mean VAF of 21.6%, significantly lower in comparison with a VAF of 29.8% for mutations detected by both sequencing methods. The remainder corresponded to 3 novel variants K634E (VAF: 3.5%), V637A (VAF: 25%), and V679M (VAF: 2.5%), all located within the catalytic SET domain of EZH2 (Figure 1A). The somatic origin of the K634E mutation was confirmed using matched remission DNA. There was no difference in the mutation frequency at diagnosis (29%; n = 70/237) and relapse (24%; n = 31/129) with detailed distribution and frequencies of the EZH2 mutations summarized in Figure 1A and Table 1 . Mutation status was not associated with overall survival of FL in the 2 cohorts studied (supplemental Figure 1).
Numbers of EZH2 mutations detected using the different sequencing approaches
Mutations | Sanger sequencing and NGS | Additional mutations by NGS only | Total |
|---|---|---|---|
| K634E | 0 | 1 | 1 |
| V637A | 1 | 0 | 1 |
| Y646N | 18 | 18 | 36 |
| Y646F | 18 | 9 | 27 |
| Y646S | 8 | 4 | 12 |
| Y646H | 6 | 3 | 9 |
| Y646C | 0 | 3 | 3 |
| V679M | 0 | 1 | 1 |
| A682G | 7 | 2 | 9 |
| A692V | 5 | 2 | 7 |
| In total | 63 | 43 | 106 |
| Mean VAF (range) | 29.78% (4-61) | 9.71% (2-31) | 21.64% (2-61) |
Mutations | Sanger sequencing and NGS | Additional mutations by NGS only | Total |
|---|---|---|---|
| K634E | 0 | 1 | 1 |
| V637A | 1 | 0 | 1 |
| Y646N | 18 | 18 | 36 |
| Y646F | 18 | 9 | 27 |
| Y646S | 8 | 4 | 12 |
| Y646H | 6 | 3 | 9 |
| Y646C | 0 | 3 | 3 |
| V679M | 0 | 1 | 1 |
| A682G | 7 | 2 | 9 |
| A692V | 5 | 2 | 7 |
| In total | 63 | 43 | 106 |
| Mean VAF (range) | 29.78% (4-61) | 9.71% (2-31) | 21.64% (2-61) |
Detaileddistribution and frequencies of the EZH2 mutations. (A) Distribution and frequency of EZH2 SET domain mutations detected in 366 diagnostic and relapsed FL cases. Altogether, 106 mutations were detected in 101 FL patients. The most prevalent variants were the mutations resulting in replacement of the tyrosine at codon 646 (Y646), followed by A682 and A692 mutations. Of note, we also detected 3 previously unreported mutations in B-cell lymphomas: K634E, V637A, and V679M of unknown significance. (B) Comparison of VAF for EZH2, CREBBP, MLL2, TNFRSF14, and MEF2B mutations identified by NGS-based targeted resequencing demonstrating i, true clonal variants with similar VAFs (less than 20% difference between VAFs for EZH2 compared with the other genes excluding VAFs of >50%) across the genes analyzed (cases 1720 and 1839), ii, clonal variants with similar, but low VAFs across the mutation targets reflecting the low tumor content within these biopsy samples (cases 1087 and 1845), and iii, true subclonal EZH2 variants with lower EZH2 VAFs as compared with the other genes (cases 1088 and 465). (C) Hierarchical clustering and heatmap of 69 FL cases (51 wild type vs 18 EZH2 mutated) with estimated high tumor content (Δβ > 0.1657) and EZH2 VAF (>17%) showing the gene expression signature of 106 differentially expressed genes. (D) Clonal representation of EZH2 mutations during transformation of FL. Illustrated are the corrected VAFs observed in sequential FL and tFL samples. The EZH2 mutations were maintained during transformation in 6 cases with relatively stable VAFs, whereas it was restricted to the FL and tFL samples in single patients (cases 13 and 18). Case 23 harbored 2 EZH2 mutations (Y646F and K634E) in monoallelic configuration.
Detaileddistribution and frequencies of the EZH2 mutations. (A) Distribution and frequency of EZH2 SET domain mutations detected in 366 diagnostic and relapsed FL cases. Altogether, 106 mutations were detected in 101 FL patients. The most prevalent variants were the mutations resulting in replacement of the tyrosine at codon 646 (Y646), followed by A682 and A692 mutations. Of note, we also detected 3 previously unreported mutations in B-cell lymphomas: K634E, V637A, and V679M of unknown significance. (B) Comparison of VAF for EZH2, CREBBP, MLL2, TNFRSF14, and MEF2B mutations identified by NGS-based targeted resequencing demonstrating i, true clonal variants with similar VAFs (less than 20% difference between VAFs for EZH2 compared with the other genes excluding VAFs of >50%) across the genes analyzed (cases 1720 and 1839), ii, clonal variants with similar, but low VAFs across the mutation targets reflecting the low tumor content within these biopsy samples (cases 1087 and 1845), and iii, true subclonal EZH2 variants with lower EZH2 VAFs as compared with the other genes (cases 1088 and 465). (C) Hierarchical clustering and heatmap of 69 FL cases (51 wild type vs 18 EZH2 mutated) with estimated high tumor content (Δβ > 0.1657) and EZH2 VAF (>17%) showing the gene expression signature of 106 differentially expressed genes. (D) Clonal representation of EZH2 mutations during transformation of FL. Illustrated are the corrected VAFs observed in sequential FL and tFL samples. The EZH2 mutations were maintained during transformation in 6 cases with relatively stable VAFs, whereas it was restricted to the FL and tFL samples in single patients (cases 13 and 18). Case 23 harbored 2 EZH2 mutations (Y646F and K634E) in monoallelic configuration.
