Key Points
Variation at 10p12.2 (PIP4K2A) and 10p14 (GATA3) influences ALL risk and tumor subtype.
GATA3 genotype is a determinant of event-free survivorship.
Abstract
Acute lymphoblastic leukemia (ALL) is the major pediatric cancer diagnosed in economically developed countries with B-cell precursor (BCP)-ALL, accounting for approximately 70% of ALL. Recent genome-wide association studies (GWAS) have provided the first unambiguous evidence for common inherited susceptibility to BCP-ALL, identifying susceptibility loci at 7p12.2, 9p21.3, 10q21.2, and 14q11.2. To identify additional BCP-ALL susceptibility loci, we conducted a GWAS and performed a meta-analysis with a published GWAS totaling 1658 cases and 4723 controls, with validation in 1449 cases and 1488 controls. Combined analysis identified novel loci mapping to 10p12.2 (rs10828317, odds ratio [OR] = 1.23; P = 2.30 × 10−9) and 10p14 marked by rs3824662 (OR = 1.31; P = 8.62 × 10−12). The single nucleotide polymorphism rs10828317 is responsible for the N215S polymorphism in exon 7 of PIP4K2A, and rs3824662 localizes to intron 3 of the transcription factor and putative tumor suppressor gene GATA3. The rs10828317 association was shown to be specifically associated with hyperdiploid ALL, whereas the rs3824662-associated risk was confined to nonhyperdiploid non–TEL-AML1 + ALL. The risk allele of rs3824662 was correlated with older age at diagnosis (P < .001) and significantly worse event-free survivorship (P < .0001). These findings provide further insights into the genetic and biological basis of inherited genetic susceptibility to BCP-ALL and the influence of constitutional genotype on disease development.
Introduction
Acute lymphoblastic leukemia (ALL) is the major pediatric cancer in developed countries with B-cell precursor (BCP)-ALL accounting for ∼70% of ALL.1 Little is known, however, about the etiology of ALL, and although there is indirect evidence for an infective origin, no specific environmental risk factors have been identified.2,3
Analysis of the Swedish family–cancer database has provided evidence for inherited predisposition to ALL, independent of the concordance in monozygotic twins (which has an in utero explanation).4 Although the heritable basis of the threefold sibling relative risk is not fully understood, recent genome-wide association studies (GWAS) have shown that common variation at IKZF1(7p12.2), CDKN2A/CDKN2B(9p21), ARID5B(10q21.2), and CEBPE(14q11.2) confer a modest but significant risk.5,6
To identify additional susceptibility loci for BCP-ALL, we conducted an independent primary scan and performed a genome-wide meta-analysis with a previously published GWAS followed by analysis of the top 8 single nucleotide polymorphisms (SNPs) not annotating known loci in an additional case-control series.5
Methods
Ethics
Collection of samples and clinicopathological information from subjects was undertaken with informed consent in accordance with the Declaration of Helsinki and with approval of the ethical review board.
Genome-wide association study
The United Kingdom (UK)-GWAS details have been previously reported.5 Briefly, this analysis, post–quality control (QC), was based on constitutional DNA (ie, remission samples) of 459 white BCP-ALL cases from the United Kingdom Childhood Cancer Study (UKCCS) (258 males; mean age at diagnosis 5.3 years); 342 cases from the UK Medical Research Council ALL 97 (99) trial (190 male; mean age of diagnosis 5.7 years) and 23 cases from Northern Institute for Cancer Research (16 males). Genotyping was performed using Illumina Human 317K arrays (Illumina, San Diego, CA). For controls, we used publicly accessible data generated by the Wellcome Trust Case Control Consortium from the 1958 British Birth Cohort. Genotyping of controls was conducted using Illumina Human 1-2M-Duo Custon_v1 Array chips. Details of genotyping, SNP calling, and QC have been previously reported (www.wtccc.org.uk).
The German GWAS was comprised of 1155 cases (620 males; mean age at diagnosis, 6 years) ascertained through the Berlin-Frankfurt-Münster (BFM) trials (1993-2004) genotyped using Illumina Human OmniExpress-12v1.0 arrays. For controls, we used genotype data on 2125 healthy individuals from the Heinz Nixdorf Recall (HNR) study; there were 704 genotyped using Illumina-HumanOmni1-Quad_v1 and 1428 on Illumina-HumanOmniExpress-12v1.0.
Quality control of GWAS datasets
DNA samples with GenCall scores <0.25 at any locus were considered “no calls.” An SNP was deemed to have failed if <95% of DNA samples generated a genotype at the locus. Cluster plots were manually inspected for SNPs considered for replication. The same quality control metrics on the German GWAS data were applied as in the UK GWAS.5 We removed individuals aged >16 years (n = 10); sex discrepancy (n = 2) and samples for whom <95% of SNPs were successfully genotyped (n = 5) (supplemental Figure 1). We computed identity-by-state (IBS) probabilities for all pairs to search for duplicates and closely related individuals among samples (defined as IBS ≥0.80, thereby excluding first-degree relatives). For all identical pairs, the sample having the highest call rate was retained, thereby eliminating 3 samples. To identify individuals who might have non-Western European ancestry, we merged our data with phase II HapMap samples (60 Western European [CEU], 60 Nigerian [YRI], 90 Japanese [JPT] and 90 Han Chinese [CHB]). For each pair of individuals, we calculated genome-wide IBS distances on markers shared between HapMap and our SNP panel, and we used these as dissimilarity measures on which to perform principal component analysis. The first 2 principal components for each individual were plotted, and 37 samples showing marked separation from the CEU cluster was excluded from the analyses. Due to the spread of the case cluster, we then performed an additional principal component analysis step making use of phase III HapMap samples (111 CEU, 88 Toscans in Italy [TSI] individuals), and we removed a further 265 cases (and 9 controls) not present in the main cluster.
We filtered out SNPs having a minor allele frequency of <1%, and a call rate of <95% in cases or controls. We also excluded SNPs showing departure from Hardy-Weinberg equilibrium at P < 10−6. For replication and validation analysis, call rates were >95% per 384-well plate for each SNP.
Replication series and genotyping
The replication series comprised of 1501 patients (794 males; mean age at diagnosis, 6.2 years) ascertained through the BFM trials (1993-2004).7 The 1516 controls (762 males; mean age, 58.2 years) were ethnically-matched healthy individuals of German origin recruited in 2004 at the Institute of Transfusion Medicine in Mannheim, Germany. As with the samples that were the subject of GWAS, immunophenotyping of diagnostic samples were undertaken using standard methods. Genotyping was performed using competitive allele-specific polymerase chain reaction KASPar chemistry (KBiosciences Ltd., Hertfordshire, UK) or Taqman (Applied Biosystems, Foster City, CA). All primers and probes that were used are available on request. Samples having SNP call rates of <90% were excluded from the analysis. To ensure quality of genotyping in all assays, at least 2 negative controls and 1% to 2% duplicates (concordance >99.99%) were genotyped.
T-ALL cases
There were 83 UK (53 males; mean age at diagnosis, 7.4 years; standard deviation 3.4) and 246 German (170 males; mean age at diagnosis, 9.0 years; standard deviation 4.5) childhood T-ALL cases were studied. The cases were ascertained through the same mechanisms and were genotyped as part of the same GWAS at each center imposing identical QC metrics.
Statistical and bioinformatic analysis
Main analyses were undertaken using R (v2.6), Stata v.10 (State College, TX) and PLINK (v1.06)8 software. The association between each SNP and risk was assessed by the Cochran-Armitage trend test. The adequacy of case-control matching and possibility of differential genotyping of cases and controls were formally evaluated using quantile-quantile plots of test statistics. The inflation factor λ was based on the 90% least significant SNPs.9 We adjusted for possible population substructure using Eigenstrat.10 The ORs and associated 95% confidence intervals (CIs) were calculated by unconditional logistic regression. Meta-analysis was conducted using standard methods under a fixed-effects model. Cochran’s Q statistic to test for heterogeneity and the I2 statistic to quantify the proportion of the total variation due to heterogeneity were calculated.11 Associations by sex and clinicopathological phenotypes were examined by logistic regression. The relationship between genotype and age were compared using a Wilcoxon-type test for trend.12
We used receiver operator characteristic curve analysis to estimate the proportion of the genetic variance on the liability scale attributable to 7p12.2, 9p21.3, 10p12.2, 10p14, 10q21.2, and 14q11.2 SNPs.13
Prediction of the untyped SNPs was carried out using IMPUTE2, based on the 1000 genomes phase 1 integrated variant set (b37) from March 2012. To filter poorly imputed SNPs, as previously recommended, we excluded variants having information scores from SNPTEST v2.3.0 < 0.4. Imputed data were analyzed using SNPTEST v2.3.0 to account for uncertainties in SNP prediction.
Linkage disequilibrium (LD) metrics were calculated in PLINK using 1000 genomes data and were plotted using SNAP. LD blocks were defined on the basis of HapMap recombination rate, as defined by using the Oxford recombination hotspots,14 and on the basis of distribution of CIs.15
Sequence conservation metrics Genomic evolutionary rate profiling (GERP) and PhastCons, as well as conserved transcription factor binding sites were obtained (http://snp.gs.washington.edu/SeattleSeqAnnotation134/ and http://genome.ucsc.edu/cgi-bin/hgGateway). GERP is an estimate of evolutionary constraint with a score that reflects the proportion of substitutions at that site rejected by selection compared with observed substitutions expected under a neutral evolutionary model, using a sequence alignment of 35 mammalian species16 ; the score per site has been standardized by UCSC to range from −12 to 6, with 6 being indicative of complete conservation. PhastCons scores reflect the probability that a given nucleotide is conserved, based on sequence alignment of 17 vertebrate species; the score ranges from 0 to 1, in which 1 is most conserved.17 To explore epigenetic profile of association signals, we used chromatin state segmentation data from the Encode Project18 lymphoblastoid cell lines data. States were inferred from ENCODE Histone Modification data (H4K20me1, H3K9ac, H3K4me3, H3K4me2, H3K4me1, H3K36me3, H3K27me3, H3K27ac, and CTCF), binarized using a multivariate Hidden Markov Model. We used RegulomeDB and HaploReg to examine if any of the SNPs annotate putative transcription factor binding/enhancer elements.
Relationship between SNP genotype and survivorship
To investigate if genotype is associated with clinical phenotype or outcome, we analyzed data on 2258 patients recruited to AIEOP-BFM 2000 (ie, from both German series).7 Briefly, patients received standard chemotherapy (ie, prednisone, vincristine, daunorubicin, l-asparaginase, cyclophosphamide, ifosfamide, cytarabine, 6-mercaptopurine, 6-thioguanine, and methotrexate) with a subset of high-risk patients treated with cranial irradiation and/or stem cell transplantation. Event-free survival (EFS) was defined as the time from diagnosis to the date of last follow-up in complete remission or to the first event. Events were resistance to therapy (nonresponse), relapse, secondary neoplasm, or death from any cause. Failure to achieve remission due to early death or nonresponse was considered as an event at time zero and patients lost to follow-up were censored at the time of their withdrawal. Patients were stratified into 3 categories: standard, intermediate, and high risk. Although minimal residual disease (MRD) analysis was the main stratification criterion, high risk was also defined by prednisone poor-response or ≥5% leukemic blasts in bone marrow on day 33, or t(9;22)/t(4;11) positivity or their molecular equivalents (BCR-ABL/MLL-AF4-fusion) independent of MRD status. Standard patients were MRD-negative on treatment day 33 (TP1) and 78 (TP2) and had no high-risk criteria. High-risk patients were defined as having residual disease (≥10−3 cells) at TP2. Intermediate patients had positive-MRD detection at either TP1 or TP2, but had a cell count of <10−3 at TP2. The Kaplan-Meier method was used to estimate survival rates, with differences compared using the log-rank test (two-sided P values). Cumulative incidences of competing events were calculated by the method of Kalbfleisch and Prentice,19 and compared by Gray’s test. 20 Cox regression analysis was used to estimate hazard ratios and 95% CIs adjusting for clinically important covariates.
Relationship between SNP genotype and messenger RNA expression
To examine for a relationship between SNP genotype and expression, we made use of publicly available expression data generated on lymphoblastoid cell lines from HapMap3, Geneva, and the Multiple Tissue Human Expression Resource pilot data using Sentrix Human-6 Expression BeadChips.21-23
Results
Genotype data from each GWAS were filtered and resulted in the use of 162 341 autosomal SNPs common to both case-control series. A total of 322 case samples from the German GWAS were removed during quality control steps for reasons that included a failure to genotype, unknown duplicates, age of diagnosis >16, being closely related individuals or non-CEU ancestry (supplemental Figures 1 and 2). Quality control steps for the UK GWAS have been previously reported.5
Quantile-quantile plots of the genome-wide χ-squared values showed minimal inflation of the test statistics, rendering substantial cryptic population substructure or differential genotype calling between cases and controls unlikely in either GWAS (genomic control inflation factor,9 λgc = 1.003 and 1.13 in UK and German GWAS, respectively) (supplemental Figure 3). For completeness, EIGENSTRAT was used for the German GWAS to determine the effects of population substructure on our findings (λcorrected = 1.05) (supplemental Figure 3). To facilitate harmonization of data, we imputed 220 435 SNPs in the UK GWAS, and using data from both GWAS, we derived joint odds ratios (ORs) and 95% CIs for each SNP, and associated P values.
In the meta-analysis association, statistics for SNPs mapping to the 4 known ALL loci 7p12.2(IKZF1), 9p21(CDKN2A/CDKN2B), 10q21.2(ARID5B), and 14q11.2(CEBPE) were genome-wide significant (ie, P < 5.0 × 10−8) (supplemental Table 1). We also identified 8 SNPs showing good evidence of association mapping to distinct loci not previously associated with ALL risk (supplemental Table 1). To validate these findings, we conducted a replication study of the 8 SNPs, genotyping an additional 1501 German BCP-ALL cases and 1516 regional controls. In the combined analysis, 2 SNPs, rs3824662 and rs10828317, showed evidence for an association with risk that was genome-wide significant (supplemental Table 2).
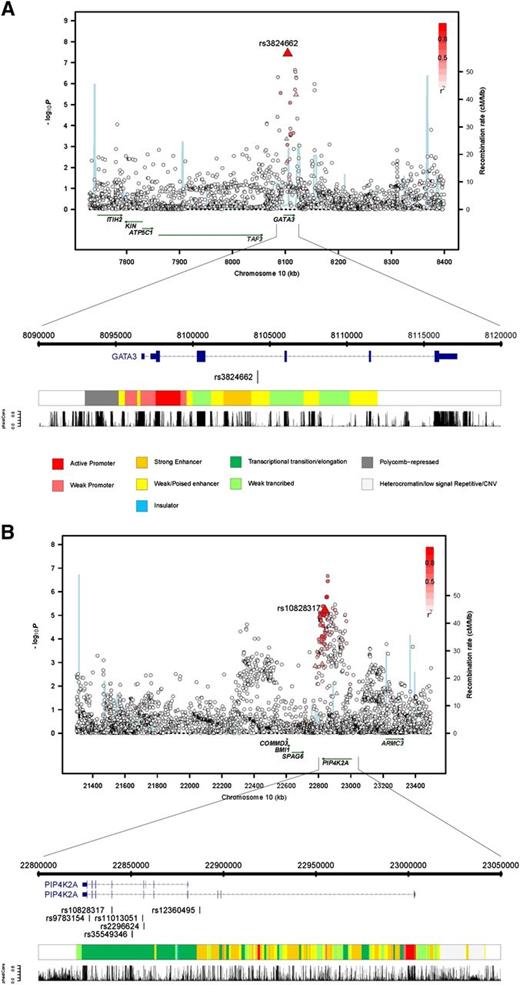
The SNP rs10828317 localizes to 10p12.2 (22 839 628bps; Pcombined = 2.30 × 10−9; OR = 1.23) (Figure 1) and is responsible for the N215S polymorphism in exon 7 of phosphatidylinositol-5-phosphate 4-kinase (PIP4K2A) (type II, α). To explore the region further, we imputed unobserved genotypes in GWAS samples using 1000 genomes data. This analysis revealed only a marginally stronger association than the typed SNP that was provided by rs11013051, which maps to intron 6 of PIP4K2A (P = 2.15 × 10−7, compared with P = 2.88 × 10−6 for rs10828317) (Figure 2). Although N215S is predicted to be benign, it resides within a highly conserved sequence (GERP score, 6.07) raising the possibility of a direct functional basis to the association. Moreover none of the highly correlated SNPs, including rs11013051 (r2 > 0.8) mapping within the association signal, are conserved (GERP < 0.62, PhastCon < 0.18) and do not reside within a functionally active domain.
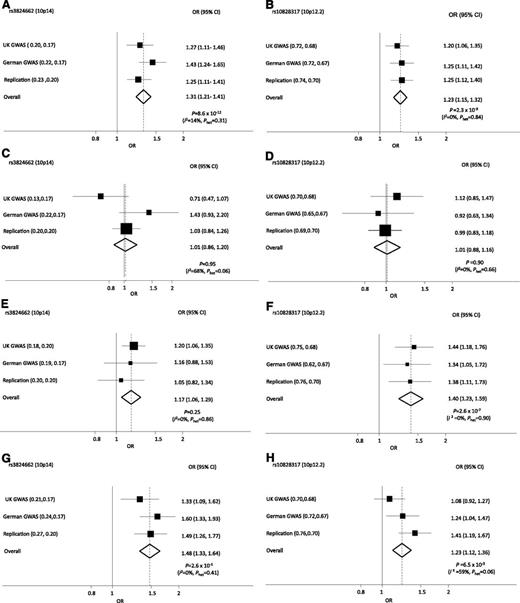
Forest plots of ORs for ALL associated with rs10828317 (PIP4K2A) and rs3824662 (GATA3) genotype. ORs for all BCP-ALL (A-B), TEL-AML ALL (C-D), HD ALL (E-F), non–HD/TEL-AML ALL (G-H).
Forest plots of ORs for ALL associated with rs10828317 (PIP4K2A) and rs3824662 (GATA3) genotype. ORs for all BCP-ALL (A-B), TEL-AML ALL (C-D), HD ALL (E-F), non–HD/TEL-AML ALL (G-H).
Regional plots of association results, recombination rates, and chromatin state segmentation track for (A) 10p12.2 and (B) 10p14 susceptibility loci. The top panel shows the association results of both genotyped (triangles) and imputed (circles) SNPs in the GWAS samples and recombination rates for rates within the two loci. For each plot, −log10P values (y-axis) of the SNPs are shown according to their chromosomal positions (x-axis). The top genotyped SNP in each combined analysis is a large triangle and is labeled by its reference SNP ID. The color intensity of each symbol reflects the extent of LD with the top genotyped SNP: white (r2 = 0) through to dark red (r2 = 1.0). Genetic recombination rates (cM/Mb), estimated using HapMap CEU samples, are shown with a light blue line. Physical positions are based on National Centre for Biotechnology Information build 36 of the human genome. Also shown are the relative positions of genes and transcripts mapping to each region of association. Genes have been redrawn to show the relative positions; therefore, maps are not to physical scale. The lower panel shows the gene of interest together with all transcripts of the gene showing exons and introns; observed SNP and any imputed SNPs showing a stronger association with ALL risk, chromatin state segmentation track (ChromHMM), and phastCons score values corresponding to the posterior probability associated with a phylogenetic hidden Markov model (HMM) inferring the most conserved state at a given base position.
Regional plots of association results, recombination rates, and chromatin state segmentation track for (A) 10p12.2 and (B) 10p14 susceptibility loci. The top panel shows the association results of both genotyped (triangles) and imputed (circles) SNPs in the GWAS samples and recombination rates for rates within the two loci. For each plot, −log10P values (y-axis) of the SNPs are shown according to their chromosomal positions (x-axis). The top genotyped SNP in each combined analysis is a large triangle and is labeled by its reference SNP ID. The color intensity of each symbol reflects the extent of LD with the top genotyped SNP: white (r2 = 0) through to dark red (r2 = 1.0). Genetic recombination rates (cM/Mb), estimated using HapMap CEU samples, are shown with a light blue line. Physical positions are based on National Centre for Biotechnology Information build 36 of the human genome. Also shown are the relative positions of genes and transcripts mapping to each region of association. Genes have been redrawn to show the relative positions; therefore, maps are not to physical scale. The lower panel shows the gene of interest together with all transcripts of the gene showing exons and introns; observed SNP and any imputed SNPs showing a stronger association with ALL risk, chromatin state segmentation track (ChromHMM), and phastCons score values corresponding to the posterior probability associated with a phylogenetic hidden Markov model (HMM) inferring the most conserved state at a given base position.
The SNP rs3824662 localizes to 10p14 (8 104 208bps; Pcombined = 8.62 × 10−12; OR = 1.31) (Figure 1) and maps within intron 3 of the transcription factor and putative tumor suppressor gene GATA3 (encoding the GATA-binding protein 3 isoform 2; MIM 131320) (Figure 2). Further evidence for variation at 10p14 being a determinant of ALL risk is provided by a previously published candidate gene study of 377 mixed ethnicity ALL cases and 448 controls, which found an association for rs3781093 that is highly correlated with rs3824662 (r2 = 0.90).24 Imputation of untyped genotypes in cases and controls did not recover a stronger association at 10p14 than that provided by rs3824662. Intriguingly, although rs3824662 is not conserved (GERP = −2.76, PhastCon = 0.0), the SNP maps within a predicted enhancer site (Figure 2).
Given the biological heterogeneity of BCP-ALL, we analyzed the association between rs10828317 and rs3824662 genotypes and the major subtypes of BCP-ALL, hyperdiploid, TEL-AML, and others (Figure 1; supplemental Table 3). A consistent association between rs10828317 and risk of hyperdiploid ALL was seen (P = 2.60 × 10−7) (Figure 1). In contrast, the association between ALL risk and rs3824662 genotype was confined to cases that were not hyperdiploid or TEL-AML–positive (Figure 1). To examine for a possible relationship between rs3824662 and other chromosomally defined forms of ALL, we examined an association with t(9;22), t(12;21), t(1;19), and (t4;11) karyotype (supplemental Table 4), but no significant association was shown.
The risk of ALL associated with rs3824662 and rs10828317 was not significantly related to gender in any of the 3 case series (Table 1). Because the incidence of ALL is strongly age-related, we examined if SNP genotype had a modifying effect on age at presentation (Table 1). Although there was no relationship with rs10828317, rs3824662 showed a strong correlation with age with homozygotes for the risk allele diagnosed on average ∼1.5 year older (Table 1).
Relationship between rs10828317 and rs3824662 genotype and age at diagnosis and sex
| SNP . | UK GWAS . | German GWAS . | Replication . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| TT . | TC . | CC . | TT . | TC . | CC . | TT . | TC . | CC . | |
| rs10828317 (10p12.2) (PIP4K2A) | |||||||||
| ALL | |||||||||
| Median age | 4 | 4 | 5 | 4 | 4 | 4 | 4 | 4 | 4 |
| Mean age (SD) | 5.2 (3.5) | 5.5 (3.6) | 5.9 (4.0) | 5.9 (4.1) | 5.6 (4.3) | 6.0 (4.4) | 5.8 (4.3) | 5.9 (4.3) | 5.5 (4.0) |
| F:M | 0.41/0.59 | 0.46/0.54 | 0.53/0.47 | 0.46/0.54 | 0.49/0.51 | 0.39/0.61 | 0.46/0.54 | 0.43/0.57 | 0.58/0.42 |
| Hyperdiploid | |||||||||
| Median age | 4 | 4 | 3 | 3 | 3 | 3 | 4 | 4 | 4 |
| Mean age (SD) | 4.8 (3.1) | 5.0 (3.2) | 4.4 (3.6) | 4.7 (3.7) | 4.7 (3.2) | 4.6 (2.5) | 5.1 (3.9) | 4.8 (3.6) | 6.0 (4.9) |
| F:M | 0.38/0.62 | 0.45/0.56 | 0.63/0.37 | 0.43/0.57 | 0.49/0.51 | 0.36/0.64 | 0.52/0.4 | 0.36/0.64 | 0.58/0.42 |
| TEL-AML | |||||||||
| Median age | 4.5 | 4 | 5 | 4 | 4 | 7 | 4 | 4 | 4 |
| Mean age (SD) | 4.9 (2.7) | 4.6 (2.1) | 4.6 (1.6) | 4.4 (2.7) | 4.6 (2.7) | 6.4 (3.1) | 4.6 (2.9) | 4.9 (2.9) | 4.7 (2.6) |
| F:M | 0.40/0.60 | 0.47/0.53 | 0.18/0.82 | 0.50/0.50 | 0.31/0.69 | 0.56/0.44 | 0.46/0.54 | 0.46/0.54 | 0.56/0.44 |
| Non–HD/TEL-AML | |||||||||
| Median | 4 | 5 | 6 | 6 | 4 | 4.5 | 6 | 6 | 3 |
| Mean age (SD) | 5.7 (4.0) | 6.2 (4.1) | 6.8 (4.4) | 6.7 (4.3) | 5.9 (4.8) | 6.7 (5.0) | 6.9 (4.8) | 7.1 (4.8) | 6.1 (5.5) |
| F:M | 0.43/0.57 | 0.46/0.54 | 0.59/0.41 | 0.46/0.54 | 0.50/0.50 | 0.32/0.68 | 0.46/0.54 | 0.46/0.54 | 0.62/0.38 |
| TT | TG | GG | TT | TG | GG | TT | TG | GG | |
| rs3824662 (10p14) (GATA3) | |||||||||
| ALL | |||||||||
| Median age | 5 | 4.5 | 4* | 7 | 4 | 4† | 6 | 4 | 4‡ |
| Mean age (SD) | 6.1 (3.8) | 5.8 (3.9) | 5.1 (3.3) | 7.0 (4.3) | 6.1(4.3) | 5.5 (4.2) | 6.8 (4.5) | 6.0 (4.4) | 5.6 (4.1) |
| F:M | 0.48/0.52 | 0.42/0.58 | 0.45/0.55 | 0.51/0.49 | 0.46/0.54 | 0.46/0.54 | 0.39/0.61 | 0.45/0.55 | 0.10/0.90 |
| Hyperdiploid | |||||||||
| Median age | 5 | 3 | 4 | 3 | 3 | 4§ | 6 | 3 | 4 |
| Mean age (SD) | 6.0 (3.6) | 4.6 (3.3) | 4.8 (3.0) | 2.8 (1.2) | 4.1 (3.0) | 5.1 (3.7) | 6.5 (4.7) | 4.6 (3.8) | 5.1 (3.8) |
| F:M | 0.43/0.57 | 0.45/0.55 | 0.42/0.58 | 0.44/0.56 | 0.52/0.48 | 0.41/0.59 | 0.46/0.54 | 0.49/0.51 | 0.37/0.63 |
| TEL-AML | |||||||||
| Median age | 6 | 5 | 4 | 3 | 4 | 4 | 4 | 4 | 4 |
| Mean age (SD) | 6.0 (2.8) | 5.2 (2.3) | 4.5 (2.3) | 3.7 (2.2) | 4.8 (2.5) | 4.9 (3.0) | 4.1 (2.0) | 4.7 (2.5) | 4.8 (3.1) |
| F:M | 0.0/1.0 | 0.33/0.67 | 0.44/0.56 | 0.50/0.50 | 0.40/0.60 | 0.44/0.56 | 0.21/0.79 | 0.43/0.57 | 0.30/0.70 |
| Non–HD/TEL-AML | |||||||||
| Median age | 5.5 | 6 | 4 | 8 | 7 | 4|| | 7.5 | 7 | 5¶ |
| Mean age (SD) | 6.2 (4.2) | 6.8 (4.4) | 5.6 (3.9) | 8.0 (4.6) | 7.2 (4.5) | 5.7 (4.4) | 8.3 (5.0) | 7.4 (5.0) | 6.4 (4.6) |
| F:M | 0.56/0.44 | 0.44/0.56 | 0.48/0.52 | 0.48/0.52 | 0.44/0.56 | 0.30/0.70 | 0.31/0.69 | 0.46/0.54 | 0.27/0.73 |
| SNP . | UK GWAS . | German GWAS . | Replication . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| TT . | TC . | CC . | TT . | TC . | CC . | TT . | TC . | CC . | |
| rs10828317 (10p12.2) (PIP4K2A) | |||||||||
| ALL | |||||||||
| Median age | 4 | 4 | 5 | 4 | 4 | 4 | 4 | 4 | 4 |
| Mean age (SD) | 5.2 (3.5) | 5.5 (3.6) | 5.9 (4.0) | 5.9 (4.1) | 5.6 (4.3) | 6.0 (4.4) | 5.8 (4.3) | 5.9 (4.3) | 5.5 (4.0) |
| F:M | 0.41/0.59 | 0.46/0.54 | 0.53/0.47 | 0.46/0.54 | 0.49/0.51 | 0.39/0.61 | 0.46/0.54 | 0.43/0.57 | 0.58/0.42 |
| Hyperdiploid | |||||||||
| Median age | 4 | 4 | 3 | 3 | 3 | 3 | 4 | 4 | 4 |
| Mean age (SD) | 4.8 (3.1) | 5.0 (3.2) | 4.4 (3.6) | 4.7 (3.7) | 4.7 (3.2) | 4.6 (2.5) | 5.1 (3.9) | 4.8 (3.6) | 6.0 (4.9) |
| F:M | 0.38/0.62 | 0.45/0.56 | 0.63/0.37 | 0.43/0.57 | 0.49/0.51 | 0.36/0.64 | 0.52/0.4 | 0.36/0.64 | 0.58/0.42 |
| TEL-AML | |||||||||
| Median age | 4.5 | 4 | 5 | 4 | 4 | 7 | 4 | 4 | 4 |
| Mean age (SD) | 4.9 (2.7) | 4.6 (2.1) | 4.6 (1.6) | 4.4 (2.7) | 4.6 (2.7) | 6.4 (3.1) | 4.6 (2.9) | 4.9 (2.9) | 4.7 (2.6) |
| F:M | 0.40/0.60 | 0.47/0.53 | 0.18/0.82 | 0.50/0.50 | 0.31/0.69 | 0.56/0.44 | 0.46/0.54 | 0.46/0.54 | 0.56/0.44 |
| Non–HD/TEL-AML | |||||||||
| Median | 4 | 5 | 6 | 6 | 4 | 4.5 | 6 | 6 | 3 |
| Mean age (SD) | 5.7 (4.0) | 6.2 (4.1) | 6.8 (4.4) | 6.7 (4.3) | 5.9 (4.8) | 6.7 (5.0) | 6.9 (4.8) | 7.1 (4.8) | 6.1 (5.5) |
| F:M | 0.43/0.57 | 0.46/0.54 | 0.59/0.41 | 0.46/0.54 | 0.50/0.50 | 0.32/0.68 | 0.46/0.54 | 0.46/0.54 | 0.62/0.38 |
| TT | TG | GG | TT | TG | GG | TT | TG | GG | |
| rs3824662 (10p14) (GATA3) | |||||||||
| ALL | |||||||||
| Median age | 5 | 4.5 | 4* | 7 | 4 | 4† | 6 | 4 | 4‡ |
| Mean age (SD) | 6.1 (3.8) | 5.8 (3.9) | 5.1 (3.3) | 7.0 (4.3) | 6.1(4.3) | 5.5 (4.2) | 6.8 (4.5) | 6.0 (4.4) | 5.6 (4.1) |
| F:M | 0.48/0.52 | 0.42/0.58 | 0.45/0.55 | 0.51/0.49 | 0.46/0.54 | 0.46/0.54 | 0.39/0.61 | 0.45/0.55 | 0.10/0.90 |
| Hyperdiploid | |||||||||
| Median age | 5 | 3 | 4 | 3 | 3 | 4§ | 6 | 3 | 4 |
| Mean age (SD) | 6.0 (3.6) | 4.6 (3.3) | 4.8 (3.0) | 2.8 (1.2) | 4.1 (3.0) | 5.1 (3.7) | 6.5 (4.7) | 4.6 (3.8) | 5.1 (3.8) |
| F:M | 0.43/0.57 | 0.45/0.55 | 0.42/0.58 | 0.44/0.56 | 0.52/0.48 | 0.41/0.59 | 0.46/0.54 | 0.49/0.51 | 0.37/0.63 |
| TEL-AML | |||||||||
| Median age | 6 | 5 | 4 | 3 | 4 | 4 | 4 | 4 | 4 |
| Mean age (SD) | 6.0 (2.8) | 5.2 (2.3) | 4.5 (2.3) | 3.7 (2.2) | 4.8 (2.5) | 4.9 (3.0) | 4.1 (2.0) | 4.7 (2.5) | 4.8 (3.1) |
| F:M | 0.0/1.0 | 0.33/0.67 | 0.44/0.56 | 0.50/0.50 | 0.40/0.60 | 0.44/0.56 | 0.21/0.79 | 0.43/0.57 | 0.30/0.70 |
| Non–HD/TEL-AML | |||||||||
| Median age | 5.5 | 6 | 4 | 8 | 7 | 4|| | 7.5 | 7 | 5¶ |
| Mean age (SD) | 6.2 (4.2) | 6.8 (4.4) | 5.6 (3.9) | 8.0 (4.6) | 7.2 (4.5) | 5.7 (4.4) | 8.3 (5.0) | 7.4 (5.0) | 6.4 (4.6) |
| F:M | 0.56/0.44 | 0.44/0.56 | 0.48/0.52 | 0.48/0.52 | 0.44/0.56 | 0.30/0.70 | 0.31/0.69 | 0.46/0.54 | 0.27/0.73 |
F, female; HD/TEL-AML, hyperdiploid TEL-AML; M, male; SD, standard deviation.
P = .04.
P = .002.
P = .003.
P = .03.
P = .0003.
P = .004.
To examine if variation at 10p12.2 and 10p14 influence T-ALL risk, we analyzed 83 UK and 246 German T-ALL cases. This analysis showed no robust association with T-ALL with either rs10828317 or rs3824662 (P = .02 and P = .85, respectively), however, this analysis is inherently limited by the small size of the datasets.
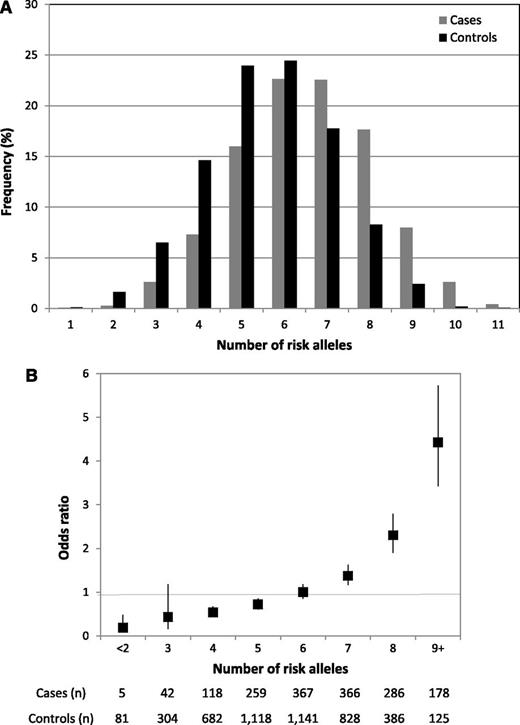
There was no evidence of significant interaction (ie, P > .05) between either rs10828317 and rs3824662 and the previously identified risk loci at 7p12.2 (rs4132601), 9p21.3 (rs3731217), 10q21.2 (rs7089424), or 14q11.2 (rs2239633), an observation compatible with each locus having an independent effect on ALL risk. The risk of ALL increases with increasing numbers of risk alleles for the 6 disease loci, counting 2 for a homozygote and 1 for a heterozygote, assuming equal weights. The proportion of cases and controls grouped according to the number of risk alleles carried is detailed in Figure 3, which shows a shift toward a higher number of risk alleles in the cases. For the 3% of the population carrying 9+ risk alleles, there is a greater than fourfold increase in risk compared with those with a median number of risk alleles (Figure 3).
Cumulative effects of the 6 BCP-ALL risk alleles (rs4132601, IKZF1; rs7089424, ARID5B; rs2239633, CEBPE; rs3731217, CDKN2A/CDKN2B; rs10828317, PIP4K2A, and rs3824662, GATA3). (A) Distribution of risk alleles in controls (black) and ALL cases (grey). (B) Plot of the increasing ORs for BCP-ALL with increasing number of risk alleles. The ORs are relative to the median number of 6 risk alleles; vertical bars correspond to 95% confidence intervals. The distribution of risk alleles follows a normal distribution in both cases and controls, with a shift toward a higher number of risk alleles in cases. Horizontal line denotes the null value (OR = 1.0).
Cumulative effects of the 6 BCP-ALL risk alleles (rs4132601, IKZF1; rs7089424, ARID5B; rs2239633, CEBPE; rs3731217, CDKN2A/CDKN2B; rs10828317, PIP4K2A, and rs3824662, GATA3). (A) Distribution of risk alleles in controls (black) and ALL cases (grey). (B) Plot of the increasing ORs for BCP-ALL with increasing number of risk alleles. The ORs are relative to the median number of 6 risk alleles; vertical bars correspond to 95% confidence intervals. The distribution of risk alleles follows a normal distribution in both cases and controls, with a shift toward a higher number of risk alleles in cases. Horizontal line denotes the null value (OR = 1.0).
To quantify the impact of the known loci on the heritability associated with common variation, we computed the receiver operator characteristic associated with 7p12.2, 9p21.3, 10p12.2, 10p14, 10q21.2, and 14q11.2 (rs4132601, rs3731217, rs10828317, rs3824662, rs7089424, and rs2239633, respectively). The area under the curve corresponding to these variants was 0.67, which translates into them contributing 16% of the genetic variance and 10% of the sibling relative risk. These estimates simply represent the additive variance and, therefore, do not include the potential impact of gene–gene interactions or dominance effects or gene-environment interactions impacting on ALL risk. Moreover, given the evidence, albeit indirect, of a role for infectious exposure in relation to ALL risk, it is possible that substantive gene-environment effects operate.
The functional basis of many GWAS signals can be ascribed to sequence changes impacting on gene expression and sequence conservation in noncoding regions has been shown to be a good predictor of cis-regulatory sequences. Although the associations identified did not show consistent statistically significant evidence of cis-acting regulatory effects in publicly accessible expression quantitative trait loci (eQTL) data (supplemental Table 5), steady-state levels of RNA in lymphocytes at a single time point and in cycling mature cells may not adequately capture the impact of differential expression in leukemogenesis.
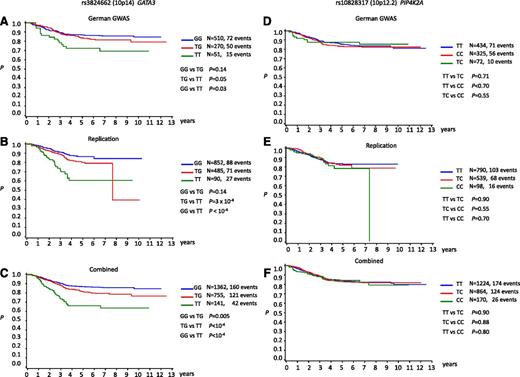
We examined for evidence of a relationship with patient outcome correlating SNP genotype with EFS (Figure 4). No association between rs10828317 genotype and EFS was shown (Figure 4). In contrast, rs3824662 showed a statistically significant association with EFS with carrier status being associated with poorer outcome and a higher rate of relapse (Figure 4; Table 2). Under the Cox proportional hazards model, the hazard ratios for TG and TT genotypes were 1.40 (95% CI, 1.10-1.77; P = .005) and 2.84 (95% CI, 2.02-3.99; P < 10−8), respectively (Table 2). The association remained statistically significant after adjustment for risk categories (Table 2). In keeping with rs3824662 genotype being a determinant of poorer prognosis, ALL risk genotype was significantly associated with high white cell count at diagnosis (P < .0001) (supplemental Table 6).
Kaplan-Meir curves of event-free survivorship in BCP-ALL patients. (A-C) Stratified by rs3824662 (10p14) genotype: (A) German GWAS, (B) replication, and (C) combined. (D-F) Stratified by rs10828317 (10p12.2) genotype: (D) German GWAS, (E) replication, and (F) combined.
Kaplan-Meir curves of event-free survivorship in BCP-ALL patients. (A-C) Stratified by rs3824662 (10p14) genotype: (A) German GWAS, (B) replication, and (C) combined. (D-F) Stratified by rs10828317 (10p12.2) genotype: (D) German GWAS, (E) replication, and (F) combined.
Cox regression hazard ratios of event-free survivorship for rs3824662 (10p14) genotype
| . | Event-free survival . | Relapse . | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | Unadjusted . | TEL-AML adjusted . | MRD adjusted . | . | ||||||||
| Genotype . | HR . | 95% CI . | P value . | HR . | 95% CI . | P value . | HR . | 95% CI . | P value . | HR . | 95% CI . | P value . |
| GG | 1.0 (ref) | — | — | 1.0 (ref) | — | — | 1.0 (ref) | — | — | 1.0 (ref) | — | — |
| TG | 1.40 | (1.10-1.77) | .005 | 1.45 | (1.14-1.86) | .003 | 1.23 | (0.96-1.58) | .10 | 1.29 | (0.98-1.70) | .06 |
| TT | 2.84 | (2.02-3.99) | 1.9 × 10−9 | 3.16 | (2.18-4.59) | 1.4 × 10−9 | 2.18 | (1.52-3.13) | 1.4 × 10−3 | 2.0 | (1.71-3.66) | 2.3 × 10−6 |
| . | Event-free survival . | Relapse . | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| . | Unadjusted . | TEL-AML adjusted . | MRD adjusted . | . | ||||||||
| Genotype . | HR . | 95% CI . | P value . | HR . | 95% CI . | P value . | HR . | 95% CI . | P value . | HR . | 95% CI . | P value . |
| GG | 1.0 (ref) | — | — | 1.0 (ref) | — | — | 1.0 (ref) | — | — | 1.0 (ref) | — | — |
| TG | 1.40 | (1.10-1.77) | .005 | 1.45 | (1.14-1.86) | .003 | 1.23 | (0.96-1.58) | .10 | 1.29 | (0.98-1.70) | .06 |
| TT | 2.84 | (2.02-3.99) | 1.9 × 10−9 | 3.16 | (2.18-4.59) | 1.4 × 10−9 | 2.18 | (1.52-3.13) | 1.4 × 10−3 | 2.0 | (1.71-3.66) | 2.3 × 10−6 |
CI, confidence interval; HR, hazard ratios; MRD, mimimal residual disease; (ref), reference group Web site links as follows:
The R suite can be found at http://www.r-project.org.
Detailed information on the tag SNP panel can be found at http://www.illumina.com.
The dbSNP can be found at http://www.ncbi.nlm.nih.gov.
HapMap can be found at http://www.hapmap.org.
1000genomes can be found at http://www.1000genomes.org.
KBioscience can be found at http://kbioscience.co.uk.
SNAP can be found at http://www.broadinstitute.org/mpg/snap.
IMPUTE can be found at https://mathgen.stats.ox.ac.uk.
EIGENSTRAT can be found at http://genetics.med.harvard.edu/reich/Reich_Lab/Software.html.
Wellcome Trust Case Control Consortium can be found at www.wtccc.org.uk.
Mendelian Inheritance In Man can be found at http://www.ncbi.nlm.nih.gov/omim.
1958 Birth Cohort can be found at http://www.cls.ioe.ac.uk/page.aspx?&sitesectionid=724&sitesectiontitle=Welcome+to+the+1958+National+Child+Development+Study.
Medical Research Council ALL 97 (Protocol 97PRT/14) can be found at http://www.thelancet.com/protocol-reviews/97PRT-14.
United Kingdom Childhood Cancer Study can be found at http://www.ukccs.org.
UCSC genome browser can be found at http://genome.ucsc.edu.
Surveillance, Epidemiology and End Results can be found at seer.cancer.gov/.
RegulomeDB can be found at http://regulome.stanford.edu/.
HaploREG can be found at http://www.broadinstitute.org/mammals/haploreg/haploreg.php.
Discussion
In a new GWAS of BCP-ALL, we have identified common variants at 10p12.2 and 10p14 that point to novel susceptibility loci. Because rs10828317 and rs3824662 localize to PIP4K2A and GATA3, there is a high likelihood that the functional basis of the associations are mediated through variation in these genes. Although the risk of ALL associated with these SNPs is modest, carrier frequencies are high and, therefore, they make a substantial contribution to the overall development of BCP-ALL. Moreover, by acting in concert with the 4 previously identified risk SNPs, they impact significantly on the risk of an individual developing ALL. As evidenced by study findings and previous observations of a relationship between 10q21.2 (ARID5B) genotype and hyperdiploid ALL, the genetic profile defining ALL predisposition increasingly appears to be subtype-specific, suggesting different etiologies.
Although we have made use of control genotypes from the analysis of adults, the prevalence of childhood ALL survivors is less than 1 in 10 000; hence, such series can be considered representative of the non-ALL population. Theoretically, different types (and quantity) of exposures could have cohort effects, however, there has been limited secular trend in the incidence of childhood leukemia in Western countries since the 1950s.25 Given such considerations, our observations should be highly robust. Support for such an assertion comes from a contemporaneous GWAS that has just reported 10p12.31 marked by rs7088318, which is highly correlated with rs10828317 (r2 = 0.79; D′=1.0), influences ALL risk.26 Although no relationship between 10p12.31 genotype and hyperdiploid status was explicitly reported, evaluation of rs10828317 in a small study of 297 cases supports our observation for the relationship.27 In contrast to rs10828317, the tumor profile associated with rs3824662 risk genotype appears to be one of high rate of relapse and MRD after remission reminiscent of what has been termed “BCR-ABL1–like” ALL.28 This observation has potential clinical ramifications, however, replication in an independent series is required to establish robustness.
PIP4K2A catalyzes the phosphorylation of PtdIns5P and through this mechanism is involved in secretion, cell proliferation, differentiation, and motility. Intriguingly, rs10828317 has previously been implicated as a risk factor for schizophrenia.29,30 Although a role in leukemogenesis has yet to be established, PIP4K2A expression has been implicated in thrombopoiesis and maturation of megakaryocytes, suggesting a role in early hematopioesis. Such an assertion would be compatible with PIP4K2A genotype influencing the development of hyperdiploid BCP-ALL.
Although the mutation of GATA3 causes dominantly inherited hypoparathyroidism, sensorineural deafness and renal dysplasia expression of GATA3 is important in hematopoietic and lymphoid cell development, acting as a master transcription factor for differentiation of Th2 cells. Moreover, GATA3 is a critical early regulator of innate lymphoid cells,31 and transcriptional repression of GATA3 is essential for early B-cell commitment.32 Inactivation of GATA3 is commonly seen in T-cell ALL,33 but GATA3 is not expressed in B-cells, hence a role in the development of BCP-ALL appears counterintuitive. Although not correlated with rs3824662, rs501764 (r2 = 0.00; D′ = 0.05), which maps 11Kb telomeric to GATA3 has previously been shown to be a risk factor for Hodgkin’s lymphoma (HL).34 Although HL is essentially a tumor of B-cell linage, an association between GATA3 variation and HL risk can be reconciled because a high proportion of the reactive infiltrate in HL tumors is composed of Th2-like cells, which can influence tumor growth. An analogous mechanism by which cognate T- and B-cell interactions underscore the association between rs3824662 with BCP-ALL is therefore plausible. Alternatively, because GATA3 is a crucial transcriptional regulator during tissue development, a basis for the association is through differential GATA3 expression in preleukemic B cells within the bone marrow. A wider impact of association at 10p14 on cancer risk in non–T-cells is also supported by observations that variation close to GATA3 influences the risk of lung cancer.35 Moreover, somatic mutations are frequently seen in a wide range of cancers, notably in invasive breast cancers (∼10%), which also display high levels of GATA3 expression.36,37
Because the SNPs marking the associations are not necessarily strong candidates for being directly functional, deciphering the underlying basis of both associations may be challenging. Although we found no evidence for a relationship between SNP and gene expression, any impact is likely to be modest and could occur at any time before diagnosis of ALL. Moreover, any expression differences may only be relevant to a subpopulation of cells that provide “targets” for leukemogenic mutations.
In summary, we have identified risk loci at 10p12.2 and 10p14 for BCP-ALL, and these findings provide additional support for the role of inherited genetic predisposition to disease etiology. Furthermore, the profile defining inherited predisposition appears to be increasingly subtype-specific, compatible with a different etiological basis. Additional studies are required to decipher the functional basis of these variants and to elucidate their role in BCP-ALL pathogenesis. Such analyses are likely to provide insight into the etiologic basis of ALL development and potentially contribute information relevant to the risk stratification of patients.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Lucy Chilton (Newcastle University), Jill Simpson (University of York), Pamela Thomson, and Adiba Hussain (University of Manchester) for assistance with data harmonization, the Leukaemia & Lymphoma Research Childhood Cancer Leukaemia Group Cell Bank for access to Medical Research Council Trial samples, the UK Cancer Cytogenetics Group for data collection and provision of samples.
This work was supported by the Leukemia Lymphoma Research, the Kay Kendall Leukemia, the Cancer Research UK (C1298/A8362), German Ministry of Education and Science and the German Research Council (DFG, Project SI236/8-1, SI236/9-1, ER 155/6-1), the Medical Faculty of the University Hospital of Essen (IFORES) (L.E.), and Children with Leukemia (P.T.); Genotyping of German Cases was funded by the Kay Kendall Leukaemia Fund; work in Germany was supported by the National Center for Tumor Diseases; the German GWAS made use of genotyping data from the population based HNR study, which is supported by the Heinz Nixdorf Foundation; and the genotyping of the Illumina HumanOmni-1 Quad BeadChips of the HNR subjects was financed by the German Centre for Neurodegenerative Disorders, Bonn.
The study made use of genotyping data on the 1958 British Birth Cohort (www.wtccc.org.uk).
The authors are grateful to investigators who contributed to this dataset. The authors are also grateful to all subjects and their clinicians for their participation.
Authorship
Contribution: R.S.H. and M.G. obtained financial support for both GWAS; R.S.H. designed the study and drafted the manuscript; G.M., F.J.H., Y.M., B.F., H.T., and M.I.d.S.F. performed bioinformatic and statistical analyses; J.V. performed validation genotyping; E.S. and S.E.K. performed curation and sample preparation of the Medical Research Council ALL-97 trial samples; T.L. and E.R. managed and maintained UKCCS sample data; P.T. performed harmonization of UKCCS samples; J.M.A. and J.A.I. performed ascertainment, curation and preparation of the Northern Institute for Cancer Research cases; A.L.S. oversaw laboratory analyses; K.H. oversaw analysis of the German cohort; R. Kumar supervised genotyping; B.F. genotyped German samples; R. Koehler, M.Z., M. Stanulla, M. Schrappe, and C.R.B. provided German DNA for analysis; K.H. supervised analysis at the DKFZ; and P.H., M.M.N., T.W.M., and L.E. provided German control data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Richard Houlston, Institute of Cancer Research, 15 Cotswold Rd, Surrey SM2 5NG United Kingdom; e-mail: richard.houlston@icr.ac.uk.
References
Author notes
G.M., B.F., and F.J.H. contributed equally to this study.