Key Points
Despite a low frequency of mutations, BCOR might be considered as a key gene in risk stratification.
Deep sequencing technologies show that BCOR mutations commonly arise after other concomitant mutations in MDS.
Abstract
Patients with low-risk myelodysplastic syndromes (MDS) that rapidly progress to acute myeloid leukemia (AML) remain a challenge in disease management. Using whole-exome sequencing of an MDS patient, we identified a somatic mutation in the BCOR gene also mutated in AML. Sequencing of BCOR and related BCORL1 genes in a cohort of 354 MDS patients identified 4.2% and 0.8% of mutations respectively. BCOR mutations were associated with RUNX1 (P = .002) and DNMT3A mutations (P = .015). BCOR is also mutated in chronic myelomonocytic leukemia patients (7.4%) and BCORL1 in AML patients with myelodysplasia-related changes (9.1%). Using deep sequencing, we show that BCOR mutations arise after mutations affecting genes involved in splicing machinery or epigenetic regulation. In univariate analysis, BCOR mutations were associated with poor prognosis in MDS (overall survival [OS]: P = .013; cumulative incidence of AML transformation: P = .005). Multivariate analysis including age, International Prognostic Scoring System, transfusion dependency, and mutational status confirmed a significant inferior OS to patients with a BCOR mutation (hazard ratio, 3.3; 95% confidence interval, 1.4-8.1; P = .008). These data suggest that BCOR mutations define the clinical course rather than disease initiation. Despite infrequent mutations, BCOR analyses should be considered in risk stratification.
Introduction
Abnormal and clonal hematopoiesis resulting in peripheral blood cytopenias, and risk of progression to acute myeloid leukemia (AML) are the main characteristics of myelodysplastic syndromes (MDS).1 The diversity of somatically mutated genes reflects a variety of pathogenic mechanisms in these disorders. Recently, mutations targeting genes whose products participate to the early steps of RNA splicing (SF3B1, SRSF2, ZRSR2, and U2AF1) have been reported.2-5 Mutations in ASXL1,6,7 DNMT3A,8 EZH2,9,10 IDH1/2,11 and TET212,13 suggest deregulation of the epigenetic control of transcription. Additional genes known to be mutated in MDS include RUNX1, TEL/ETV6, TP53, and NRAS.14 The alteration of some of these genes may carry prognostic value.4,14,15 Around 25% of MDS patients with International Prognostic Scoring System (IPSS) low or intermediate-1 risk are exempt of these recurrent mutations, underscoring the need for further molecular characterization.4 Next-generation sequencing analyses of AML and related disorders identified mutations in 2 chromosome X genes, BCOR and BCORL1, which code for related transcriptional corepressors.3,16,17 Both proteins interact with histone deacetylases but also have specific properties. BCOR interacts with BcL6, and constitutional inactivating mutations of the gene have been described in the oculofacicocardiodental syndrome.18,19 BCOR is also affected by acquired mutations or translocations in retinoblastoma and sarcoma.20,21 It has also been identified as a novel fusion partner of retinoic acid receptor α in a t(X;17)(p11;q12) variant of acute promyelocytic leukemia.22 BCORL1 has been implicated in chromosomal rearrangements in hepatocellular carcinoma.23,24
To identify gene mutations that might carry prognostic value, we compared the coding sequence of mononuclear bone marrow (BM) cells with T cells of an IPSS intermediate-1 MDS patient who progressed to AML 5 months after diagnosis and died 8 months later. A total of 8 somatic mutations could be identified including BCOR, RUNX1, and STAG2, which have recently been identified in hematopoietic malignancies.3,25 Because BCOR and BCORL1 mutations have been described mostly in AML, we analyzed their coding sequences in a cohort of 354 MDS, 54 chronic myelomonocytic leukemia (CMML), and 22 AML with myelodysplasia-related changes (AML-MRC) patients. BCOR and BCORL1 mutations were found in 4.2% and 0.8% of MDS patients, respectively, and 2 patients presented with concomitant inactivating mutations of BCOR and BCORL1.
Patients, materials, and methods
Patients
The 57-year-old male MDS index patient selected for whole-exome sequencing was enrolled on the prospective noninterventional multicenter MDS04 trial (EUDRACT: 2010-A00033-36). He presented with 4 × 109/L white blood cells (1.8 × 109/L neutrophils), 13.8 g/dL hemoglobin, and 43 × 109/L platelets at diagnosis. The BM report revealed 6% of blast infiltration and the patient was classified as a RAEB-1 disease. Cytogenetic analysis showed a normal karyotype. A cohort of 430 patients including 354 MDS (including the MDS index patient), 54 CMML, and 22 AML-MRC was included in this study. The 222 French MDS samples were collected at diagnosis in multicenter clinical trials in France between 1999 and 2011 in Paris Cochin (n = 135) and Marseille (n = 87). The Asian cohort of 208 patients with various myeloid malignancies was collected in Chang Gung Memorial Hospital, the University of Tokyo, and collaborating centers. According to World Heath Organization (WHO) classification, this cohort includes 132 MDS patients, 54 CMML, and 22 AML-MRC. A flowchart of the study is provided in supplemental Figure 1. Clinical and hematologic data were recorded following informed consent in accordance with the Declaration of Helsinki; this study was approved by institutional review boards of Paris Centre, Marseille, and the University of Tokyo. Treatment details for the French cohort were previously reported7,26 and are summarized in supplemental Table 1. Distributions of 2008 WHO subtypes, IPSS risk groups, and cytogenetic risk groups27 are shown in Table 1 and in supplemental Tables 2-4.
Clinical characteristics of 222 MDS patients of the French cohort according to BCOR and BCORL1 mutation status
| Characteristic . | BCOR mutated (n = 11) . | BCOR wild-type (n =211) . | P . | BCORL1 mutated (n = 2) . | BCORL1 wild-type (n = 220) . |
|---|---|---|---|---|---|
| Age, y | .92 | ||||
| Median | 72.5 | 72 | 69 | 88 | |
| Range | 57-95 | 42-95 | |||
| Sex | .53 | ||||
| Male, no. (%) | 5 (45) | 125 (59) | 1 (50) | 129 (58) | |
| Female, no. (%) | 6 (55) | 86 (41) | 1 (50) | 91 (42) | |
| WHO subtype | .81 | ||||
| RA, no. (%) | 0 (0) | 34 (16) | 0 (0) | 34 (15) | |
| RARS, no. (%) | 2 (18) | 25 (12) | 2 (100) | 25 (11) | |
| RCMD, no. (%) | 1 (9) | 30 (14) | 0 (0) | 31 (14) | |
| RCMD-RS, no. (%) | 1 (9) | 7 (3) | 0 (0) | 8 (4) | |
| RAEB-1, no. (%) | 4 (36) | 52 (25) | 0 (0) | 56 (25) | |
| RAEB-2, no. (%) | 2 (18) | 43 (20) | 0 (0) | 45 (20) | |
| del (5q), no. (%) | 0 (0) | 2 (1) | 0 (0) | 2 (1) | |
| RARS-T, no. (%) | 0 (0) | 6 (3) | 0 (0) | 6 (3) | |
| MDS-U, (%) | 1 (9) | 12 (6) | 0 (0) | 13 (6) | |
| Missing data, no. (%) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |
| Ring sideroblasts | .4 | ||||
| Present, no. (%) | 3 (30) | 38 (18) | 2 (100) | 39 (18) | |
| Karyotype risk* | .45 | ||||
| Low, no. (%) | 9 (81) | 150 (72) | 1 (50) | 158 (72) | |
| Intermediate, no. (%) | 1 (9) | 36 (17) | 1 (50) | 36 (17) | |
| High, no. (%) | 0(0) | 19 (9) | 0 (0) | 19 (9) | |
| Missing data, no. (%) | 1 (9) | 5 (2) | 0 (0) | 7 (3) | |
| Bone marrow blasts | .95 | ||||
| Median (%) | 2.5 | 4 | 1.5 | 6 | |
| Range | 1-19 | 0-19 | |||
| Normal karyotype | .75 | ||||
| Yes, no. (%) | 7 (70) | 128 (61) | 0 (0) | 135 (61) | |
| Missing data, no. (%) | 1 (10) | 5 (2) | 0 (0) | 6 (3) | |
| Hemoglobin | .79 | ||||
| Median (g/dL) | 10 | 9.9 | 9 | 10 | |
| Range (g/dL) | 7.7-13.9 | 6-15 | |||
| White blood cell count | .58 | ||||
| Median, × 109/L | 4.4 | 4.2 | 6 | 5 | |
| Range, × 109/L | 1.3-36.7 | 0.9-20 | 5 | 0.9-36.7 | |
| Neutrophil count | .73 | ||||
| Median, × 109/L | 1.8 | 2.2 | 3.1 | 2.1 | |
| Range, × 109/L | 0.3-23.1 | 0.3-17.5 | 5 | 0.3-23.1 | |
| Platelet count | .17 | ||||
| Median, × 109/L | 147 | 158 | 258 | 197 | |
| Range, × 109/L | 27-276 | 5-1398 | |||
| IPSS | .89 | ||||
| Low risk (%) | 3 (27) | 71 (34) | 1 (50) | 73 (33) | |
| Intermediate-1 (%) | 5 (45) | 88 (42) | 1 (50) | 92 (42) | |
| Intermediate-2 (%) | 2 (18) | 24 (11) | 0 (0) | 26 (12) | |
| High (%) | 1 (9) | 24 (11) | 0 (0) | 25 (11) | |
| Missing data, no. (%) | 0 (0) | 3 (1) | 0 (0) | 4 (2) | |
| Cytology | |||||
| Multilineage dysplasia, no. (%) | 5 (50) | 110 (52) | 1 | 0 (0) | 115 (52) |
| Dyserythropoesis, no. (%) | 5 (50) | 113 (54) | .46 | 1 (50) | 137 (62) |
| Dysgranulopoesis, no. (%) | 9 (90) | 131 (62) | .06 | 2 (100) | 138 (63) |
| Dysmegalopoesis, no. (%) | 5 (50) | 126 (60) | .5 | 0 (0) | 131 (59) |
| Transfusion dependence | .74 | ||||
| Yes, no. (%) | 5 (55) | 89 (48) | 2 (100) | 92 (42) | |
| Missing data, no. (%) | 2 (20) | 23 (11) | 0 (0) | 26 (12) |
| Characteristic . | BCOR mutated (n = 11) . | BCOR wild-type (n =211) . | P . | BCORL1 mutated (n = 2) . | BCORL1 wild-type (n = 220) . |
|---|---|---|---|---|---|
| Age, y | .92 | ||||
| Median | 72.5 | 72 | 69 | 88 | |
| Range | 57-95 | 42-95 | |||
| Sex | .53 | ||||
| Male, no. (%) | 5 (45) | 125 (59) | 1 (50) | 129 (58) | |
| Female, no. (%) | 6 (55) | 86 (41) | 1 (50) | 91 (42) | |
| WHO subtype | .81 | ||||
| RA, no. (%) | 0 (0) | 34 (16) | 0 (0) | 34 (15) | |
| RARS, no. (%) | 2 (18) | 25 (12) | 2 (100) | 25 (11) | |
| RCMD, no. (%) | 1 (9) | 30 (14) | 0 (0) | 31 (14) | |
| RCMD-RS, no. (%) | 1 (9) | 7 (3) | 0 (0) | 8 (4) | |
| RAEB-1, no. (%) | 4 (36) | 52 (25) | 0 (0) | 56 (25) | |
| RAEB-2, no. (%) | 2 (18) | 43 (20) | 0 (0) | 45 (20) | |
| del (5q), no. (%) | 0 (0) | 2 (1) | 0 (0) | 2 (1) | |
| RARS-T, no. (%) | 0 (0) | 6 (3) | 0 (0) | 6 (3) | |
| MDS-U, (%) | 1 (9) | 12 (6) | 0 (0) | 13 (6) | |
| Missing data, no. (%) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |
| Ring sideroblasts | .4 | ||||
| Present, no. (%) | 3 (30) | 38 (18) | 2 (100) | 39 (18) | |
| Karyotype risk* | .45 | ||||
| Low, no. (%) | 9 (81) | 150 (72) | 1 (50) | 158 (72) | |
| Intermediate, no. (%) | 1 (9) | 36 (17) | 1 (50) | 36 (17) | |
| High, no. (%) | 0(0) | 19 (9) | 0 (0) | 19 (9) | |
| Missing data, no. (%) | 1 (9) | 5 (2) | 0 (0) | 7 (3) | |
| Bone marrow blasts | .95 | ||||
| Median (%) | 2.5 | 4 | 1.5 | 6 | |
| Range | 1-19 | 0-19 | |||
| Normal karyotype | .75 | ||||
| Yes, no. (%) | 7 (70) | 128 (61) | 0 (0) | 135 (61) | |
| Missing data, no. (%) | 1 (10) | 5 (2) | 0 (0) | 6 (3) | |
| Hemoglobin | .79 | ||||
| Median (g/dL) | 10 | 9.9 | 9 | 10 | |
| Range (g/dL) | 7.7-13.9 | 6-15 | |||
| White blood cell count | .58 | ||||
| Median, × 109/L | 4.4 | 4.2 | 6 | 5 | |
| Range, × 109/L | 1.3-36.7 | 0.9-20 | 5 | 0.9-36.7 | |
| Neutrophil count | .73 | ||||
| Median, × 109/L | 1.8 | 2.2 | 3.1 | 2.1 | |
| Range, × 109/L | 0.3-23.1 | 0.3-17.5 | 5 | 0.3-23.1 | |
| Platelet count | .17 | ||||
| Median, × 109/L | 147 | 158 | 258 | 197 | |
| Range, × 109/L | 27-276 | 5-1398 | |||
| IPSS | .89 | ||||
| Low risk (%) | 3 (27) | 71 (34) | 1 (50) | 73 (33) | |
| Intermediate-1 (%) | 5 (45) | 88 (42) | 1 (50) | 92 (42) | |
| Intermediate-2 (%) | 2 (18) | 24 (11) | 0 (0) | 26 (12) | |
| High (%) | 1 (9) | 24 (11) | 0 (0) | 25 (11) | |
| Missing data, no. (%) | 0 (0) | 3 (1) | 0 (0) | 4 (2) | |
| Cytology | |||||
| Multilineage dysplasia, no. (%) | 5 (50) | 110 (52) | 1 | 0 (0) | 115 (52) |
| Dyserythropoesis, no. (%) | 5 (50) | 113 (54) | .46 | 1 (50) | 137 (62) |
| Dysgranulopoesis, no. (%) | 9 (90) | 131 (62) | .06 | 2 (100) | 138 (63) |
| Dysmegalopoesis, no. (%) | 5 (50) | 126 (60) | .5 | 0 (0) | 131 (59) |
| Transfusion dependence | .74 | ||||
| Yes, no. (%) | 5 (55) | 89 (48) | 2 (100) | 92 (42) | |
| Missing data, no. (%) | 2 (20) | 23 (11) | 0 (0) | 26 (12) |
RA, refractory anemia; RAEB-1, refractory anemia with excess blasts-1; RAEB-2, refractory anemia with excess blasts-2; RARS, refractory anemia with ringed sideroblats; RARS-T, refractory anemia with ringed sideroblats and thrombocytosis; RCMD, refractory cytopenia with multilineage dysplasia; RCMD-RS, refractory cytopenia with multilineage dysplasia and ringed sideroblasts; MDS-U, myelodysplastic syndrome unclassified.
Karyotype risk groups according to Greenberg et al.27
Cytogenetic and mutational analyses
Cytogenetic analysis was performed by G- and R-banding analysis. Genomic DNA was extracted from BM mononuclear cells using the DNA/RNA Kit (Qiagen, Hilden, Germany) and amplified using the REPLI-G Kit (Qiagen). For all French MDS patients, the coding sequences of BCOR (ENST00000378444) and BCORL1 (ENST00000218147) were analyzed by Sanger sequencing (see supplemental Table 5 for primer sequences). Candidate mutations were confirmed using native genomic DNA. Mutational analyses of ASXL1, CBL, DNMT3A, ETV6, EZH2 IDH1/2, JAK2, NRAS, RUNX1, SF3B1, SRSF2, TET2, TP53, U2AF1, and ZRSR2 were previously reported.4 For patients from the Asian cohort, SureSelct-captured gene targets, including BCOR and the above-mentioned genes, were analyzed for gene mutations on HiSequation 2000 at the mean depth of 415, followed by mutation calling (supplemental Methods). Mutations of BCORL1 were screened by deep sequencing of pooled DNA followed by deep sequencing validation, as previously described.3 Nontumoral material from buccal swabs (n = 3) or peripheral blood CD3+ T cells (n = 7) was analyzed for the presence of the identified variations when available.
Whole-exome analysis
Tumor DNA was extracted from patient BM mononuclear cells. For germline control, DNA was obtained from paired CD3+ T cells. Purity of CD3+ cells controlled by flow cytometry was at least superior to 95%. Whole-exome capture was accomplished based on liquid phase hybridization of sonicated genomic DNA having 150 to 200 bp of mean length to the bait complementary RNA library synthesized on magnetic beads (SureSelect, SureSelect Human All Exon 50 Mb; Agilent Technology, Santa Clara, CA) according to the manufacturer’s protocol. The captured targets were subjected to sequencing using HiSequation 2000 with the pair-end 75-bp to 108-bp read option, according to the manufacturer’s instructions. Data processing and mutation calling were performed using Genomon-exome (http://genomon.hgc.jp/exome/en/index.html) as previously described.3
Mutation quantification by deep sequencing
Short fragments of 100 to 200 bp were polymerase chain reaction (PCR) amplified from genomic DNA in 10 BCOR-mutated patients by PCR (primers are listed in supplemental Table 6). After quality controls by gel electrophoresis and Bioanalyzer (Agilent), PCR fragments were purified using the QIAquick PCR Purification Kit (Qiagen), quantified by Qubit (Life Technologies, Carlsbad, CA), and subsequently pooled in equimolar amounts for library construction using the Ion Xpress Plus Fragment Library Kit (Life Technologies). Template preparation was performed with the OneTouch System v37 (Life Technologies). Bar-coded libraries were run on a 100 Mb chip on an Ion PGM Sequencer (Life Technologies). Analysis of acquired data was performed with the Ion torrent v2.2 software (Life Technologies). Only high-quality reads with a Phred score ≥ Q20 were included for further analysis.
Expression analysis of BCOR mRNA transcript levels
Trizol messenger RNA (mRNA) extraction from diagnostic BM mononuclear cells of 35 MDS patients and 20 non-MDS patients was followed by reverse transcription (Thermo Scientific, Waltham, MA). BCOR mRNA levels were quantified in triplicate by SYBR Green real-time PCR using the Light Cycler 480 system (Qiagen). A 200 bp fragment including partially exons 1 and 2 of BCOR was amplified using forward primer 5′-TTCACAGCTGGATGAACAGC-3′ and reverse primer 5′-CGTTGTGGTTCAAGGGATTC-3′. B2M was used as a housekeeping gene for normalization (primers 5′-ATTTGGGTCGCGGTTCTTG-3′ and 5′-TGCCTTGACATTCTCGATGGT-3′). Quantification of samples was performed using the ΔΔCt methods, using the average of U937 cell line ΔCt as a calibrator.
Statistical analysis
Overall survival (OS) end points, measured from date of diagnosis, were deceased (failure) and alive at last contact (censored). Cumulative incidence of AML transformation was measured from date of MDS diagnosis to time of AML diagnosis, considering death as a competing event. AML transformation was defined according to the 2008 WHO classification and patients known to be alive without report of AML transformation were censored at the date of last examination confirming the absence of transformation, with refreshment in February 2013 when available. The median follow-up time for patients alive was calculated according to the method of Korn.28 Primary analysis was performed on OS and AML transformation. The Kaplan-Meier method was used to estimate OS. Differences in OS were tested with the log-rank test and Cox proportional hazards models for univariate and multivariate analyses, respectively. The Fine and Gray test was used for cumulative incidence of AML. Two-group comparisons were performed by Mann-Whitney test for continuous and by 2-sided Fisher’s exact or χ2 tests for categorical variables. For multivariate analysis, a Cox proportional hazards model was constructed for OS and AML transformation adjusting for potential confounding covariates.29 Variables considered for model inclusion were IPSS risk group, transfusion dependence, age (below vs above median), and mutation status of all 18 analyzed genes (mutated vs wild-type). Variables with P ≤ .1 in univariate analysis were included in the model. The statistical analyses were performed with the statistical software package SPSS 19.0 (SPSS Science, Chicago, IL), R 2.14.1, or STATA v12 (STATA Corporation).
Results
Identification of somatic mutations in the MDS index patient
The whole-exome sequencing produced in total approximately 276 million mapped reads (135 million from the tumor DNA and 141 million from the control DNA) of 27.6 billion nucleotides. After removal of low-quality and clonal reads, the mean depth of the covered exome was 98-fold (tumor) and 114-fold (normal), respectively, and with 99% and 98% of the target exome being covered by at least 2 reads and 97% and 93% by at least 10 reads for tumor and normal DNA, respectively. Bioinformatical analyses allowed the identification of 8 somatically acquired mutations including ACOX1, BCOR, DPF2, EIF5B, GPR179, RUNX1, STAG2, and ZNF354C (Table 2), which were confirmed by Sanger sequencing (data not shown).
List of somatic mutations identified in the MDS index patient (UPN11)
| Gene . | Exonic function . | Amino acid change . | Chromosome . | Position . | Reference . | Observed . | Tumor (reads Ref/Obs) . | CD3+ T cells (reads Ref/Obs) . |
|---|---|---|---|---|---|---|---|---|
| ACOX1 | Nonsynonymous SNV | NM_001185039:c.G1534T:p.A512S | 17 | 73945378 | C | A | 100/49 | 195/1 |
| ZNF354C | Nonsynonymous SNV | NM_014594:c.G821T:p.C274F | 5 | 178506254 | G | T | 74/27 | 152/0 |
| DPF2 | Stopgain SNV | NM_006268:c.C661T:p.R221X | 11 | 65113160 | C | T | 9/8 | 70/0 |
| RUNX1 | Nonsynonymous SNV | NM_001001890:c.C341T:p.S114L | 21 | 36252940 | G | A | 40/24 | 44/0 |
| BCOR | Stopgain SNV | NM_001123384:c.C3433T:p.R1163X | X | 39923604 | G | A | 2/8 | 17/0 |
| GPR179 | Nonsynonymous SNV | NM_001004334:c.G5980T:p.A1994S | 17 | 36483472 | C | A | 147/14 | 629/15 |
| STAG2 | Stopgain SNV | NM_006603:c.C775T:p.R259X | X | 123181311 | C | T | 19/9 | 29/0 |
| EIF5B | Nonsynonymous SNV | NM_015904:c.G895A:p.A299T | 2 | 99978259 | G | A | 32/9 | 32/0 |
| Gene . | Exonic function . | Amino acid change . | Chromosome . | Position . | Reference . | Observed . | Tumor (reads Ref/Obs) . | CD3+ T cells (reads Ref/Obs) . |
|---|---|---|---|---|---|---|---|---|
| ACOX1 | Nonsynonymous SNV | NM_001185039:c.G1534T:p.A512S | 17 | 73945378 | C | A | 100/49 | 195/1 |
| ZNF354C | Nonsynonymous SNV | NM_014594:c.G821T:p.C274F | 5 | 178506254 | G | T | 74/27 | 152/0 |
| DPF2 | Stopgain SNV | NM_006268:c.C661T:p.R221X | 11 | 65113160 | C | T | 9/8 | 70/0 |
| RUNX1 | Nonsynonymous SNV | NM_001001890:c.C341T:p.S114L | 21 | 36252940 | G | A | 40/24 | 44/0 |
| BCOR | Stopgain SNV | NM_001123384:c.C3433T:p.R1163X | X | 39923604 | G | A | 2/8 | 17/0 |
| GPR179 | Nonsynonymous SNV | NM_001004334:c.G5980T:p.A1994S | 17 | 36483472 | C | A | 147/14 | 629/15 |
| STAG2 | Stopgain SNV | NM_006603:c.C775T:p.R259X | X | 123181311 | C | T | 19/9 | 29/0 |
| EIF5B | Nonsynonymous SNV | NM_015904:c.G895A:p.A299T | 2 | 99978259 | G | A | 32/9 | 32/0 |
Ref/Obs, reference/observed; SNV, single-nucleotide variant.
Mutation status of BCOR and BCORL1 among 354 MDS, 54 CMML, and 22 AML-MRC patients
Among the 354 MDS patients, a total of 15 patients displayed 15 BCOR alterations (4.2%; 95% confidence interval [CI], 2.6-6.9) and 3 patients were mutated for BCORL1 (0.8%; 95% CI, 0.8-2.5) (Table 3). A single patient (UPN4) showed both an acquired nonsense and a validated germline missense of BCOR (Table 3). These changes spread all over the entire coding regions (Figure 1A). Among the 15 BCOR truncating mutations, 8 were frameshift, 5 were nonsense, and 2 were splice site mutation. Only alterations affecting the most conserved 2 nucleotides at exon/intron junctions have been considered as splice-site mutations. Nontumoral DNA was available in 10 of the 15 patients with BCOR mutations, and the somatic nature of the mutations was confirmed in all 10 cases by direct sequencing. One frameshift and 2 nonsense mutations were identified for BCORL1. The somatic origin could be verified for 2 truncating BCORL1 mutations. Two patients showed a concomitant inactivation of BCOR and BCORL1 (UPN 9 and UPN 10, Table 3). No significant differences in age, sex, karyotype, blood counts, or BM blasts between BCORmut and BCORwt patients was observed. Mutations were observed in all IPSS risk groups and WHO subtypes (Table 1 and supplemental Table 2). BCOR mutation frequencies did not differ between the 222 French and the 132 Asian MDS patients (5% vs 3%; P = .59). Comparison of cytological BM reports revealed a trend for a higher rate of dysgranulopoiesis in BCORmut patients (P = .06, Table 3). BCOR mutations were significantly enriched in RUNX1mut patients (17% vs 3% in RUNX1wt patients; P = .002) and DNMT3Amut patients (13% vs 3% in DNMT3Awt patients; P = .015; Figure 1B).
Main characteristics of 30 patients with myelodysplasia related disorders harboring a BCOR (transcript ENST00000378444) or a BCORL1 (transcript ENST00000218147) mutation
| Patient . | BCOR or BCORL1 variation . | Finding in nontumor or DNA . | Age . | Gender . | Disease . | WHO type . | IPSS score . | Karyotype . | Status . |
|---|---|---|---|---|---|---|---|---|---|
| UPN1 | BCOR: Ex12-Splice-site | Somatic | 95 | F | MDS | RAEB-2 | High risk | 46, XX [20] | Alive |
| UPN2 | BCOR: W534fs | Somatic | 68 | M | MDS | RARS | Low risk | 45, X, -Y [20] | Dead |
| UPN3 | BCOR: V1616fs | Somatic | 71 | F | MDS | RAEB-1 | Intermediate-1 | 46, XX [32] | Alive |
| UPN4 | BCOR: R1514X, A1695P | Somatic, germline | 85 | F | MDS | RCMD | Intermediate-2 | N/A | Dead |
| UPNS | BCOR: G1376X | Somatic | 73 | M | MDS | RAEB-1 | Intermediate-1 | 4B, XY [13] | Dead |
| UPN6 | BCOR: Ex13-Splice-site | N/A | 78 | F | MDS | MD5-U | Low risk | 46, XY [20] | Alive |
| UPN7 | BCOR: L1296fs | N/A | 63 | F | MDS | RAEB-2 | Intermediate-2 | 46, XX [20] | Dead |
| UPN8 | BCOR: T5fs | N/A | 66 | F | MDS | RCM D-RS | Low risk | 46, XX [20] | Dead |
| UPN9 | BCOR: D1040fs, BCORL1: C1467fs | Somatic somatic | 70 | M | MDS | RARS | Intermediate-1 | 46, XY, t(4; 12)(q28; p12)[5]/46, XY [15] | Alive |
| UPN10 | BCOR: W534X, BCORL1: R1196X | Somatic somatic | 75 | F | MDS | RAEB-1 | Intermediate-1 | 47, XX, +6[14]/46, XX[6] | Dead |
| UPN11 | BCOR: R1163X | Somatic | 57 | M | MDS | RAEB-1 | Intermediate-1 | 46, XY[13] | Dead |
| CMML-058 | BCOR: Q995X | N/A | 81 | M | CMML | CMML2 | Intermediate-2 | 46, XY | Dead |
| CMML-072 | BCOR: C1606fs | N/A | 74 | M | CMML | CMML2 | Intermediate-2 | 46, XY | Alive |
| CMML-076 | BCOR: K941X | N/A | 66 | M | CMML | CMML2 | Intermediate-2 | 46, XY | Dead |
| CMML-083 | BCOR: Q1220X | Somatic | 83 | M | CMML | CMML2 | N/A | 47, XY, +14 | Alive |
| MDS-176 | BCOR: R861fs | N/A | 76 | M | MDS | RAEB2 | Intermediate-2 | 46, XY | Dead |
| MDS-187 | BCOR: E1108X | N/A | 75 | M | MDS | RAEB2 | Intermediate-2 | 46, XY | Dead |
| M DS-204 | BCOR: D1541fs | N/A | 51 | M | MDS | RAEB2 | Intermediate-2 | 46, XY | Dead |
| MDS-217 | BCOR: V1385fs | Somatic | 78 | F | MDS | RAEB2 | N/A | 46, XX | Alive |
| CMML-055 | BCORL1: R72fs | N/A | 90 | M | CMML | CMML2 | N/A | N/A | Dead |
| MDS-174 | BCORL1: R1196X | N/A | 48 | F | MDS | RAEB2 | High risk | 4B, XX, dup(1)(q21q32) | Dead |
| AML-MRC-016 | BCORL1: I754fs | N/A | 34 | M | AML-MRC | AML/MRC | N/A | 46.XY | Dead |
| AML-MRC-021 | BCORL1: S1679fs | N/A | 57 | M | AML-MRC | AML/MRC | N/A | 46.XY | Alive |
| Patient . | BCOR or BCORL1 variation . | Finding in nontumor or DNA . | Age . | Gender . | Disease . | WHO type . | IPSS score . | Karyotype . | Status . |
|---|---|---|---|---|---|---|---|---|---|
| UPN1 | BCOR: Ex12-Splice-site | Somatic | 95 | F | MDS | RAEB-2 | High risk | 46, XX [20] | Alive |
| UPN2 | BCOR: W534fs | Somatic | 68 | M | MDS | RARS | Low risk | 45, X, -Y [20] | Dead |
| UPN3 | BCOR: V1616fs | Somatic | 71 | F | MDS | RAEB-1 | Intermediate-1 | 46, XX [32] | Alive |
| UPN4 | BCOR: R1514X, A1695P | Somatic, germline | 85 | F | MDS | RCMD | Intermediate-2 | N/A | Dead |
| UPNS | BCOR: G1376X | Somatic | 73 | M | MDS | RAEB-1 | Intermediate-1 | 4B, XY [13] | Dead |
| UPN6 | BCOR: Ex13-Splice-site | N/A | 78 | F | MDS | MD5-U | Low risk | 46, XY [20] | Alive |
| UPN7 | BCOR: L1296fs | N/A | 63 | F | MDS | RAEB-2 | Intermediate-2 | 46, XX [20] | Dead |
| UPN8 | BCOR: T5fs | N/A | 66 | F | MDS | RCM D-RS | Low risk | 46, XX [20] | Dead |
| UPN9 | BCOR: D1040fs, BCORL1: C1467fs | Somatic somatic | 70 | M | MDS | RARS | Intermediate-1 | 46, XY, t(4; 12)(q28; p12)[5]/46, XY [15] | Alive |
| UPN10 | BCOR: W534X, BCORL1: R1196X | Somatic somatic | 75 | F | MDS | RAEB-1 | Intermediate-1 | 47, XX, +6[14]/46, XX[6] | Dead |
| UPN11 | BCOR: R1163X | Somatic | 57 | M | MDS | RAEB-1 | Intermediate-1 | 46, XY[13] | Dead |
| CMML-058 | BCOR: Q995X | N/A | 81 | M | CMML | CMML2 | Intermediate-2 | 46, XY | Dead |
| CMML-072 | BCOR: C1606fs | N/A | 74 | M | CMML | CMML2 | Intermediate-2 | 46, XY | Alive |
| CMML-076 | BCOR: K941X | N/A | 66 | M | CMML | CMML2 | Intermediate-2 | 46, XY | Dead |
| CMML-083 | BCOR: Q1220X | Somatic | 83 | M | CMML | CMML2 | N/A | 47, XY, +14 | Alive |
| MDS-176 | BCOR: R861fs | N/A | 76 | M | MDS | RAEB2 | Intermediate-2 | 46, XY | Dead |
| MDS-187 | BCOR: E1108X | N/A | 75 | M | MDS | RAEB2 | Intermediate-2 | 46, XY | Dead |
| M DS-204 | BCOR: D1541fs | N/A | 51 | M | MDS | RAEB2 | Intermediate-2 | 46, XY | Dead |
| MDS-217 | BCOR: V1385fs | Somatic | 78 | F | MDS | RAEB2 | N/A | 46, XX | Alive |
| CMML-055 | BCORL1: R72fs | N/A | 90 | M | CMML | CMML2 | N/A | N/A | Dead |
| MDS-174 | BCORL1: R1196X | N/A | 48 | F | MDS | RAEB2 | High risk | 4B, XX, dup(1)(q21q32) | Dead |
| AML-MRC-016 | BCORL1: I754fs | N/A | 34 | M | AML-MRC | AML/MRC | N/A | 46.XY | Dead |
| AML-MRC-021 | BCORL1: S1679fs | N/A | 57 | M | AML-MRC | AML/MRC | N/A | 46.XY | Alive |
Patients UPN9 and UPN10 harbor mutations in both genes.
NA, not applicable.
BCOR and BCORL1 truncating mutations in MDS and related disorders. (A) Localization of mutations identified in BCOR and BCORL1 genes. Each mutation is shown with an arrow. Only frameshift, nonsense, and splice site mutations are indicated. Confirmed somatic mutations are discriminated by an asterisk. The domain structures are shown in colored boxes as indicated. (B) Co-occurrence of BCOR mutations with other genes studied in 354 MDS patients (Circos Graph made on http://circos.ca/; TM, truncating mutations). The P value from Fisher’s exact test is shown, and the gene appears in red for statistically significant associations
BCOR and BCORL1 truncating mutations in MDS and related disorders. (A) Localization of mutations identified in BCOR and BCORL1 genes. Each mutation is shown with an arrow. Only frameshift, nonsense, and splice site mutations are indicated. Confirmed somatic mutations are discriminated by an asterisk. The domain structures are shown in colored boxes as indicated. (B) Co-occurrence of BCOR mutations with other genes studied in 354 MDS patients (Circos Graph made on http://circos.ca/; TM, truncating mutations). The P value from Fisher’s exact test is shown, and the gene appears in red for statistically significant associations
Among the 54 CMML patients, 3 BCOR nonsense and 1 frameshift mutations were identified (4/54 = 7.4%; 95% CI, 2.9-17.6; Table 3 and Figure 1). All 4 BCORmut CMML patients were classified as CMML-2 disease type at diagnosis (P = .047; supplemental Table 3). The most common concomitant mutations found in BCORmut CMML patients affected U2AF1 in 3 cases (P = .003) and RUNX1 in 2 patients, while CBL, DNMT3A, NRAS, and TET2 mutations were found in single cases each. One CMML patient showed a BCORL1 mutation (1/54 = 1.8%; 95% CI, 0.3-9.8). Although no BCOR mutations were identified in patients with AML-MRC, this disease entity exhibited the highest frequency of BCORL1 mutations, with 2 frameshift cases (2/22 = 9.1%; 95% CI, 2.5-27.8). BCORL1 mutations were the fourth most frequent molecular aberration after TP53 (40.9%), RUNX1 (27.3%), and U2AF35 (18.2%) mutations in AML-MRC.
Mutant sequence burden of BCOR mutated patients
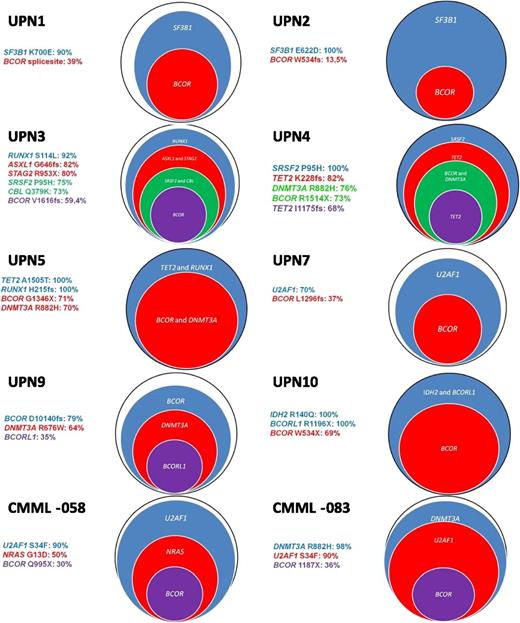
To get insights into the hierarchy of somatic mutations in BCOR-mutated patients, we first evaluated mutant-copy burden in flow-sorted BM cells from a female patient UPN3 (Table 3 and Figure 2) using deep-sequencing technologies. To determine the estimated cell frequency, the mutated read count was multiplied by 2 according to the 2 gene copies per cell expected for normal karyotype. For male patients with mutations affecting genes located on the X chromosome (eg, BCOR, BCORL1, or STAG2), the quantified read count directly translated into the estimated cell frequency. BCOR V1616fs and other concomitant mutations of UPN3 (ASXL1, CBL, RUNX1, STAG2, and SRSF2) were quantified in flow-sorted CD3+ T cells, CD19+ B-cells, CD34+CD38− progenitors, and in the bulk of mononuclear BM cells. The purity of each sorted fraction was superior to 95%. Every mutation was covered with a median of 6415 reads (range, 1160-27 766 reads). The BCOR V1616fs mutation was identified in 7317 of 24 600 reads (29.7%), which corresponds to an estimated totality of 59.4% of cells carrying this mutation in the bulk of BM cells. RUNX1, ASXL1, STAG2, SRSF2, and CBL mutations were found with decreasing frequencies ranging from 92% to 73% of total BM cells (Figure 2, UPN3). In concordance with these results, the BCOR mutation was detected at lower frequencies in CD34+CD38− early hematopoietic progenitors (25% of cells compared with 90% of cells harboring RUNX1, STAG2, and ASXL1 mutations) and was absent in T and B lymphocytes. Next, we investigated all identified mutations of 10 BCOR mutated patients in bulky BM cells. The quantified frequency of BCOR mutations affected mononuclear BM cells with a range of 13.5% to 79% (Figure 2 and supplemental Table 7). With the exception of 1 patient in whom the BCOR mutation occurred before the DNMT3A and the BCORL1 mutation (Figure 2: UPN9), BCOR mutations presented at low frequency, suggesting they were not involved in disease-initiating events. The mutations detected at the highest frequency affected the splicing machinery (SF3B1, SRSF2, and U2AF1) in 6 patients, while 2 patients showed an early dominance for RUNX1 and RUNX1 and TET2, respectively.
Repartition of BCOR and other mutations by targeted deep resequencing. For each gene, the percentage is representing the estimated number of cells carrying the unique mutation. All UPN are MDS cases.
Repartition of BCOR and other mutations by targeted deep resequencing. For each gene, the percentage is representing the estimated number of cells carrying the unique mutation. All UPN are MDS cases.
BCOR mRNA expression levels according to the BCOR mutation status
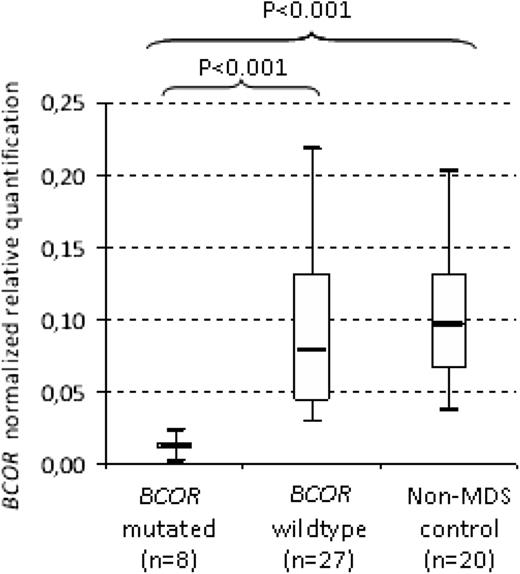
We also evaluated the BCOR mRNA levels from 8 BCORmut patients, 27 BCORwt patients, and 20 non-MDS controls. The distribution of BCOR transcript levels in MDS patients with BCORwt was equivalent to that of non-MDS control samples. BCOR normalized transcript levels did not differ with respect to gender (supplemental Figure 2). BCOR expression levels were significantly lower in BCORmut patients (P < .001; median, 0.012; range, 0.008-0.017) compared with BCORwt patients (median, 0.078; range, 0.071-0.11; Figure 3) and non-MDS controls (P < .001; median, 0.095; range, 0.082-0.120). In addition, BCORwt patients showed a BCOR expression level similar to non-MDS control (P ≤ .001). These data suggest a control of the mutated transcript by the nonsense-mediated decay system.
BCOR mRNA expression levels of BMMC according to the BCOR mutation status of 35 MDS patients and 20 non-MDS patients.P value from Student t test of Δ Ct values.
BCOR mRNA expression levels of BMMC according to the BCOR mutation status of 35 MDS patients and 20 non-MDS patients.P value from Student t test of Δ Ct values.
Prognostic impact of BCOR mutations
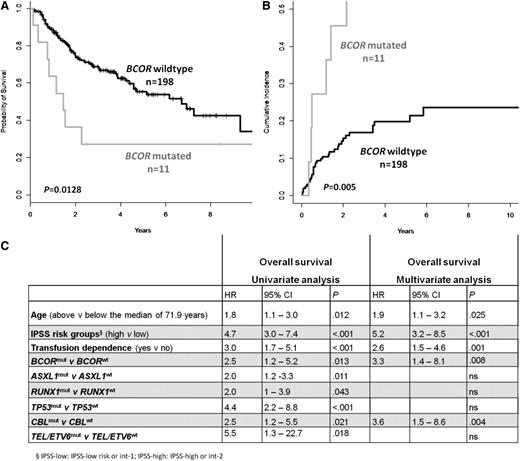
The prognostic impact of BCOR mutations was evaluated in French MDS patients for whom follow-up information was available (n = 209). The median follow-up of patients alive was 33.0 months by February 2013. In univariate analysis, an inferior OS (P = .013) and a higher cumulative incidence of AML transformation (P = .005) was observed for 11 patients with truncating BCOR mutations (Figures 4A-B). The negative impact of BCOR mutations on OS is observed in patients with low-risk (IPSS and intermediate-1 patients; P = .05) and in patients with high-risk MDS (IPSS intermediate-2 and high patients; P = .003; supplemental Figure 3A-B). The low number of BCORL1-mutated patients precludes any statistical analyses, but clinical follow-up was available for 3 MDS patients. All died 7, 17, and 28 months after MDS diagnosis. In univariate analyses, ASXL1, CBL, RUNX1, TEL/ETV6, and TP53 were associated with a poor prognosis for OS. Multivariate analysis including age, IPSS risk groups, transfusion dependence, mutational status for ASXL1, RUNX1, TEL/ETV6, TP53, and CBL revealed the presence of a BCOR mutation as an independent unfavorable prognostic factor for OS (hazard ratio, 3.3; 95% CI, 1.4-8.1; P = .008; Figure 4C).
Negative impact on OS and on the cumulative incidence of AML transformation of BCOR truncating mutations in MDS patients. (A) Kaplan-Meier curve for OS and (B) cumulative incidence of AML transformation according to BCOR mutation status (Fine & Gray test). (C) Univariate and multivariate analyses for OS in MDS patients.
Negative impact on OS and on the cumulative incidence of AML transformation of BCOR truncating mutations in MDS patients. (A) Kaplan-Meier curve for OS and (B) cumulative incidence of AML transformation according to BCOR mutation status (Fine & Gray test). (C) Univariate and multivariate analyses for OS in MDS patients.
Discussion
Next-generation sequencing allowed the identification of a BCOR inactivating mutation in an IPSS intermediate-1 low-risk patient who suffered from an aggressive disease course with AML progression within 5 months after diagnosis and death 8 months later. To define the prevalence of BCOR and BCORL1 mutations, we investigated a large cohort of 354 well-characterized MDS patients. A total of 15 truncating BCOR (4.2%) and 3 BCORL1 (0.8%) alterations were identified all over the coding regions (Figure 1A). The somatic nature of truncating BCOR mutations was confirmed whenever nontumor DNA was available (10 cases). Two patients (UPN9 and UPN10, Table 3) showed concomitant truncating mutations of BCOR and BCORL1. Mutations were found in patients of all IPSS risk groups and WHO subtypes. BCOR and BCORL1 mutations were found in 7.4% and 1.8% of 54 CMML patients. In line with the initial report in which 4 of the 10 identified BCORL1 mutations were observed in AML-MRC,17 BCORL1 mutations were most often found in patients with AML-MRC disease (9.1%).