The majority of EZH2 mutations represent clonal events
Although novel EZH2 inhibitors hold great promise, it is critical for the success of these therapies that the actionable mutations are clonally present within the tumor population. To decide whether EZH2 mutations were clonal or subclonal events in FL, we compared EZH2 VAFs with those of other mutation targets (CREBBP, MLL2, TNFRSF14, and MEF2B) in 43 FLs with EZH2 mutations (VAF range: 3.1% to 49.1%; supplemental Table 3). Although the direct comparison may often be complicated by presence of acquired uniparental disomy or changes in copy number leading to VAFs of >50%,21,22 we were able to discriminate 3 different patterns for EZH2 mutations (Figure 1B). The majority of EZH2 variants (81%; 35/43) represented true clonal events with similar VAFs for other genes mutated in the same sample. These included rare cases (4/43) characterized by low VAFs across all the mutational targets, which is probably a reflection of low tumor content within these biopsy samples. True subclonal EZH2 mutations, with lower EZH2 VAFs compared with the other genes, represented 19% (8/43) of all the EZH2 variants tested. The dominance of clonal EZH2 variants was also supported using our previous array-based methylation profiling data,23 which allowed us to rank the samples based on their tumor content as described in the supplemental Information (supplemental Figures 2 and 3).
We next tested whether EZH2 mutation status defined a particular subgroup of FL patients, based on global gene expression profiles. Although we failed to identify an EZH2 gene expression signature using the entire cohort of 181 cases (125 wt vs 56 EZH2 mutated), we were able to define a weak EZH2 signature of 106 differentially expressed genes (Figure 1C; supplemental Table 4) by restricting the analysis to cases with estimated high tumor content (Δβ > 0.1657, supplemental Figure 2) and EZH2 VAF (>17%) (EZH2 mutated; n = 18 and EZH2 wt; n = 51). The relatively small number of differentially expressed genes and the low fold changes observed in our signature are consistent with the findings of McCabe et al, reporting only 35 common loci reactivated in 4 cell lines on treatment with the EZH2 inhibitor GSK126 highlighting the complexity and diversity of the EZH2 mediated epigenetic deregulation in individual lymphoma samples.13
EZH2 mutations are maintained during transformation of FL and represent an early event in FL
To determine the clonal representation of EZH2 mutations during the disease progression, 33 sequential FL-tFL cases were screened for EZH2 mutations using NGS. Of the 33 cases, 8 carried EZH2 Y646 mutations (24.2%) with an average VAF of 27.6%. EZH2 mutation was detected in both the FL and tFL biopsy in 6 patients, and in 2 additional individuals the EZH2 mutation was restricted to either the FL or tFL biopsy (supplemental Table 5). Green and colleagues reported significant clonal diversity in genes that are recurrently mutated in FL highlighting CREBBP mutations as an early driver event in the disease evolution, based on their clonal nature at diagnosis and their maintained presence between diagnosis and relapses.24 Our data demonstrate that EZH2 mutations are also present at relatively high allelic frequencies and in the majority of cases are maintained through transformation of the disease (Figure 1D), implying that they may also represent early mutations in this lymphoma.
In conclusion, our observations demonstrate a higher frequency of EZH2 mutations in FL than previously reported.4,5 Mutations cluster to 3 codons, Y646, A682, and A692, are clonal in the majority of cases, and are stable during disease progression. The variable tumor content in FL biopsies supports the use of more sensitive and quantitative approaches during routine screening of FL to select patients with clonal EZH2 mutations, as these will be better suited for treatment with EZH2 inhibitors.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the Leukaemia Lymphoma Research UK (10036; J.F.), Partner fellowship (2009/01; C.B.) awarded by European Hematology Association, Kay Kendall Leukaemia Fund Junior Clinical Research Fellowship (KKL 557; J.O.), OTKA K-76204 grant (C.B. and A.M.), and Cancer Research UK Programme award (C15966/A15968; J.F.). C.B. was supported by the European Union and the State of Hungary, cofinanced by the European Social Fund in the framework of TÁMOP 4.2.4. A/-11-1-2012-0001 National Excellence Program.
Authorship
Contribution: C.B. and J.F. designed the study, performed research, analyzed data, and wrote the manuscript; C.B., V.G., A.K., and T.H. performed the mutation analysis; N.P., J.O., C.O., K.T., S.A., A.M.L., A.C., and H.R. performed research and analyzed data; S.M., T.A.L., and J.G. selected patients for the study; S.I. and J.M. provided clinical information; A.R., G.O., E.C., L.M.R., E.B.S., W.C.C., R.M.B., L.M.S., G.W., A.M., and R.D.G. provided samples; J.M., J.W., C.C., O.E., and R.H. performed bioinformatical analyses; and all other authors read and approved the final manuscript.
Conflict-of-interest disclosure: V.G. and A.K. are employed by MLL Munich Leukemia Laboratory GmbH. T.H. has equity ownership of MLL Munich Leukemia Laboratory GmbH. The remaining authors declare no competing financial interests.
Correspondence: Csaba Bödör, Centre for Haemato-Oncology, Barts Cancer Institute, Queen Mary University of London, Charterhouse Square, London EC1M 6BQ, United Kingdom; e-mail: c.bodor@qmul.ac.uk.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal