Key Points
GATA1-dependent chromatin looping at the TAL1 locus accounts for many biological and evolutionary features of TAL1 expression.
Chromatin looping also regulates expression of TAL1 flanking genes and brings TAL1 and STIL into close proximity at T-ALL breakpoints.
Abstract
TAL1 is an important regulator of hematopoiesis and its expression is tightly controlled despite complexities in its genomic organization. It is frequently misregulated in T-cell acute lymphoblastic leukemia (T-ALL), often due to deletions between TAL1 and the neighboring STIL gene. To better understand the events that lead to TAL1 expression in hematopoiesis and in T-ALL, we studied looping interactions at the TAL1 locus. In TAL1-expressing erythroid cells, the locus adopts a looping “hub” which brings into close physical proximity all known TAL1 cis-regulatory elements including CTCF-bound insulators. Loss of GATA1 results in disassembly of the hub and loss of CTCF/RAD21 from one of its insulators. Genes flanking TAL1 are partly dependent on hub integrity for their transcriptional regulation. We identified looping patterns unique to TAL1-expressing T-ALL cells, and, intriguingly, loops occurring between the TAL1 and STIL genes at the common TAL1/STIL breakpoints found in T-ALL. These findings redefine how TAL1 and neighboring genes communicate within the nucleus, and indicate that looping facilitates both normal and aberrant TAL1 expression and may predispose to structural rearrangements in T-ALL. We also propose that GATA1-dependent looping mechanisms may facilitate the conservation of TAL1 regulation despite cis-regulatory remodeling during vertebrate evolution.
Introduction
The TAL1 gene encodes a basic helix loop helix transcription factor that is a key regulator during vertebrate hematopoiesis.1,2 Although TAL1 is not normally expressed in lymphoid cells, ectopic T-cell expression of TAL1 occurs in 60% of cases of T-cell acute lymphoblastic leukemia (T-ALL).3,4 In ∼6% to 26% of T-ALL cases, TAL1 is expressed via a fusion messenger RNA (mRNA) resulting from a microdeletion (∼74-82 kb) involving TAL1 and its neighboring STIL gene, bringing TAL1 under the control of STIL expression.5,6 Of these deletions, the majority have common breakpoints within TAL1 promoter 1b and within STIL intron 1 (the latter known as the TALd breakpoint). The mechanisms which regulate TAL1 expression in T-ALL and predispose TAL1 and STIL to undergo structural rearrangements are not well understood.
The importance of TAL1 in normal hematopoiesis and in T-ALL has fueled systematic dissection of the TAL1 locus and led to the identification of multiple cis-acting regulatory elements capable of directing TAL1 expression.7-10 In addition to TAL1 promoters 1a and 1b, these elements include enhancers and CTCF-bound elements (supplemental Figure 1, available on the Blood Web site). Collectively, these cis-regulatory elements span ∼88 kb in humans, a genomic region which contains the entire TAL1 “regulon” defined by CTCF-bound elements at either end.9 This regulon also contains the 3′ segment of STIL, and the entirety of PDZK1IP1, the latter being juxtaposed between TAL1 and its erythroid enhancer (supplemental Figure 1). PDZK1IP1 is expressed in all hematopoietic cell types where TAL1 is expressed11 (albeit at low levels; supplemental Figure 2), suggesting that their transcription is coregulated. The basis for this coregulation is unclear, although others have suggested that TAL1 and PDZK1IP1 share cis-regulatory elements.12 However, this is unlikely to occur given that a CTCF-bound element with enhancer-blocking activity9 is positioned between the PDZK1IP1 promoter and the majority of the TAL1 cis-regulatory circuitry (supplemental Figure 1B). Similarly, the location of this CTCF-bound element makes it difficult to envisage how the TAL1 erythroid enhancer communicates with its promoters to regulate TAL1 expression in the erythroid lineage (supplemental Figure 1C). Finally, the presence of a CTCF-bound element within the 3′ portion of the transcribed region of the STIL gene presents an impediment to STIL transcription (supplemental Figure 1D).
Because of this genomic organization, we hypothesized that context-dependent chromatin looping would be required at the TAL1 locus to facilitate appropriate communication between genes and their cis-regulatory circuitry. We rationalized that such looping interactions may also predispose the TAL1 locus to be expressed and undergo structural rearrangement in T-ALL (the latter by bringing sequences which are prone to rearrange into close physical proximity). We report here transcription-associated looping models of the TAL1 locus as detected by 3C- and 4C-based methods. We discuss how these models could account for a range of biological and evolutionary features associated with the expression of TAL1 and its flanking genes and how they could predispose to, or facilitate, TAL1 expression in cases of T-ALL.
Methods
Cell sources
K562, HPB-ALL, Jurkat, MEL (C88), and BW5147 cell lines were cultured as described previously.9 Primary murine lymphocytes were isolated from spleens of wild-type ICR mice (Harlan-Olac Ltd.) by density centrifugation (lympholyte-mammal; VH-Bio). Nuclei from primary murine erythroblasts were obtained as described previously.13
GATA1 siRNA knockdown
K562 cells (5 × 106) were transfected with 20nM double-stranded small interfering RNA (siRNA) oligonucleotides targeted to GATA1 (sense: 5′-GGAUGGUAUUCAGACUCGAdTdT-3′) or firefly luciferase (sense: 5′-CUUACGCUGAGUACUUCGAdTdT-3) using the Amaxa Nucleofector (T-016 program). Cells were monitored for growth arrest (trypan blue; Sigma-Aldrich), morphologic changes (Stain Quick-Staining kit; Lamb), and apoptosis (annexin V-phycoerythrin; BD Biosciences) by microscopy.
RNA/protein extraction and expression analysis
Total cellular RNA from cultured cells was purified (TRIzol; Invitrogen) and reverse-transcribed (Superscript II kit; Invitrogen). SyBr green quantitative polymerase chain reaction (PCR) was performed (Roche) (supplemental Table 1) on an Mx3000P QPCR Systems PCR machine (Agilent Technologies). Protein was extracted and quantified (Protein Assay Kit II; Bio-Rad). Western detection used the anti-GATA1 antibody (M-20; Santa Cruz Biotechnology).
ChIP quantitative PCR
Chromatin immunoprecipitation (ChIP) from multiple bioreplicate samples was performed as described elsewhere9 (supplemental Table 2). ChIP enrichment levels were determined by SyBr green PCR as noted in the previous paragraph (supplemental Table 3).
3C and 4C analysis
3C libraries were prepared as described previously.14 Bacterial artificial clones for the human TAL1 locus (RP1-18D14; Invitrogen) and the murine Tal1 locus (RP23-453H14; BPRC) were used to prepare mock 3C libraries. PCR conditions for each 3C assay (supplemental Table 4) are available on request. Quantification of PCR band intensities was performed with background correction (AIDA; Raytest). 3C-relative ligation frequencies were obtained by normalizing band intensities obtained from 3C libraries with those from mock 3C libraries. Statistical significance within or between samples was determined by the Student t test (P < .05) and via normalization strategies as described previously.15 4C libraries were prepared as described previously16 (supplemental Table 5). PCR cycling conditions are available on request. Microarray analysis of fluorescently labeled 4C and genomic DNAs, hybridized to a TAL1 tiling path microarray, were performed as described previously.9 3C and 4C data were derived from at least 2 independent biological replicates.
Results
We performed 3C analysis17,18 (supplemental Figure 3) in human and murine cell types to determine (1) whether cis-regulatory elements at the TAL1 locus showed enhanced frequencies of ligation between each other in 3C libraries, relative to appropriate controls, and (2) whether there were differences in looping frequencies of these elements between TAL1-expressing and nonexpressing cell types.
Context-dependent looping between the TAL1 promoters and its enhancers
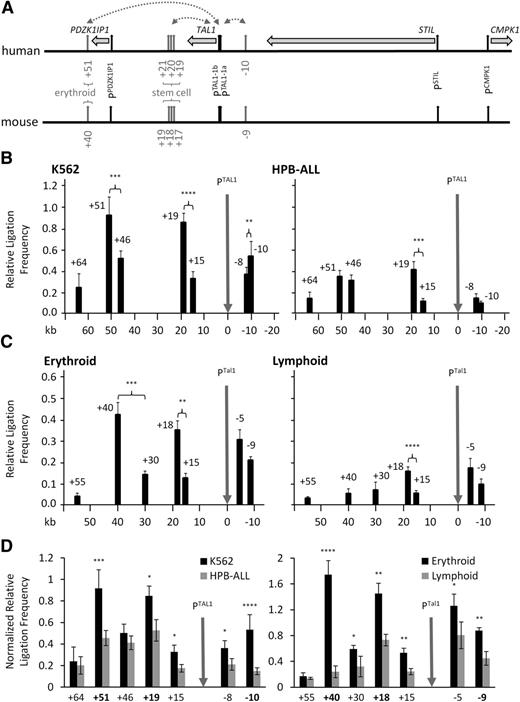
We first asked whether specific looping interactions occurred between the 2 human TAL1 promoters (1a and 1b) in combination with the erythroid enhancer (+51), the stem cell enhancer (+19/+20/+21), or a 5′ proximal enhancer (−10) (Figure 1A). In the TAL1-expressing erythroid19 K562 cell line, we detected looping between the TAL1 promoters and both the erythroid (+51) and stem cell enhancers (+19) (Figure 1B). These looping interactions were also observed in equivalent murine erythroid cell types at +40 and +18, respectively (Figure 1C; supplemental Figure 4). There did, however, appear to be differences in the levels of looping between the promoters and the 5′ proximal enhancer in humans and mice (−10 and −9, respectively) relative to their cognate control regions (−8 and −5, respectively). These differences may be explained by the complexity of multiple cis-regulatory elements upstream of TAL1.7,9,20,21 This is supported by comparisons of ligation frequencies at the TAL1/Tal1 locus in both erythroid and lymphoid cells normalized against those from the ERCC3/Ercc3 control locus (supplemental Figure 5) which showed that all 3 enhancers, as well as the control regions upstream of TAL1, had significantly higher levels of looping with the TAL1/Tal1 promoters when the gene was transcribed (Figure 1D; supplemental Figure 4). Furthermore, additional control regions (eg, +15 in human; +30, +15 in mouse) showed higher interaction frequencies in erythroid cells than in lymphoid cells, suggesting that a number DNA segments of the downstream of the TAL1/Tal1 gene were also in close contact with its promoters when the gene was transcribed. Although we did see some evidence for elevated ligation frequencies between the TAL1/Tal1 promoters and the human +19 stem cell enhancer (mouse +18) in lymphoid cells (Figure 1B-C; supplemental Figure 4), these occurred at relatively low levels compared with those in TAL1/Tal1-expressing cell types. These data provided our first lines of evidence that there are transcription-associated looping interactions which occur between TAL1 cis-regulatory elements.
Looping interactions involving TAL1 promoters and enhancers in erythroid and lymphoid cells. (A) Schematic organization of the human and murine TAL1 loci. Locations and directions of transcription of TAL1 and its flanking genes PDZK1IP1, STIL, and CMPK1 are shown (horizontal gray arrows). Locations of promoters (vertical black arrows) and enhancers of the TAL1 gene studied here (vertical gray arrows) are shown. The erythroid and the stem cell enhancers are highlighted. Elements are named according to their distance (in kb) from TAL1 promoter 1a. Looping interactions tested in this study are denoted by dotted gray lines with arrowheads. (B) Bar diagram of interaction patterns across the human TAL1 locus in erythroid (K562) and lymphoid (HPB-ALL) cell lines determined by 3C. (C) Bar diagrams of interaction patterns across the murine Tal1 locus in primary erythroblasts (Erythroid) and lymphocytes (Lymphoid) determined by 3C. Interactions, measured as relative ligation frequencies (black bars) at various locations across the locus, are shown with standard errors (SEs). Location of 3C “bait” region (TAL1 promoter 1b = PTAL1; PTal1 in mouse) is shown (vertical gray arrows). P values are indicated for relative ligation frequencies which are significantly higher for test regions when compared with those of control regions (controls defined as regions located between the “bait” and test regions). Scales (in kb) are shown at the bottom of panels B-C. (D) Comparison of interaction patterns at the TAL1 locus in human and murine cell types normalized against ERCC3 ligation frequencies. (Left panel) Human K562 and HPB-ALL cell lines. (Right panel) Primary murine erythroblasts and lymphocytes. P values are indicated for interactions which are significantly higher in the TAL1-expressing cell type. *P < .05; **P < .01; ***P < .001; ****P < .0001.
Looping interactions involving TAL1 promoters and enhancers in erythroid and lymphoid cells. (A) Schematic organization of the human and murine TAL1 loci. Locations and directions of transcription of TAL1 and its flanking genes PDZK1IP1, STIL, and CMPK1 are shown (horizontal gray arrows). Locations of promoters (vertical black arrows) and enhancers of the TAL1 gene studied here (vertical gray arrows) are shown. The erythroid and the stem cell enhancers are highlighted. Elements are named according to their distance (in kb) from TAL1 promoter 1a. Looping interactions tested in this study are denoted by dotted gray lines with arrowheads. (B) Bar diagram of interaction patterns across the human TAL1 locus in erythroid (K562) and lymphoid (HPB-ALL) cell lines determined by 3C. (C) Bar diagrams of interaction patterns across the murine Tal1 locus in primary erythroblasts (Erythroid) and lymphocytes (Lymphoid) determined by 3C. Interactions, measured as relative ligation frequencies (black bars) at various locations across the locus, are shown with standard errors (SEs). Location of 3C “bait” region (TAL1 promoter 1b = PTAL1; PTal1 in mouse) is shown (vertical gray arrows). P values are indicated for relative ligation frequencies which are significantly higher for test regions when compared with those of control regions (controls defined as regions located between the “bait” and test regions). Scales (in kb) are shown at the bottom of panels B-C. (D) Comparison of interaction patterns at the TAL1 locus in human and murine cell types normalized against ERCC3 ligation frequencies. (Left panel) Human K562 and HPB-ALL cell lines. (Right panel) Primary murine erythroblasts and lymphocytes. P values are indicated for interactions which are significantly higher in the TAL1-expressing cell type. *P < .05; **P < .01; ***P < .001; ****P < .0001.
GATA1 is essential for a TAL1 erythroid “hub”
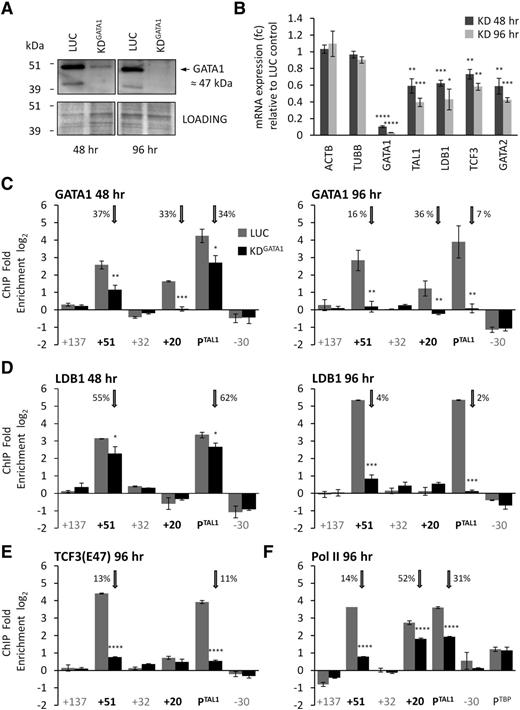
Our previous work demonstrated that the human +51 erythroid enhancer and TAL1 promoter 1a were both occupied with RNA polymerase II (Pol II) and components of the TAL1 erythroid complex (TEC) containing GATA1, TAL1, LMO2, TCF3, and LDB.9 We hypothesized that these proteins were involved in looping between these elements. We optimized an siRNA assay for a core component of the TEC, GATA1, in K562 cells (Figure 2A). At 48 hours after knockdown, GATA1 protein expression was reduced by 87% and was accompanied by a reduction in TAL1 mRNA expression to 60% of its wild-type level (Figure 2B). Maximum knockdown of GATA1 was achieved 96 hours after transfection (98% decrease in protein; Figure 2A) and resulted in a further decrease in TAL1 expression to 40% of its wild-type level (Figure 2B). Transient knockdown of GATA1 for 96 hours in K562 cells did not adversely affect cell viability and morphology compared with controls (supplemental Figure 6).
Effect of GATA1 knockdown on gene expression of TAL1 and recruitment of members of the TEC and Pol II. (A) GATA1 protein levels in K562 cells in a representative GATA1 siRNA knockdown (KDGATA1) and luciferase siRNA control (LUC) at 48 and 96 hours after transfection. The immunoblot was stained in Bradford reagent (Bio-Rad) and imaged to use as a loading control (also shown). GATA1 protein is ∼47 kDa (black arrow). (B) TAL1 transcript levels (with SEs) in GATA1 knockdown samples at 48 and 96 hours after transfection are shown relative to levels in luciferase control samples. mRNA levels for controls (ACTB and TUBB), GATA1, LDB1, TCF3, and GATA2 levels are also shown. Occupancies of GATA1 (C) or LDB1 (D) at the TAL1 erythroid enhancer (+51), the TAL1 stem cell enhancer (+20), and TAL1 promoter 1a (PTAL1) 48 or 96 hours after transfection with siRNA for GATA1 or luciferase. Occupancies of TCF3 (E47 isoform) (E) or Pol II (F) at the TAL1 erythroid enhancer (+51), the TAL1 stem cell enhancer (+20), and TAL1 promoter 1a (PTAL1) 96 hours after transfection with siRNA for GATA1 or luciferase. Positive control for Pol II occupancy was the promoter of TBP (PTBP). For the bar diagrams in panels C-F, ChIP enrichments (log2) are shown with SEs. Annotation of test and negative control regions is denoted in black and gray text, respectively. Luciferase (LUC) and GATA1 knockdown treated samples (KDGATA1) are shown as gray and black bars, respectively, as per the key in panel C. Vertical gray arrows highlight the occupancy data for the +51 enhancer, the +20 stem cell enhancer, and the TAL1 promoter 1a (PTAL1). The percentage (%) occupancies of each protein in the GATA1 knockdown relative to the levels found in the luciferase control are also shown. *P < .05; **P < .01; ***P < .001; ****P < .0001.
Effect of GATA1 knockdown on gene expression of TAL1 and recruitment of members of the TEC and Pol II. (A) GATA1 protein levels in K562 cells in a representative GATA1 siRNA knockdown (KDGATA1) and luciferase siRNA control (LUC) at 48 and 96 hours after transfection. The immunoblot was stained in Bradford reagent (Bio-Rad) and imaged to use as a loading control (also shown). GATA1 protein is ∼47 kDa (black arrow). (B) TAL1 transcript levels (with SEs) in GATA1 knockdown samples at 48 and 96 hours after transfection are shown relative to levels in luciferase control samples. mRNA levels for controls (ACTB and TUBB), GATA1, LDB1, TCF3, and GATA2 levels are also shown. Occupancies of GATA1 (C) or LDB1 (D) at the TAL1 erythroid enhancer (+51), the TAL1 stem cell enhancer (+20), and TAL1 promoter 1a (PTAL1) 48 or 96 hours after transfection with siRNA for GATA1 or luciferase. Occupancies of TCF3 (E47 isoform) (E) or Pol II (F) at the TAL1 erythroid enhancer (+51), the TAL1 stem cell enhancer (+20), and TAL1 promoter 1a (PTAL1) 96 hours after transfection with siRNA for GATA1 or luciferase. Positive control for Pol II occupancy was the promoter of TBP (PTBP). For the bar diagrams in panels C-F, ChIP enrichments (log2) are shown with SEs. Annotation of test and negative control regions is denoted in black and gray text, respectively. Luciferase (LUC) and GATA1 knockdown treated samples (KDGATA1) are shown as gray and black bars, respectively, as per the key in panel C. Vertical gray arrows highlight the occupancy data for the +51 enhancer, the +20 stem cell enhancer, and the TAL1 promoter 1a (PTAL1). The percentage (%) occupancies of each protein in the GATA1 knockdown relative to the levels found in the luciferase control are also shown. *P < .05; **P < .01; ***P < .001; ****P < .0001.
There was substantial loss of GATA1 from both TAL1 promoter 1a and the +51 erythroid enhancer after 48 hours of GATA1 knockdown (Figure 2C). Surprisingly, GATA1 occupancy showed the most significant decrease at the stem cell enhancer (+20); at 48 hours of knockdown, its level was indistinguishable from background. GATA1 was not known previously to bind to the stem cell enhancer in K562 cells. By 96 hours of knockdown, GATA1 occupancies at both the TAL1 promoter 1a and the +51 erythroid enhancer were also reduced to background levels. Occupancy of other members of the TEC (LDB1 and TCF3) also decreased in GATA1 knockdown cells from both the TAL1 promoter 1a and the +51 enhancer (Figure 2D-E). GATA1 knockdown also resulted in decreased transcription of LDB1, TCF3, and GATA2 (Figure 2B), all known to be downstream targets of GATA1 in hematopoietic cells.19,22-25 However, decreased expression of LDB1 and TCF3 could not account for the much larger drop in occupancies for these transcription factors at both +51 and TAL1 promoter 1a, and supported that loss of GATA1 directly impaired recruitment of LDB1 and TCF3 to these elements. Impairment of Pol II recruitment at TAL1 cis-regulatory elements was also evident by 96 hours of GATA1 knockdown (Figure 2F). These data demonstrated a direct relationship between TAL1 transcription, the kinetics of GATA1 depletion from its cis-regulatory elements, and the dependence of other factors on GATA1 occupancy at these sites.
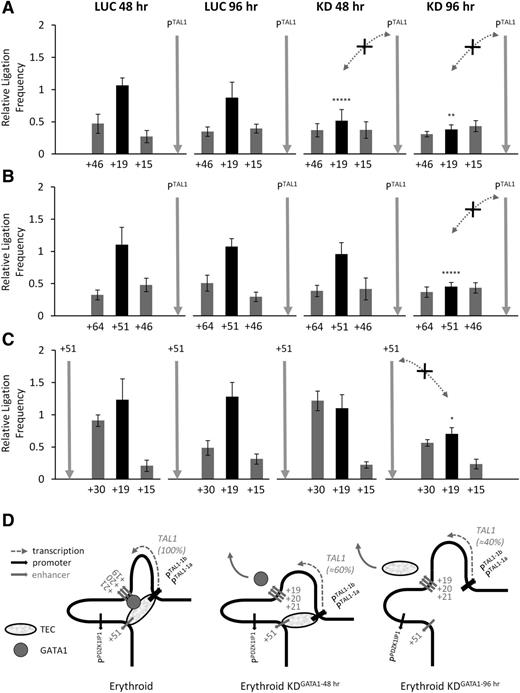
To determine whether GATA1 was required for looping between TAL1 regulatory elements, we performed 3C on material obtained from the GATA1 knockdown cells (Figure 3). After 48 hours of knockdown, we observed significant loss in the looping interaction between the +19 stem cell enhancer and the TAL1 promoters (Figure 3A), coinciding with the complete loss of GATA1 from +19 and a reduction in TAL1 transcription. Only modest decreases in looping between the +51 erythroid enhancer with either the TAL1 promoters or with the +19 enhancer were observed at 48 hours. However, at the 96-hour time point, we observed a substantial loss of the looping interactions between these elements (Figure 3B-C), coinciding with complete loss of GATA1 from TAL1 promoter 1a and +51 and a further drop in TAL1 transcription. All of these data supported a model where the TAL1 promoters and the stem cell and erythroid enhancers were in contact in a transcriptionally active erythroid “hub” (Figure 2D). The kinetics of looping interactions within this hub were in agreement with the kinetics of GATA1 depletion and decreases in TAL1 transcription, supporting a model whereby this hub structure was linked to GATA1-dependent TAL1 expression in erythroid cells. We could not, however, rule out the possibility that reductions in TCF3, LDB1, and GATA2 expression also impact upon these looping interactions and TAL1 expression.
Loss of GATA1 results in the loss of looping interactions within the TAL1 active hub. Bar diagrams of interaction patterns across the human TAL1 locus after siRNA transfection (48 hours and 96 hours) with GATA1 (KD) or with luciferase (LUC) as determined by 3C. (A) Interaction between TAL1 promoter 1b (PTAL) and the stem cell enhancer (+19). (B) Interaction between TAL1 promoter 1b (PTAL) and the erythroid enhancer (+51). (C) Interaction between the TAL1 erythroid enhancer (+51) and the stem cell enhancer (+19). Interactions, measured as relative ligation frequencies, are shown with SEs. The +19 and +51 enhancers are highlighted as black bars in the histograms. Locations of 3C “bait” regions are denoted by vertical gray arrows. P values are indicated for relative ligation frequencies which are significantly reduced between the bait and enhancers in KD conditions when compared with the corresponding LUC controls. Significant loss of interactions due to GATA1 knockdown are shown by the dotted gray lines with arrowheads (and marked with an “X”). Control regions are as in Figure 1. (D) Schematic model of looping interactions at the TAL1 locus, and during GATA1 siRNA knockdown, in K562 cells. Loss of GATA1 and the TEC from TAL1 cis-regulatory elements (Figure 2) is accompanied by loss of TAL1 transcription (percentage of mRNA remaining relative to control shown in brackets) and loss of looping interactions, initially most evident between +19 and the TAL1 promoters (48 hours GATA1 knockdown), and then between +19, +51 and the TAL1 promoters (96 hours GATA1 knockdown). *P < .05; **P < .01; *****P < .00001.
Loss of GATA1 results in the loss of looping interactions within the TAL1 active hub. Bar diagrams of interaction patterns across the human TAL1 locus after siRNA transfection (48 hours and 96 hours) with GATA1 (KD) or with luciferase (LUC) as determined by 3C. (A) Interaction between TAL1 promoter 1b (PTAL) and the stem cell enhancer (+19). (B) Interaction between TAL1 promoter 1b (PTAL) and the erythroid enhancer (+51). (C) Interaction between the TAL1 erythroid enhancer (+51) and the stem cell enhancer (+19). Interactions, measured as relative ligation frequencies, are shown with SEs. The +19 and +51 enhancers are highlighted as black bars in the histograms. Locations of 3C “bait” regions are denoted by vertical gray arrows. P values are indicated for relative ligation frequencies which are significantly reduced between the bait and enhancers in KD conditions when compared with the corresponding LUC controls. Significant loss of interactions due to GATA1 knockdown are shown by the dotted gray lines with arrowheads (and marked with an “X”). Control regions are as in Figure 1. (D) Schematic model of looping interactions at the TAL1 locus, and during GATA1 siRNA knockdown, in K562 cells. Loss of GATA1 and the TEC from TAL1 cis-regulatory elements (Figure 2) is accompanied by loss of TAL1 transcription (percentage of mRNA remaining relative to control shown in brackets) and loss of looping interactions, initially most evident between +19 and the TAL1 promoters (48 hours GATA1 knockdown), and then between +19, +51 and the TAL1 promoters (96 hours GATA1 knockdown). *P < .05; **P < .01; *****P < .00001.
Looping between CTCF elements compartmentalizes the transcribed TAL1 regulon
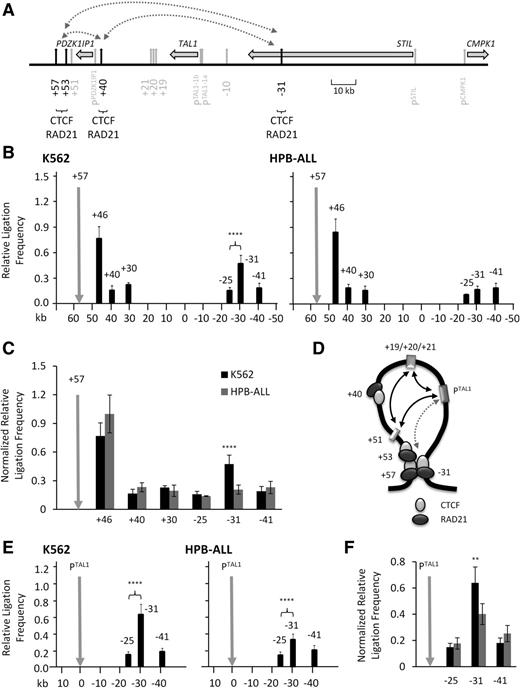
We investigated whether the 4 CTCF-bound elements at the human TAL1 locus (+57, +53, +40, and −31) were involved in looping to facilitate the formation of the TAL1 erythroid hub. Each of these elements acts as enhancer-blocking insulators in reporter assays9 and is also occupied by RAD21 and SMC3, components of the cohesin complex (supplemental Figure 7). We considered all possible looping interactions between these elements (Figure 4A; supplemental Figure 8) using 3C. In TAL1-expressing K562 cells, looping interactions were identified between the +57 and −31 elements (Figure 4B) and between +53 and −31 (supplemental Figure 9) in a transcription-associated manner when compared with levels found in HPB-ALL (Figure 4C; supplemental Figure 10). We did not detect looping interactions between +40 and any of +53, +57, or −31 in either K562 or in HPB-ALL which we considered to be biologically relevant (supplemental Figures 9-10). These data would support a configuration where the TAL1 promoters and the stem cell and erythroid enhancers were compartmentalized in a single chromatin loop in TAL1-expressing erythroid cells, but not in TAL1-nonexpressing lymphoid cells (Figure 4D). Based on this, we predicted that the interaction between +51 and the TAL1 promoters would bring the +57/+53/−31 interaction into close proximity to the TAL1 promoters. Using 3C, we were able to demonstrate that the TAL1 promoters and −31 were in close proximity in the K562 cell line (Figure 4E). This proximity was significantly increased in K562 when compared with levels in HPB-ALL (Figure 4E-F), linking it to TAL1 transcription.
Compartmentalization of the TAL1 regulon via looping interactions between CTCF/RAD21-bound elements. (A) Schematic organization of CTCF elements at the human TAL1 locus. Locations and directions of transcription of TAL1 and its flanking genes PDZK1IP1, STIL, and CMPK1 are shown by the horizontal gray arrows. Locations of promoters and enhancers are shown with vertical gray arrows. CTCF/RAD21-occupied elements studied here (+57, +53, +40, and −31) are highlighted (vertical black arrows). Scale (in kb) is shown. Looping interactions tested in this study are denoted by dotted gray lines with arrowheads. (B) Bar diagrams of looping interactions between CTCF/RAD21-bound elements determined by 3C in human erythroid (K562) and lymphoid (HPB-ALL) cell lines. (C) Comparison of interaction patterns between CTCF/RAD21-bound elements at the TAL1 locus in human K562 and HPB-ALL cells normalized against ERCC3 ligation frequencies. (D) Schematic diagram depicted the hypothesized proximity (gray dotted arrowhead) between the TAL1 promoters (PTAL1) and the −31 element based on the data described in the text, shown here, and in Figures 1 and 3. CTCF (light gray sphere) and RAD21 (dark gray sphere) occupancies are depicted. (E) Bar diagram of looping interactions, measured as relative ligation frequencies (black bars), between the −31 element and TAL1 promoter 1b (PTAL1) in K562 and HPB-ALL cell lines. (F) Comparison of interaction patterns between the TAL1 promoters and the −31 insulator element in human K562 and HPB-ALL cells normalized against ERCC3 ligation frequencies. Color coding of the cell lines is the same as in panel C. For the bar diagrams in panels B and E, interactions, measured at relative ligation frequencies (black bars), are shown with SEs. Locations of 3C “bait” regions are denoted by vertical gray arrows. P values are indicated for relative ligation frequencies which are significantly higher for test regions when compared with those of control regions. (B,E) Scales (in kb) are shown at the bottom of panels. (C,F) P values are indicated for interactions which are significantly higher in K562 (TAL1 expressing). **P < .01; ****P < .0001.
Compartmentalization of the TAL1 regulon via looping interactions between CTCF/RAD21-bound elements. (A) Schematic organization of CTCF elements at the human TAL1 locus. Locations and directions of transcription of TAL1 and its flanking genes PDZK1IP1, STIL, and CMPK1 are shown by the horizontal gray arrows. Locations of promoters and enhancers are shown with vertical gray arrows. CTCF/RAD21-occupied elements studied here (+57, +53, +40, and −31) are highlighted (vertical black arrows). Scale (in kb) is shown. Looping interactions tested in this study are denoted by dotted gray lines with arrowheads. (B) Bar diagrams of looping interactions between CTCF/RAD21-bound elements determined by 3C in human erythroid (K562) and lymphoid (HPB-ALL) cell lines. (C) Comparison of interaction patterns between CTCF/RAD21-bound elements at the TAL1 locus in human K562 and HPB-ALL cells normalized against ERCC3 ligation frequencies. (D) Schematic diagram depicted the hypothesized proximity (gray dotted arrowhead) between the TAL1 promoters (PTAL1) and the −31 element based on the data described in the text, shown here, and in Figures 1 and 3. CTCF (light gray sphere) and RAD21 (dark gray sphere) occupancies are depicted. (E) Bar diagram of looping interactions, measured as relative ligation frequencies (black bars), between the −31 element and TAL1 promoter 1b (PTAL1) in K562 and HPB-ALL cell lines. (F) Comparison of interaction patterns between the TAL1 promoters and the −31 insulator element in human K562 and HPB-ALL cells normalized against ERCC3 ligation frequencies. Color coding of the cell lines is the same as in panel C. For the bar diagrams in panels B and E, interactions, measured at relative ligation frequencies (black bars), are shown with SEs. Locations of 3C “bait” regions are denoted by vertical gray arrows. P values are indicated for relative ligation frequencies which are significantly higher for test regions when compared with those of control regions. (B,E) Scales (in kb) are shown at the bottom of panels. (C,F) P values are indicated for interactions which are significantly higher in K562 (TAL1 expressing). **P < .01; ****P < .0001.
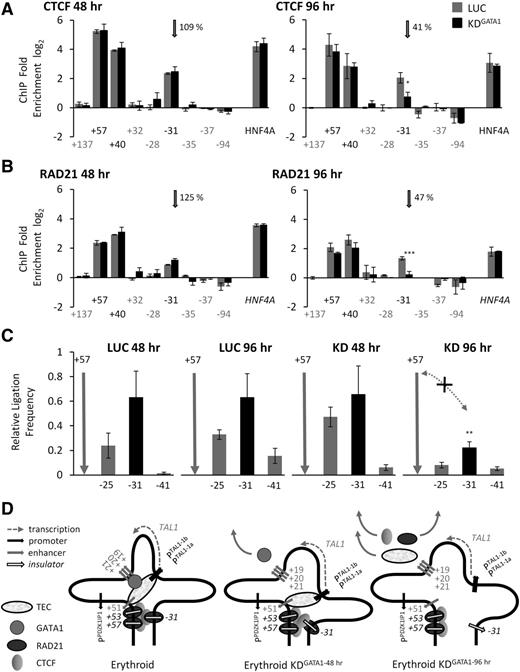
CTCF and RAD21 eviction from −31 accompanies disassembly of the TAL1 hub
We asked whether disassembly of the TAL1 erythroid hub during GATA1 knockdown was accompanied by loss of CTCF-associated loops. Forty-eight hours after transfection, CTCF and RAD21 occupancies at +57, +40, and −31 remained unchanged in GATA1 knockdown cells when compared with controls (Figure 5A-B). However, there was considerable loss of both CTCF and RAD21 (≤47% remaining) from the −31 element at the 96-hour time point. This loss was accompanied by a significant decrease in looping between +57 and −31 (Figure 5C). Thus, while GATA1 loss from TAL1 regulatory elements began early in our perturbation time course and was accompanied by partial disassembly of the hub, loss of CTCF and RAD21 from −31 only occurred once the hub was completely disassembled, and GATA1 was completely depleted from cis-regulatory elements in the hub (Figure 5D). This demonstrated a hub-dependent recruitment of CTCF and RAD21 to the −31 insulator element when TAL1 is transcribed. Surprisingly, this recruitment is GATA1-mediated, despite GATA1 not being directly bound to −31. We identified several features of the −31 element in K562 cells which distinguished it from other CTCF/RAD21-bound elements at the TAL1 locus (supplemental Figures 7 and 11). We also showed that RAD21 recruitment to −31 is substantially reduced when the +57/+53/−31 loop is not formed in TAL1-nonexpressing HPB-ALL cells (supplemental Figure 12).
Eviction of CTCF and RAD21 from the −31 element accompanies dissolution of the TAL1 active hub and loss of proximity between the +57 and −31 elements. (A) CTCF occupancy at the +57, +40, and −31 elements 48 and 96 hours after siRNA transfection with GATA1 (KD) or with luciferase (LUC) in K562 cells. (B) RAD21 occupancy at the +57, +40, and −31 elements 48 and 96 hours after siRNA transfection with GATA1 (KD) or with luciferase (LUC) in K562 cells. Gray vertical arrows in panels A and B highlight CTCF and RAD21 occupancy levels at −31. ChIP enrichments (log2) are shown with SEs. Annotation of test and negative control regions is denoted in black and gray text, respectively. Positive control is a CTCF/RAD21-bound element at the HNF4A locus. The percentage (%) occupancies of each protein in the GATA1 knockdown relative to the level found in the luciferase control are also shown. (C) Bar diagrams of looping interactions, measured as relative ligation frequencies (black bars), between the +57 and −31 elements determined by 3C at 48 and 96 hours after siRNA transfection with GATA1 (KD) or with luciferase (LUC) in K562 cells. Locations of 3C “bait” regions are denoted by vertical gray arrows. Loss of interactions due to GATA1 knockdown at 96 hours is shown by the dotted gray lines with arrowheads (and marked with an “X”). P values are indicated for relative ligation frequencies which are significantly reduced between the +57 and −31 elements in KD conditions when compared with the corresponding LUC controls. (D) Schematic model of looping interactions between CTCF/RAD21-bound elements at the TAL1 locus, and during GATA1 siRNA knockdown, in K562 cells. Loss of GATA1 is accompanied by (1) loss of CTCF and RAD21 from the −31 element and (2) loss of looping interactions between +57 and −31 (96 hours GATA1 knockdown) as the TAL1 erythroid hub is disassembled. *P < .05; **P < .01; ***P < .001.
Eviction of CTCF and RAD21 from the −31 element accompanies dissolution of the TAL1 active hub and loss of proximity between the +57 and −31 elements. (A) CTCF occupancy at the +57, +40, and −31 elements 48 and 96 hours after siRNA transfection with GATA1 (KD) or with luciferase (LUC) in K562 cells. (B) RAD21 occupancy at the +57, +40, and −31 elements 48 and 96 hours after siRNA transfection with GATA1 (KD) or with luciferase (LUC) in K562 cells. Gray vertical arrows in panels A and B highlight CTCF and RAD21 occupancy levels at −31. ChIP enrichments (log2) are shown with SEs. Annotation of test and negative control regions is denoted in black and gray text, respectively. Positive control is a CTCF/RAD21-bound element at the HNF4A locus. The percentage (%) occupancies of each protein in the GATA1 knockdown relative to the level found in the luciferase control are also shown. (C) Bar diagrams of looping interactions, measured as relative ligation frequencies (black bars), between the +57 and −31 elements determined by 3C at 48 and 96 hours after siRNA transfection with GATA1 (KD) or with luciferase (LUC) in K562 cells. Locations of 3C “bait” regions are denoted by vertical gray arrows. Loss of interactions due to GATA1 knockdown at 96 hours is shown by the dotted gray lines with arrowheads (and marked with an “X”). P values are indicated for relative ligation frequencies which are significantly reduced between the +57 and −31 elements in KD conditions when compared with the corresponding LUC controls. (D) Schematic model of looping interactions between CTCF/RAD21-bound elements at the TAL1 locus, and during GATA1 siRNA knockdown, in K562 cells. Loss of GATA1 is accompanied by (1) loss of CTCF and RAD21 from the −31 element and (2) loss of looping interactions between +57 and −31 (96 hours GATA1 knockdown) as the TAL1 erythroid hub is disassembled. *P < .05; **P < .01; ***P < .001.
Transcriptional regulation of flanking genes is dependent on the TAL1 hub
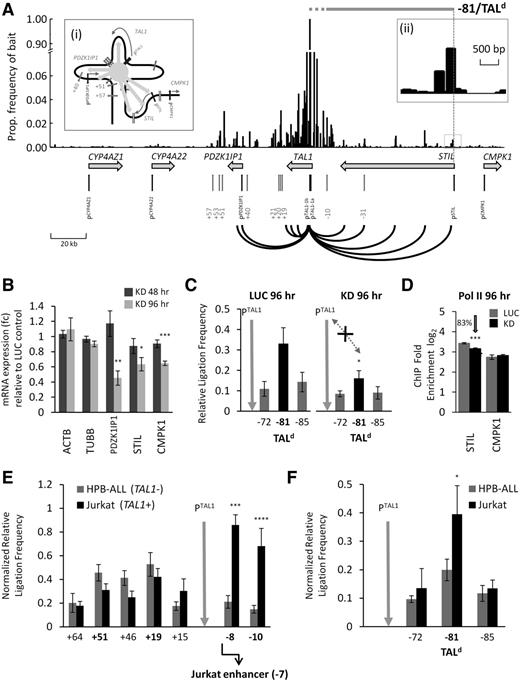
Based on our erythroid model (Figures 3D and 5D), we predicted that expression of genes flanking TAL1 may be dependent on the hub for transcriptional regulation because of their increased proximity to the TAL1 promoters. We used 4C26 (supplemental Figure 13) in combination with a TAL1 genomic tiling microarray9 to determine whether the TAL1 hub showed transcription-associated proximities with its flanking genes.
In addition to detecting interactions between the TAL1 promoters, its enhancers, and various points along the gene body of TAL1, our 4C-microarray analysis revealed that both PDZK1IP1 and STIL showed increased frequencies of interactions with the TAL1 promoters in TAL1-expressing cells (K562), compatible with the idea that the hub was bringing these flanking genes into close contact with the TAL1 promoters (Figure 6Ai). These interactions were found to be distributed at discrete points throughout the PDZK1IP1 and STIL genes (shown in red in Figure 6A). Levels of these interactions were noticeably higher than those detected by 4C in TAL1-nonexpressing HPB-ALL cells (supplemental Figure 14), and this was confirmed for one of these interactions by 3C (supplemental Figure 15; referred to as −81/TALd in Figure 6A, see the next section). mRNA levels of PDZK1IP1, STIL, and CMPK1 in GATA1 knockdown cells, all showed statistically significant decreases after 96 hours of GATA1 knockdown (Figure 6B) correlating with the complete dissolution of the TAL1 looping hub and loss of contact between the TAL1 promoters and the −81/TALd looping interaction (Figure 6C). Furthermore, Pol II recruitment at the STIL promoter was reduced during GATA1 knockdown, although this was not the case for CMPK1 (Figure 6D) or PDZK1IPI (which did not have detectable levels of Pol II at its promoter9 ). All of these data correlate well with a model whereby the TAL1 flanking genes have some degree of dependence on the TAL1 hub in erythroid cells for their expression.
Looping of genes flanking TAL1 suggest coregulation with TAL1 expression and involvement in T-ALL. (A) 4C-microarray interaction patterns obtained across the human TAL1 locus in K562 cells using TAL1 promoter 1b as the “bait”. Y-axis is the frequency of interactions expressed as a proportion of the “bait” signal for each microarray tile. Bars on the x-axis show the location of each tile across the TAL1 locus and its flanking genes. Looping interactions between TAL1 promoter 1b and the gene bodies of the TAL1, PDZK1IP1, and STIL are highlighted (black lines). The scale (in kb) is shown at the bottom left. (i) Schematic organization of the TAL1 hub with schematic interactions between the hub core, TAL1, and its flanking genes highlighted by the light gray arrows. (ii) The 4C interaction peak (−81) located in STIL intron 1 at the site of the TALd breakpoints found in patients with T-ALL. The extents of known TALd deletions are also shown by the horizontal gray bar (dotted gray portion represents a region of 8 kb known to contain breakpoints near the 5′ end of TAL1). (B) PDZK1IPI, STIL, and CMPK1 transcript levels (with SEs) in GATA1 knockdown samples 48 and 96 hours after transfection are shown relative to levels in the luciferase control samples. ACTB and TUBB were used as gene expression controls. (C) Bar diagrams of TAL1 promoter/STIL intron 1 (−81/TALd) looping interactions detected by 3C in K562 cells 96 hours after siRNA transfection with either GATA1 (KD) or with luciferase (LUC). P value is indicated for relative ligation frequencies which are significantly reduced between PTAL1 and TALd at 96 hour of GATA1 knockdown when compared with the corresponding LUC control. (D) ChIP-quantitative PCR occupancy for Pol II at the STIL and CMPK1 promoters 96 hours after transfection with GATA1 (black) or luciferase (gray) siRNA. Control was the TBP promoter (Figure 2F). (E-F) Comparison of looping interactions at the TAL1 locus in 2 T-ALL cell lines: HPB-ALL (TAL1 nonexpressing) and Jurkat (TAL1 expressing). Ligation frequencies were normalized against ERCC3 ligation frequencies. The location of the Jurkat −7 enhancer21 ∼500 bp downstream of −8 is shown in panel E. P values are indicated for interactions which are significantly higher in Jurkat cells. (C,E,F) Interaction frequencies are shown with SEs. Location of the 3C “bait” region (TAL1 promoter 1b) is denoted by vertical gray arrows. *P < .05; **P < .01; ***P < .001; ****P < .0001.
Looping of genes flanking TAL1 suggest coregulation with TAL1 expression and involvement in T-ALL. (A) 4C-microarray interaction patterns obtained across the human TAL1 locus in K562 cells using TAL1 promoter 1b as the “bait”. Y-axis is the frequency of interactions expressed as a proportion of the “bait” signal for each microarray tile. Bars on the x-axis show the location of each tile across the TAL1 locus and its flanking genes. Looping interactions between TAL1 promoter 1b and the gene bodies of the TAL1, PDZK1IP1, and STIL are highlighted (black lines). The scale (in kb) is shown at the bottom left. (i) Schematic organization of the TAL1 hub with schematic interactions between the hub core, TAL1, and its flanking genes highlighted by the light gray arrows. (ii) The 4C interaction peak (−81) located in STIL intron 1 at the site of the TALd breakpoints found in patients with T-ALL. The extents of known TALd deletions are also shown by the horizontal gray bar (dotted gray portion represents a region of 8 kb known to contain breakpoints near the 5′ end of TAL1). (B) PDZK1IPI, STIL, and CMPK1 transcript levels (with SEs) in GATA1 knockdown samples 48 and 96 hours after transfection are shown relative to levels in the luciferase control samples. ACTB and TUBB were used as gene expression controls. (C) Bar diagrams of TAL1 promoter/STIL intron 1 (−81/TALd) looping interactions detected by 3C in K562 cells 96 hours after siRNA transfection with either GATA1 (KD) or with luciferase (LUC). P value is indicated for relative ligation frequencies which are significantly reduced between PTAL1 and TALd at 96 hour of GATA1 knockdown when compared with the corresponding LUC control. (D) ChIP-quantitative PCR occupancy for Pol II at the STIL and CMPK1 promoters 96 hours after transfection with GATA1 (black) or luciferase (gray) siRNA. Control was the TBP promoter (Figure 2F). (E-F) Comparison of looping interactions at the TAL1 locus in 2 T-ALL cell lines: HPB-ALL (TAL1 nonexpressing) and Jurkat (TAL1 expressing). Ligation frequencies were normalized against ERCC3 ligation frequencies. The location of the Jurkat −7 enhancer21 ∼500 bp downstream of −8 is shown in panel E. P values are indicated for interactions which are significantly higher in Jurkat cells. (C,E,F) Interaction frequencies are shown with SEs. Location of the 3C “bait” region (TAL1 promoter 1b) is denoted by vertical gray arrows. *P < .05; **P < .01; ***P < .001; ****P < .0001.
TAL1 expression in T-ALL is linked to looping interactions
The −81/TALd interaction with the TAL1 promoters was of particular interest to us because it was associated with TAL1 transcription (in our K562 model), was also present in lymphoid cells, and was mapped to the sites of the common TAL1/STIL deletion breakpoints in patients with T-ALL (supplemental Figure 16). We hypothesized that this interaction, and possibly others at the TAL1 locus, could be related to ectopic expression of TAL1 in T-ALL either through (1) predisposing the TAL1 locus to TAL1/STIL deletions or (2) alterations in TAL1 looping patterns in the lymphoid lineage in T-ALL cases where TAL1/STIL deletions were absent. We tested the latter of these 2 hypotheses by performing 3C on a TAL1-expressing T-ALL cell line (Jurkat) and compared this to looping interactions in a TAL1-nonexpressing T-ALL cell line (HPB-ALL). Jurkat cells showed high-frequency interactions between the TAL1 promoters and sequences located at 8 and 10 kb upstream (supplemental Figure 16). One of these interactions (at −8) was located ∼500 bp from a site where the TAL1, RUNX1, and GATA3 proteins had previously been shown to bind and enhance TAL1 transcription in Jurkat cells (“the Jurkat enhancer”).21 The second of these was at the −10 enhancer known to be in contact with the TAL1 promoters in K562 cells (Figure 1). One or both of these upstream interactions was at significantly higher frequencies in Jurkat when compared with HPB-ALL (Figure 6E) or K562 (supplemental Figure 16). Furthermore, interaction of the TAL1 promoters with the TALd STIL breakpoint region (−81/TALd) was significantly higher in Jurkat cells than in HPB-ALL (Figure 6F). These data strongly suggest that TAL1 expression in Jurkat cells may be regulated by at least 3 transcription-associated looping interactions involving the TAL1 promoters and sequences upstream of TAL1 including the −81/TALd breakpoint region.
Discussion
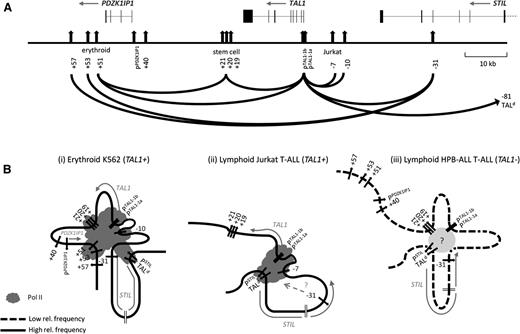
We describe here looping models of TAL1 transcriptional control (Figure 7). In our erythroid model, the transcriptionally active hub contains multiple loops which bring together the TAL1 promoters, several enhancers, and CTCF elements flanking the TAL1 regulon. In this model, contact between the erythroid enhancer and the TAL1 promoters reflects the activity of an enhancer known to be active in the erythroid lineage.8,9 Central to this hub is the presence of Pol II and the TEC (GATA1, TAL1, TCF3, LDB1, and LMO2). In Jurkat cells, where TAL1 is inappropriately expressed in T-ALL, looping between the TAL1 promoters and elements upstream are linked to TAL1 expression. In our lymphoid model, the transcriptionally inactive hub contains the TAL1 promoters, the stem cell enhancer and the −31 element, but these interactions occur at a significantly reduced frequency than in either erythroid or Jurkat cells and may reflect residual looping activity when TAL1 was expressed at some point prior to commitment to the lymphoid lineage. In all 3 models, interaction of the TAL1 promoters with the TALd breakpoint region is shown, as our data support that this interaction is present in all 3 cell types, but is elevated when TAL1 is transcribed. These models are consistent for data obtained from human cell lines, murine cell lines, and murine primary cells, thus providing substantial proof that these looping structures are found in hematopoietic cells in vivo. Recently, 2 other studies have described looping models for TAL1 expression.27,28 Both are highly compatible with our models and also account for the proximities of +5128 and +5327 to the TAL1 promoters. These studies also demonstrate requirements for Mediator27 and hSET128 in TAL1 looping interactions, suggesting further that GATA1 is not the only requirement for loop formation at the TAL1 locus.
Looping organization of TAL1 regulatory hubs. (A) Schematic organization of the human TAL1 locus and looping interactions between its regulatory elements identified in this study across all cell types. The scale (in kb) is shown to the right. Exon-intron structures (joined-up black bars) of TAL1 and its flanking genes PDZK1IP1, STIL, and CMPK1 and directions of transcription (gray arrows) are shown at the top of the panel. Promoters, enhancers, and CTCF binding sites are the vertical black arrows; the Jurkat −7 enhancer and −81/TALd are also shown. Interactions between TAL1 promoter 1b and its enhancers, between enhancers, and between CTCF/RAD21-bound elements are shown with black loops. (B) Schematic models of looping hubs at the TAL1 locus in 3 different cell types: (i) TAL1-expressing erythroid cells (K562) (assuming the −10 enhancer is also present in the active hub), (ii) TAL1-expressing lymphoid T-ALL cells (Jurkat) (assuming loops detected in this study are occurring in the same nuclei), and (iii) TAL1-nonexpressing lymphoid T-ALL cells (HPB-ALL) (assuming loops detected in this study are occurring in the same nuclei). Locations of promoters, enhancers, and CTCF/RAD21-bound elements are depicted as in the preceding figures. Direction of transcription of relevant genes (gray arrows) and Pol II machinery (dark gray lobules) are also shown and detailed in the key. Unknown factors mediating the interaction of the stem cell enhancer, the TAL1 promoters, and the −31 element in HPB-ALL are represented by the light gray ball. The looping interactions of CTCF/RAD21-bound elements in Jurkat cells were not studied here; thus, the location of the −31 element in the TAL1-expressing hub in Jurkat cells is not known and is denoted by a question mark (?). In all 3 models, contacts between TALd (and PSTIL located ∼1 kb away) and the TAL1 hubs are shown. Note: Interaction frequencies between TAL1 cis-regulatory elements in HPB-ALL were overall at lower levels than in either K562 or Jurkat, as depicted in the models.
Looping organization of TAL1 regulatory hubs. (A) Schematic organization of the human TAL1 locus and looping interactions between its regulatory elements identified in this study across all cell types. The scale (in kb) is shown to the right. Exon-intron structures (joined-up black bars) of TAL1 and its flanking genes PDZK1IP1, STIL, and CMPK1 and directions of transcription (gray arrows) are shown at the top of the panel. Promoters, enhancers, and CTCF binding sites are the vertical black arrows; the Jurkat −7 enhancer and −81/TALd are also shown. Interactions between TAL1 promoter 1b and its enhancers, between enhancers, and between CTCF/RAD21-bound elements are shown with black loops. (B) Schematic models of looping hubs at the TAL1 locus in 3 different cell types: (i) TAL1-expressing erythroid cells (K562) (assuming the −10 enhancer is also present in the active hub), (ii) TAL1-expressing lymphoid T-ALL cells (Jurkat) (assuming loops detected in this study are occurring in the same nuclei), and (iii) TAL1-nonexpressing lymphoid T-ALL cells (HPB-ALL) (assuming loops detected in this study are occurring in the same nuclei). Locations of promoters, enhancers, and CTCF/RAD21-bound elements are depicted as in the preceding figures. Direction of transcription of relevant genes (gray arrows) and Pol II machinery (dark gray lobules) are also shown and detailed in the key. Unknown factors mediating the interaction of the stem cell enhancer, the TAL1 promoters, and the −31 element in HPB-ALL are represented by the light gray ball. The looping interactions of CTCF/RAD21-bound elements in Jurkat cells were not studied here; thus, the location of the −31 element in the TAL1-expressing hub in Jurkat cells is not known and is denoted by a question mark (?). In all 3 models, contacts between TALd (and PSTIL located ∼1 kb away) and the TAL1 hubs are shown. Note: Interaction frequencies between TAL1 cis-regulatory elements in HPB-ALL were overall at lower levels than in either K562 or Jurkat, as depicted in the models.
We demonstrated that GATA1 is important for both the transcription of TAL1 and the maintenance of the active erythroid hub. The kinetics of hub disassembly and decreases in TAL1 transcription correlate well with the loss of GATA1 or complexes containing GATA1 from TAL1 cis-regulatory elements during GATA1 knockdown experiments, although we cannot exclude the possibility that other factors such as Mediator, hSET1, and CTCF may influence the kinetics of hub disassembly during GATA1 knockdown. GATA1 could facilitate the formation of such a hub via bridging and/or recruitment of other factors, such as LDB1, which may directly mediate looping.15,29,30 Alternatively, the transcription factory model of gene expression13 would predict that local concentrations of Pol II, GATA1, and other factors are sufficient to provide a loose scaffold that maintains the proximity between cis-regulatory elements. Furthermore, the ability of a single GATA factor to regulate the integrity of the TAL1 active hub has significant evolutionary implications for the TAL1 locus, which has undergone cis-regulatory remodeling over the last 360 million years while maintaining highly conserved GATA sites (supplemental Figure 17).
It is easy to envisage why the TAL1 erythroid enhancer is present in a transcriptionally active erythroid hub but it is less easy to understand why the stem cell enhancer, bound with GATA1, is also present, and why loss of this enhancer from the hub is accompanied by significant reductions in TAL1 transcription in K562 cells. The stem cell enhancer has previously been shown to be active only in hematopoietic progenitors by recruiting GATA2.31 However, exchange between GATA1 and GATA2 occupancy at the stem cell enhancer during lineage fate decisions could regulate its activity as has previously been described for other cis-regulatory elements.32 It is also plausible that K562 is a cell line which has attributes of both stem cell and erythroid function although our data from primary murine erythroblasts would argue that the stem cell enhancer is in contact with the TAL1 promoters in the erythroid lineage. Our data suggest a possible new function for this element in hub formation and enhancing TAL1 transcription in the erythroid lineage.
The roles of CTCF-bound elements in transcription-dependent looping at the TAL1 locus were also studied here. While all 4 CTCF elements at the TAL1 locus are bound by both CTCF and RAD21 in both erythroid and lymphoid cells, looping between 3 of these elements (+57 and/or +53 in contact with −31) occurs only in erythroid cells where TAL1 is expressed. This looping configuration facilitates compartmentalization of the TAL1 regulon and assembly of the erythroid hub because all TAL1 cis-regulatory circuitry is located between +57 and −31. Furthermore, this looping pattern overcomes issues created by the juxtaposition of the +40 CTCF element located between +51 erythroid enhancer and the TAL1 promoters (Introduction and supplemental Figure 1) because +40 does not participate in local insulator-based looping.
The −31 element appears to have a crucial role in determining looping interactions at the TAL1 locus in erythroid cells because both RAD21 and CTCF are lost from this element upon disassembly of the transcriptional hub in GATA1 knockdown cells. Furthermore, −31 appears to be dependent on GATA1 binding at other TAL1 cis-regulatory elements for the recruitment of CTCF and RAD21 to −31. This relationship may reflect the cooperative nature of GATA1, bound at TAL1 promoters and enhancers, in facilitating looping between TAL1 insulators through the recruitment of CTCF and RAD21 to −31 (as all of these elements are in close proximity in the hub). This mechanism is reminiscent of GATA1 and CTCF function at the HBBa locus in erythroid cells where they bind to different cis-regulatory elements but act cooperatively to enable looping.33
We demonstrated here that the TAL1 erythroid hub also coregulates, to some degree, the transcription of PDZK1IP1, STIL, and CMPK1 through their interaction with the hub via looping (supplemental Figure 11C). Thus, the TAL1 regulon definition which we previously proposed9 no longer adequately describes the regulatory influence that TAL1 imposes on its flanking genes because the majority of STIL and the entirety of CMPK1 reside outside of the predicted regulon boundaries. We propose that there are 2 models which could explain the coregulation of genes at the TAL1 locus, both of which rely on activity of the TAL1 hub and are consistent with our data (supplemental Figure 18). Importantly, in both models, expression of STIL relies on the dynamic assembly and disassembly of CTCF and RAD21 at −31. However, we cannot rule out the possibly that these, or other models, may account for transcription of genes flanking TAL1 in a coregulated way. Nevertheless, our data would suggest that conventional views of gene regulation may not apply to TAL1.
The occurrence of TAL1/STIL deletions (TALd deletions), in cases of T-ALL, is well documented.5,6,34-36 How these deletions occur with such frequency in T-ALL is remarkable and somewhat of a paradox. Although recombination by aberrant immunoglobulin/T-cell receptor (Ig/TCR) recombinase activity could be involved in mediating these deletions, sequences at these breakpoints are relatively poor substrates for this recombinase.34 Our 3C and 4C data show that regulatory hubs which control TAL1 transcription bring the TAL1/STIL common breakpoint regions (TALd) into close proximity in both TAL1-expressing cells (K562 and Jurkat) and TAL1-nonexpressing cells (HPB-ALL). Thus, physical proximity between these regions in early hematopoietic progenitors, or even in committed lymphoid cells, may predispose to TALd deletions by increasing the likelihood that they will recombine and give rise to an expressed TAL1/STIL fusion mRNA. Recent evidence suggests that chromosomal proximity may be a key determinant in double-stranded breakage and rejoining which lead to structural alterations in cancer genomes.37
Moreover, in cases of T-ALL where TALd deletions are not found, our data also support that ectopic expression of TAL1 is linked to changes in TAL1 regulation, mediated by aberrant looping between TAL1 cis-regulatory elements upstream of TAL1. To this end, loops involving the TALd STIL breakpoint and the TAL1 promoters may also be relevant to TAL1 transcription in T-ALL, in addition to any possible roles these loops may have in mediating deletion events. Our view of the events leading to TAL1 expression in T-ALL demonstrates that local transcription-associated looping topology may be deterministic in pathobiology via multiple mechanisms. Further analysis into these events is ongoing in our laboratory.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by a University of Glasgow studentship (Y.Z.).
Authorship
Contribution: Y.Z. and R.S. performed 3C analysis; Y.Z. performed 4C analysis, ChIP, and siRNA studies; S.K. developed the 4C procedure in our laboratory; M.V. and A.M.M. provided primary murine lymphocytes; P.S. provided bioinformatic support; D.V. and A.G.W. planned the study; and Y.Z., A.G.W., and D.V. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for S.K. is Department of Animal Sciences, School of Life Sciences, University of Hyderabad, Hyderabad, India.
The current affiliation for P.S. is lllumina United Kingdom, Chesterford Research Park, Little Chesterford, United Kingdom.
The current affiliation for R.S. is Department of Physiology Development and Neuroscience, University of Cambridge, Downing Site, Cambridge, United Kingdom.
Correspondence: David Vetrie, Epigenetics Unit, Institute of Cancer Sciences, University of Glasgow, Glasgow, United Kingdom G12 8QQ; e-mail: david.vetrie@glasgow.ac.uk.