At least 1 mutation in any of the 18 investigated genes was found in 252 of the 354 MDS patients (71.2%). Isolated BCOR or BCORL1 mutations were detected in only 3 patients. Truncating BCOR mutations were significantly more frequent in RUNX1mut and DNMT3Amut MDS patients in the global cohort. Although not reaching statistical significance, BCOR mutations were exclusive with mutations in TP53 and ZRSR2, the only 2 putative tumor suppressor genes in the investigated gene set. Our data support the idea that BCOR mutations associate preferentially with a group of molecular aberrations including RUNX1 and DNMT3A mutations as reported in AML.16 However, in CMML, the main observed association was BCORmut/U2AF1mut. BCORmut MDS patients showed lower BCOR mRNA levels, suggesting the activation nonsense-mediate decay pathways. Furthermore, mutant-copy quantification in BM mononuclear cells revealed that BCOR mutations rarely occur at a frequency higher than the other mutations found in the same patient. In all 10 investigated patients, more than 75% of BM cells are affected by the initiating mutation, confirming previous observations of an early clonal dominance in MDS pathogenesis.30,31 We also performed mutant-allele burden quantification of sorted cells from patient UPN3 (supplemental Table 7 and Figure 2). The BCOR V1616fs mutation was absent in T and B lymphocytes and affected 25% of CD34+CD38− progenitors and 60% of bulky BM cells. Other driver mutations such as ASXL1, SRSF2, and STAG2 were detected at higher frequencies in the stem cell compartment but were also found in 35% of B lymphocytes, indicating an appearance of these mutations at an early multipotent stage of hematopoietic differentiation. These data suggests that BCOR mutations are not involved in disease-initiating events but define the clinical course.
BCORmut MDS patients had an inferior OS and a higher cumulative incidence of AML transformation. Multivariate analysis confirmed the independent importance of BCOR mutations for OS. In this cohort mainly composed of MDS patients with low/intermediate-1 IPSS, the impact of BCOR mutations on the response to treatment like azacitidine could not be studied. Our univariate analysis confirms the importance in 4 out of 5 prognostically relevant genes reported in MDS.14 Surprisingly, some of these established molecular markers in MDS lost their significance from the univariate in the subsequent multivariate analysis for OS such as TP53 mutations (Figure 4C and supplemental Table 8). This might be explained by strong associations of variables (eg, TP53 mutations and IPSS risk score). TP53 mutations were found in 22% of IPSS intermediate-2/ high-risk but only in 1.2% of IPSS low-risk/intermediate-1 patients (P < .001). With respect to recently published studies on novel candidate genes in MDS, data on SF3B1, SRSF2, and U2AF35 mutations appear somehow immature and/or inconsistent. While SF3B1 mutations were initially shown to be associated with a favorable prognosis,2,15 4 different MDS cohorts failed to reproduce these data.32-34 In contrast to Graubert et al, whose analysis of 150 MDS patients suggested an increased probability of secondary AML progression for patients with U2AF35 mutation,35 subsequent studies could not confirm this observation.4,36 Also, the reports on SRSF2 mutations are conflicting. Some studies suggest a negative impact on patient outcome,36 whereas others find no independent influence on survival.4,34 This fact may be at least partially related to the heterogeneity of investigated cohorts and also to the importance of preferential associations between distinct gene mutations (eg, SF3B1 and DNMT3A, or U2AF35 and ASXL1).4,34 Considering these data together, we propose an algorithm for the molecular workup of MDS (supplemental Figure 4).
In summary, truncating mutations of BCOR were independently associated with a worse OS in MDS. Our study warrants confirmation but suggests consideration of the BCOR gene in the diagnostic workup of MDS patients.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the investigators of the Groupe Francophone des Myélodysplasies (Dr B. Slama, CH Avignon; Dr A. Stamatoulas-Bastard, Centre de Lutte Contre le Cancer, CHU Rouen; Dr M. Gardembas; Dr Aline Schmidt-Tanguy; Dr M. Dib, Angers; Dr S. Natarajan-Ame, Strasbourg; Dr E. Gyan, Tours; Dr L. Legros, Nice; Dr Odile Beyne-Rauzy, Toulouse; Dr C. Soussain, CH René Huguenin, Saint-Cloud; Dr E. Raffoux, Hôpital Saint-Louis, Paris) for including patients and M. Bourmaud and A. Gauthier for technical assistance.
This study was supported by grants from the Deutsche Krebshilfe (#109686, F. Damm), by the Ministère de la Recherche (V.C.), association “Laurette Fugain” (O.A.B. and F. Dreyfus); Association pour la Recherche sur le Cancer (no. 4992, 2010) (D.B.), “Direction de la recherche clinique Assistance Publique-Hôpitaux de Paris (PHRC MDS-04),” the Cancer Research and Personalized Medicine Center, “INCa génomique et fonction des gènes dans les cancers-valorisation des ressources biologiques 2008” (M.F.), and INCa (O.A.B.), and a labilization from the Ligue Nationale Contre le Cancer. S.O.is supported by grant-in-aids from the Ministry of Health, Labor and Welfare of Japan and KAKENHI (23249052, 22134006, and 21790907) and by the project for development of innovative research on cancer therapies (p-direct).
Authorship
Contribution: F. Damm, O.K., O.A.B., and M.F. designed the research; F. Damm, V.C., Y.N., K.Y., Y.O., O.K., V.G.-B., A.R., and L.S. performed the research; F. Dreyfus, S.P., A.G.-B., B.Q., E.S., S.C., C.R., T.P., S.d.B., L.-Y.S., and N.V. contributed patient samples and clinical data; F. Damm, Y.N., Y.K., Y.O., O.K., Y.S., M.S., S.M., V.G.-B., V.C., R.I., D.B., and A.R. analyzed the data; F. Damm, O.K., S.O., O.A.B., and M.F. wrote the paper. All authors read and agreed to the final version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Olivier Kosmider, Service d’Hématologie Biologique, GH Broca-Cochin-Hôtel-Dieu, and Institut Cochin, 27 rue du Fg St Jacques 75014 Paris, France; e-mail: olivier.kosmider@cch.aphp.fr; and Michaela Fontenay, Service d’Hématologie Biologique, GH Broca-Cochin-Hôtel-Dieu, and Institut Cochin, 27 rue du Fg St Jacques 75014 Paris, France; e-mail: michaela.fontenay@inserm.fr.
References
Author notes
F. Damm, V.C., and Y.N. contributed equally to this work.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal