Key Points
Myc amplifies BCR signaling and increases its own levels via upregulation of miR-17∼92 and subsequent targeting of ITIM proteins.
Inhibition of miR-17∼92 in DLBCL cell lines dampens the BCR response; DLBCL tumors of the BCR subtype have elevated levels of miR-17∼92.
Abstract
The c-Myc oncoprotein regulates >15% of the human transcriptome and a limited number of microRNAs (miRNAs). Here, we establish that in a human B-lymphoid cell line, Myc-repressed, but not Myc-stimulated, genes are significantly enriched for predicted binding sites of Myc-regulated miRNAs, primarily those comprising the Myc-activated miR-17∼92 cluster. Notably, gene set enrichment analysis demonstrates that miR-17∼92 is a major regulator of B-cell receptor (BCR) pathway components. Many of them are immunoreceptor tyrosine inhibitory motif (ITIM)-containing proteins, and ITIM proteins CD22 and FCGR2B were found to be direct targets of miR-17∼92. Consistent with the propensity of ITIM proteins to recruit phosphatases, either MYC or miR-17∼92 expression was necessary to sustain phosphorylation of spleen tyrosine kinase (SYK) and the B-cell linker protein (BLNK) upon ligation of the BCR. Further downstream, stimulation of the BCR response by miR-17-92 resulted in the enhanced calcium flux and elevated levels of Myc itself. Notably, inhibition of the miR-17∼92 cluster in diffuse large B-cell lymphoma (DLBCL) cell lines diminished the BCR response as measured by SYK and BLNK phosphorylation. Conversely, human DLBCLs of the BCR subtype express higher Myc and mir17hg transcript levels than other subtypes. Hence, the Myc-miR-17-92-BCR axis, frequently affected by genomic rearrangements, constitutes a novel lymphomagenic feed-forward loop.
Introduction
Myc is a noncanonical transcription factor that regulates >15% of the human transcriptome.1 Consistent with these broad effects on gene expression, Myc is known to regulate many facets of tumorigenesis including cell cycle, apoptosis, metabolism, and angiogenesis. This regulation entails activation or repression of thousands of protein-coding and noncoding RNAs. Although initial and some recent studies emphasized promoter-dependent regulatory mechanisms,2,3 posttranscriptional mechanisms are now coming to the fore. The discovery that Myc regulates microRNAs (miRNAs) provided a breakthrough in the field.
miRNAs are small (18-22 nt) noncoding RNAs that negatively regulate gene expression through the inhibition of translation and destabilization of messenger RNAs (mRNAs).4 Mature miRNAs are able to target hundreds of mRNAs involved in virtually all cellular processes, resembling in this respect the Myc family oncoproteins. Thus, it is not surprising that Myc can both directly activate5,6 and repress7 miRNA expression. miRNA deregulation could in principle account for posttranscriptional effects of Myc; for instance, upregulation of the miR-17∼92 cluster comprising miR-17, miR-18a, miR-19a/b, miR-20, and mir-92 could lead to downregulation of genes whose mRNAs have target sites for any of these 6 miRs. Indeed, subsequent experiments demonstrated that Myc-stimulated expression of miR-18a and miR-19a/b results in direct targeting of the thrombospondin-1 3′ untranslated region (UTR) in colon cancer cells, providing a definitive molecular mechanism for thrombospondin-1 mRNA destabilization and ensuing angiogenesis.8 Similarly, the Myc-repressed miR-15a/16 cluster largely accounts for upregulation of another nuclear oncoprotein c-Myb and erythroid differentiation.9,10 However, it stands to reason that other miR-controlled cell phenotypes are due to deregulation of multiple targets acting in frequently overlapping pathways. In this study, we aimed to characterize the role of miRNAs in global Myc-mediated gene regulation.
Materials and methods
Cell lines, western blotting, and quantitative PCR
Details of these standard analyses are available in supplemental Methods (available on the Blood Web site). Institutional animal care and use committee (IACUC) approval was provided by the Children’s Hospital of Philadelphia (#902).
BCR ligation
Cells were harvested after the indicated times and flash-frozen for western blotting after ligation of the B-cell receptor (BCR). In P493-6 cells, human α-immunoglobulin M (α-IgM) (Southern Biotech) was added to cells in the amounts indicated in the text for soluble α-IgM experiments. Human α-IgM was immobilized by incubating cell-culture plates with 5 μg/mL human α-IgM in phosphate-buffered saline at 4°C overnight. For the diffuse large B-cell lymphoma (DLBCL) cell lines, ligation of the BCR (α-BCR) was performed by treating cells with 5 µg of soluble α-IgM and α-IgG (Southern Biotech).
Luciferase reporter constructs and sensor assays
Luciferase reporter plasmids were constructed and luciferase sensor assays were performed essentially as described previously.10 3′UTR sequences are available in the supplemental Methods.
Microarray analysis
RNAs were harvested from triplicate cultures of P493-6 cells. Amplified complementary DNAs were hybridized to the Agilent Human GE 4x44K v2 microarray. Median intensities of each element on the array were captured with Agilent Feature Extraction (Version 9.53; Agilent Technologies). For statistical analysis, genes were called differentially expressed using the significance analysis of microarray 1 class response package with a false discovery rate (FDR) of 20%.
SigTerms analysis
The SigTerms Microsoft Excel macro was downloaded from http://sigterms.sourceforge.net/. Monte Carlo simulations were used to determine the average enrichment from random gene sets.
GSEA analysis
Gene set enrichment analysis (GSEA)11 was performed on microarray data from the miR-17∼92 mimic experiment. A nominal P value of < .05, in combination with an FDR q value of < 0.25, was considered significant.
Cytosolic calcium measurements
A full description is available in the supplemental Methods. Briefly, P439-6 cells were loaded with the calcium indicator fura-2 AM (3μM; Invitrogen) and then were allowed to adhere to Poly-l-lysine–coated cover slips. Ca2+ measurements were performed using a Leica DMI 6000 fluorescence microscope controlled by Metafluor software (Molecular Dynamics). The fluorescence emission ratio at 510 nm was recorded following excitation at 340 and 380 nm with a monochromator (TILL Photonics). Intracellular Ca2+ images were captured and represented as the Fura-2 fluorescence emission ratio. After 100 seconds of baseline recording, cells were stimulated by addition of anti-IgM antibody (5 μg/mL) to the imaging chamber. Mean Fura-2 ratio values were plotted (Origin Laboratory, Microcal). Pseudocolor ratio images at resting and peak cytosolic Ca2+ concentrations shown are representative of 3 individual experiments.
DLBCL gene expression and GSEA
Data from the Montes-Moreno et al12 and Lenz et al13 studies were downloaded from the Gene Expression Omnibus gateway (http://www.ncbi.nlm.nih.gov/geo/) using accession numbers GSE21849 and GSE10846, respectively. Consensus clustering classification (CCC) of the 233 rituximab, cyclophosphamide, doxorubicin dydrochloride (Hydroxydaunomycin), vincristine sulfate (Oncovin), and prednisone–treated patients in the Lenz study was previously established.14 Gene expression of patient tumors was normalized; and the average value of each DLBCL subtype was plotted ± the SEM. Analysis of variance of Myc and mir17hg expression in human DLBCL tumors was performed using GraphPad Prism 6. A P value < .05 was considered significant. For GSEA in Figure 5E, gene set analysis (GSA) was implemented. The average expression value of miR-17, miR-18a, miR-19a, miR-19b, miR-20a, and miR-92a for each sample was used as a quantitative response variable, and a gene set list was constructed using 41 genes in the Shipp BCR signature and 99 randomly selected gene sets of the same size. Restandardization was performed using the entire expression data, and the maxmean statistic was used. The significance of the enrichment for the Shipp BCR signature was calculated based on 1000 permutations.
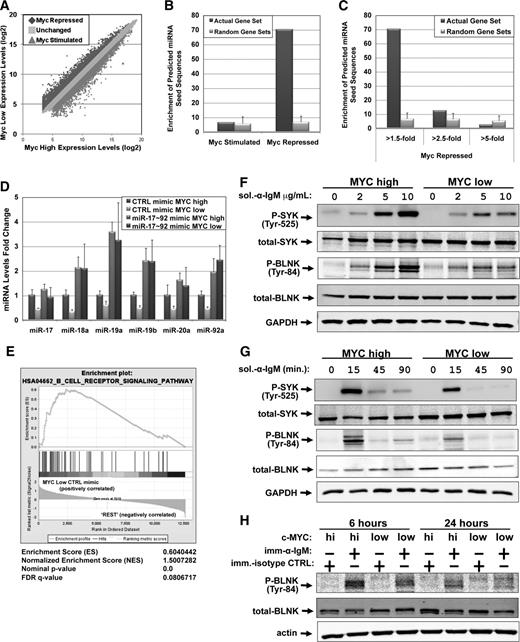
Myc-repressed transcripts are enriched for miR-17∼92 targets and BCR signaling components. (A) mRNA profiling was performed on P493-6 cells grown in the absence (MYCHIGH) or presence (MYCLOW) of 1μM doxycycline for 48 hours. Log2 gene expression levels were plotted for MYCHIGH vs MycLOW. Genes repressed or stimulated >1.5-fold by Myc are denoted. (B) SigTerms analysis comparing the enrichment of predicted miRNA seed sequences in Myc-stimulated vs Myc-repressed genes. Predictions are based on the TargetScan algorithm. One hundred random gene sets of the same sample size were used as a control. The average and SD of those random gene sets are plotted. (C) SigTerms analysis (as in panel B) comparing genes repressed by Myc >1.5-fold, >2.5-fold, and >5-fold. (D) P493-6 cells grown in the absence (MYCHIGH) or presence (MYCLOW) of 1μM doxycycline were treated with control mimic or miR-17∼92 mimic mix for 48 hours. RNA was isolated and qPCR was performed to quantitate changes in miRNA levels. RNU6B was used as an endogenous control and values are relative to MYC high/CTRL mimic. The average of 4 repetitions is reported. Error bars represent the SD. (E) mRNA profiling was performed on the cells from (D). GSEA was performed comparing the MYCLOW/CTRL mimic-treated cells to the “REST” (MYCLOW/miR-17∼92 mimic, MYCHIGH/CTRL mimic, and MYCHIGH /miR-17∼92 mimic). MYCLOW/CTRL mimic-treated cells are enriched for components of the BCR signaling pathway. The GSEA enrichment score, normalized enrichment score, P value, and FDR are indicated. For panels F through H, P493-6 cells were grown in the absence (MYCHIGH) or presence (MYCLOW) of 1μM doxycycline for 48 hours prior to various treatments with anti-IgM. Western blotting was performed for P-spleen tyrosine kinase (P-SYK), total-SYK, P-B–cell linker protein (BLNK), total-BLNK, and either GAPDH or actin. (F) P493-6 cells were treated with increasing amounts of soluble anti-IgM for 15 minutes to ligate the BCR. (G) P493-6 cells were treated with 10 μg/mL soluble anti-IgM to ligate the BCR. Samples were harvested at times indicated. (H) P493-6 cells were treated with immobilized anti-IgM (imm-α-IgM) or immobilized isotype control (imm-isotype-CTRL) for 6 or 24 hours. GAPDH, glyceraldehyde-3-phosphate dehydrogenase; qPCR, quantitative PCR.
Myc-repressed transcripts are enriched for miR-17∼92 targets and BCR signaling components. (A) mRNA profiling was performed on P493-6 cells grown in the absence (MYCHIGH) or presence (MYCLOW) of 1μM doxycycline for 48 hours. Log2 gene expression levels were plotted for MYCHIGH vs MycLOW. Genes repressed or stimulated >1.5-fold by Myc are denoted. (B) SigTerms analysis comparing the enrichment of predicted miRNA seed sequences in Myc-stimulated vs Myc-repressed genes. Predictions are based on the TargetScan algorithm. One hundred random gene sets of the same sample size were used as a control. The average and SD of those random gene sets are plotted. (C) SigTerms analysis (as in panel B) comparing genes repressed by Myc >1.5-fold, >2.5-fold, and >5-fold. (D) P493-6 cells grown in the absence (MYCHIGH) or presence (MYCLOW) of 1μM doxycycline were treated with control mimic or miR-17∼92 mimic mix for 48 hours. RNA was isolated and qPCR was performed to quantitate changes in miRNA levels. RNU6B was used as an endogenous control and values are relative to MYC high/CTRL mimic. The average of 4 repetitions is reported. Error bars represent the SD. (E) mRNA profiling was performed on the cells from (D). GSEA was performed comparing the MYCLOW/CTRL mimic-treated cells to the “REST” (MYCLOW/miR-17∼92 mimic, MYCHIGH/CTRL mimic, and MYCHIGH /miR-17∼92 mimic). MYCLOW/CTRL mimic-treated cells are enriched for components of the BCR signaling pathway. The GSEA enrichment score, normalized enrichment score, P value, and FDR are indicated. For panels F through H, P493-6 cells were grown in the absence (MYCHIGH) or presence (MYCLOW) of 1μM doxycycline for 48 hours prior to various treatments with anti-IgM. Western blotting was performed for P-spleen tyrosine kinase (P-SYK), total-SYK, P-B–cell linker protein (BLNK), total-BLNK, and either GAPDH or actin. (F) P493-6 cells were treated with increasing amounts of soluble anti-IgM for 15 minutes to ligate the BCR. (G) P493-6 cells were treated with 10 μg/mL soluble anti-IgM to ligate the BCR. Samples were harvested at times indicated. (H) P493-6 cells were treated with immobilized anti-IgM (imm-α-IgM) or immobilized isotype control (imm-isotype-CTRL) for 6 or 24 hours. GAPDH, glyceraldehyde-3-phosphate dehydrogenase; qPCR, quantitative PCR.
Myc stimulates the BCR response in a miR-17∼92–dependent manner. (A-D) Western blotting was performed for P-SYK, total-SYK, P-BLNK, total-BLNK, and GAPDH. (A) P493-6 cells were grown in the presence (MYCLOW) of 1μM doxycycline and treated with control mimic or miR-17∼92 mimic mix for 48 hours prior to addition of 10 μg/mL soluble anti-IgM for 15 minutes. (B) P493-6 cells were grown in the presence (MYCLOW) of 1μM doxycycline and treated with control mimic or miR-17∼92 mimic mix for 48 hours prior to treatment with immobilized anti-IgM (imm-α-IgM) or immobilized isotype control (imm-isotype-CTRL) for 6 hours. (C) P493-6 cells were grown in the absence (MYCHIGH) of 1μM doxycycline and treated with control short-hairpin inhibitor or miR-17∼92 short-hairpin inhibitor mix for 48 hours prior to addition of 10 μg/mL soluble anti-IgM for 15 minutes. (D) P493-6 cells were grown in the absence (MYCHIGH) of 1μM doxycycline and treated with control short-hairpin inhibitor or miR-17∼92 short-hairpin inhibitor mix for 48 hours prior to treatment with immobilized anti-IgM (imm-α-IgM) or immobilized isotype control (imm-isotype-CTRL) for 6 hours.
Myc stimulates the BCR response in a miR-17∼92–dependent manner. (A-D) Western blotting was performed for P-SYK, total-SYK, P-BLNK, total-BLNK, and GAPDH. (A) P493-6 cells were grown in the presence (MYCLOW) of 1μM doxycycline and treated with control mimic or miR-17∼92 mimic mix for 48 hours prior to addition of 10 μg/mL soluble anti-IgM for 15 minutes. (B) P493-6 cells were grown in the presence (MYCLOW) of 1μM doxycycline and treated with control mimic or miR-17∼92 mimic mix for 48 hours prior to treatment with immobilized anti-IgM (imm-α-IgM) or immobilized isotype control (imm-isotype-CTRL) for 6 hours. (C) P493-6 cells were grown in the absence (MYCHIGH) of 1μM doxycycline and treated with control short-hairpin inhibitor or miR-17∼92 short-hairpin inhibitor mix for 48 hours prior to addition of 10 μg/mL soluble anti-IgM for 15 minutes. (D) P493-6 cells were grown in the absence (MYCHIGH) of 1μM doxycycline and treated with control short-hairpin inhibitor or miR-17∼92 short-hairpin inhibitor mix for 48 hours prior to treatment with immobilized anti-IgM (imm-α-IgM) or immobilized isotype control (imm-isotype-CTRL) for 6 hours.
miR-17∼92 directly targets the ITIM-containing CD22 and FCGR2B to stimulate the BCR response. (A) P493-6 cells grown in the absence (MYCHIGH) or presence (MYCLOW) of 1μM doxycycline were treated with control mimic or miR-17∼92 mimic mix for 48 hours. RNA was isolated and qPCR was performed to quantitate changes in the FCRL4, FCGR2B, CD22, and CD72 mRNAs. GAPDH was used as an endogenous control and values are relative to MYC high/CTRL mimic. The average of 4 repetitions is reported. Error bars represent the SD. *P < .05 and **P < .01 by Student t test. (B) Changes in FCGR2B and CD22 protein levels were examined by western blotting on cells from Figure 4A. GAPDH was used as a loading control. (C-D) Dual luciferase assays performed in HCT-116 Dicer hypomorph cells cotransfected with CD22 3′UTR or FCGR2B 3′UTR luciferase sensors (panels C and D, respectively) and either control or miRNA mimics. Schematic of predicted 3′UTR-miRNA interactions are depicted (top panel). Shaded nucleotides indicate regions mutated to disrupt the mRNA-miRNA interaction in the mut 3′UTR constructs. Changes in luciferase activity upon treatment with miRNA mimics are plotted (bottom panel). Values were normalized to luciferase activity from control mimic-transfected cells. Error bars represent SD of 3 independent experiments. *P < .05 and **P < .01 by Student t test. (E) P493-6 cells grown in the presence (MYCLOW) of 1μM doxycycline and treated with control, anti-CD22, or anti-FCGR2B small interfering RNA (siRNA) for 48 hours prior to addition of 10 μg/mL soluble anti-IgM for 15 minutes to ligate the BCR. Western blotting was performed for CD22, FCGR2B, P-BLNK, total-BLNK, and actin.
miR-17∼92 directly targets the ITIM-containing CD22 and FCGR2B to stimulate the BCR response. (A) P493-6 cells grown in the absence (MYCHIGH) or presence (MYCLOW) of 1μM doxycycline were treated with control mimic or miR-17∼92 mimic mix for 48 hours. RNA was isolated and qPCR was performed to quantitate changes in the FCRL4, FCGR2B, CD22, and CD72 mRNAs. GAPDH was used as an endogenous control and values are relative to MYC high/CTRL mimic. The average of 4 repetitions is reported. Error bars represent the SD. *P < .05 and **P < .01 by Student t test. (B) Changes in FCGR2B and CD22 protein levels were examined by western blotting on cells from Figure 4A. GAPDH was used as a loading control. (C-D) Dual luciferase assays performed in HCT-116 Dicer hypomorph cells cotransfected with CD22 3′UTR or FCGR2B 3′UTR luciferase sensors (panels C and D, respectively) and either control or miRNA mimics. Schematic of predicted 3′UTR-miRNA interactions are depicted (top panel). Shaded nucleotides indicate regions mutated to disrupt the mRNA-miRNA interaction in the mut 3′UTR constructs. Changes in luciferase activity upon treatment with miRNA mimics are plotted (bottom panel). Values were normalized to luciferase activity from control mimic-transfected cells. Error bars represent SD of 3 independent experiments. *P < .05 and **P < .01 by Student t test. (E) P493-6 cells grown in the presence (MYCLOW) of 1μM doxycycline and treated with control, anti-CD22, or anti-FCGR2B small interfering RNA (siRNA) for 48 hours prior to addition of 10 μg/mL soluble anti-IgM for 15 minutes to ligate the BCR. Western blotting was performed for CD22, FCGR2B, P-BLNK, total-BLNK, and actin.
miR-17∼92 stimulates Myc stabilization and Ca2+flux upon ligation of the BCR. (A-B) Western blotting was performed for P-PLCγ2, total-PLCγ2, and actin. Bands were quantified and the ratio of P-PLCγ2 /total-PLCγ2 is shown. (A) P493-6 cells were grown in the presence (MYCLOW) of 1μM doxycycline and treated with control mimic or miR-17∼92 mimic mix for 48 hours prior to addition of 10 μg/mL soluble anti-IgM for 2 minutes. (B) P493-6 cells were grown in the absence (MYCHIGH) of 1μM doxycycline and treated with control short-hairpin inhibitor or miR-17∼92 short-hairpin inhibitor mix for 48 hours prior to addition of 10 μg/mL soluble anti-IgM for 2 minutes. (C-D) Following BCR crosslinking by anti-IgM, cytosolic Ca2+ levels in P493-6 cells were monitored by fluorescence microscopy using the ratiometric Ca2+ indicator dye Fura-2. Mean traces of 3 independent experiments are plotted (top); *P < .05, **P < .01. Independent experiments are plotted in supplemental Figure 2A-B. Representative images at basal cytosolic Ca2+ and maximum amplitude are displayed (bottom). (C) P493-6 cells were grown in the presence (MYCLOW) of 1μM doxycycline and treated with control mimic or miR-17∼92 mimic mix for 48 hours prior to addition of anti-IgM. (D) P493-6 cells (MYCHIGH) were treated with control short-hairpin inhibitor or miR-17∼92 short-hairpin inhibitor mix for 48 hours prior to addition of anti-IgM. (E) P493-6 cells grown in the presence (MYCLOW) of 1μM doxycycline and treated with control mimic or miR-17∼92 mimic mix for 48 hours prior to addition of 10 μg/mL soluble anti-IgM to ligate the BCR. Samples were harvested at times indicated. Western blotting was performed for P-Akt, pan-Akt, Pten, P-GSK-3β, total-GSK-3β, myc, and actin. (F) Model of Myc-miR-17∼92-BCR signaling feed-forward loop. Ligation of the BCR leads to phosphorylation of SYK and PI3K. ITIMs in CD22 and FCGR2B recruit the Src homology region 2 domain containing phosphatase (SHP)-1 and SHIP phosphatases, which target SYK and PI3K to dampen BCR signaling. miR-17∼92 represses cd22 and fcgr2b either directly and amplifies BCR signaling. This results in increased phospho-AKT, phospho-GSK-3β, Myc, and therefore, miR-17∼92.
miR-17∼92 stimulates Myc stabilization and Ca2+flux upon ligation of the BCR. (A-B) Western blotting was performed for P-PLCγ2, total-PLCγ2, and actin. Bands were quantified and the ratio of P-PLCγ2 /total-PLCγ2 is shown. (A) P493-6 cells were grown in the presence (MYCLOW) of 1μM doxycycline and treated with control mimic or miR-17∼92 mimic mix for 48 hours prior to addition of 10 μg/mL soluble anti-IgM for 2 minutes. (B) P493-6 cells were grown in the absence (MYCHIGH) of 1μM doxycycline and treated with control short-hairpin inhibitor or miR-17∼92 short-hairpin inhibitor mix for 48 hours prior to addition of 10 μg/mL soluble anti-IgM for 2 minutes. (C-D) Following BCR crosslinking by anti-IgM, cytosolic Ca2+ levels in P493-6 cells were monitored by fluorescence microscopy using the ratiometric Ca2+ indicator dye Fura-2. Mean traces of 3 independent experiments are plotted (top); *P < .05, **P < .01. Independent experiments are plotted in supplemental Figure 2A-B. Representative images at basal cytosolic Ca2+ and maximum amplitude are displayed (bottom). (C) P493-6 cells were grown in the presence (MYCLOW) of 1μM doxycycline and treated with control mimic or miR-17∼92 mimic mix for 48 hours prior to addition of anti-IgM. (D) P493-6 cells (MYCHIGH) were treated with control short-hairpin inhibitor or miR-17∼92 short-hairpin inhibitor mix for 48 hours prior to addition of anti-IgM. (E) P493-6 cells grown in the presence (MYCLOW) of 1μM doxycycline and treated with control mimic or miR-17∼92 mimic mix for 48 hours prior to addition of 10 μg/mL soluble anti-IgM to ligate the BCR. Samples were harvested at times indicated. Western blotting was performed for P-Akt, pan-Akt, Pten, P-GSK-3β, total-GSK-3β, myc, and actin. (F) Model of Myc-miR-17∼92-BCR signaling feed-forward loop. Ligation of the BCR leads to phosphorylation of SYK and PI3K. ITIMs in CD22 and FCGR2B recruit the Src homology region 2 domain containing phosphatase (SHP)-1 and SHIP phosphatases, which target SYK and PI3K to dampen BCR signaling. miR-17∼92 represses cd22 and fcgr2b either directly and amplifies BCR signaling. This results in increased phospho-AKT, phospho-GSK-3β, Myc, and therefore, miR-17∼92.
Inhibition of miR-17∼92 blunts the BCR response in DLBCL cell lines. (A-C) The DLBCL cell lines HBL-1, TMD8, U2932, OCI-LY10, OCI-LY1, SUDHL4, Pfeiffer, and Karpas 422 were treated with either control short-hairpin inhibitor or miR-17∼92 short-hairpin inhibitor mix for 48 hours. (A) The DLBCL cell lines were harvested and western blotting was performed for CD22 (two exposures shown), FCGR2B, FCRL4, and actin. (B) The DLBCL cell lines were treated with treated with 5 μg/mL soluble anti-IgM and anti-IgG or 10 μg/mL soluble isotype control for 15 minutes. Western blotting was performed for P-SYK, total-SYK, P-BLNK, total-BLNK, and actin. (C-D) The average (mean) normalized Myc (C) and mir17hg (D) expression in the various DLBCL subtypes classified by the consensus signature. Error bars represent the SEM. One-way analysis of variance P values are denoted. (E) GSA demonstrating a positive correlation between Shipp BCR signature genes and averaged mature miR-17∼92 expression. The heatmap shows the expression of the 41 genes in columns with each tumor sample in rows. Orange indicates high expression; blue indicates low expression. For each gene, the gene score from the GSA is indicated in the plot above the heatmap, with positive scores indicating positive association with miR-17∼92 expression. The averaged mature miR-17∼92 expression for each sample is shown in the graph to the right of the heatmap.
Inhibition of miR-17∼92 blunts the BCR response in DLBCL cell lines. (A-C) The DLBCL cell lines HBL-1, TMD8, U2932, OCI-LY10, OCI-LY1, SUDHL4, Pfeiffer, and Karpas 422 were treated with either control short-hairpin inhibitor or miR-17∼92 short-hairpin inhibitor mix for 48 hours. (A) The DLBCL cell lines were harvested and western blotting was performed for CD22 (two exposures shown), FCGR2B, FCRL4, and actin. (B) The DLBCL cell lines were treated with treated with 5 μg/mL soluble anti-IgM and anti-IgG or 10 μg/mL soluble isotype control for 15 minutes. Western blotting was performed for P-SYK, total-SYK, P-BLNK, total-BLNK, and actin. (C-D) The average (mean) normalized Myc (C) and mir17hg (D) expression in the various DLBCL subtypes classified by the consensus signature. Error bars represent the SEM. One-way analysis of variance P values are denoted. (E) GSA demonstrating a positive correlation between Shipp BCR signature genes and averaged mature miR-17∼92 expression. The heatmap shows the expression of the 41 genes in columns with each tumor sample in rows. Orange indicates high expression; blue indicates low expression. For each gene, the gene score from the GSA is indicated in the plot above the heatmap, with positive scores indicating positive association with miR-17∼92 expression. The averaged mature miR-17∼92 expression for each sample is shown in the graph to the right of the heatmap.
Results
Myc-repressed transcripts are enriched for miR-17∼92 targets
To determine how many and which Myc-regulated genes are regulated through miRNA intermediates, we chose the P493-6 cell line. P493-6 cells are an Epstein-Barr virus-immortalized lymphoblastoid cell line with a tet-repressible myc gene.15 Grown in the absence of doxycycline, high levels of myc mRNA are transcribed mimicking an Ig-myc translocation. This is accompanied by excessive alterations to miRNA expression.5,7 Additionally, mRNA profiling of P493-6 cells identified 3171 genes whose expression increased >1.5-fold (Myc-stimulated) and 3779 genes whose expression decreased >1.5-fold (Myc-repressed) with MycHIGH compared with MycLOW (Figure 1A).
We then used the SigTerms algorithm16 to identify which Myc-regulated miRNAs had the greatest influence on the Myc signature. After importing the Myc-stimulated gene list into SigTerms and searching for the enrichment of predicted miRNA binding sites, we found no enrichment compared with simulated random gene sets. However, Myc-repressed genes exhibited a 10-fold (P < .05) enrichment of predicted miRNA binding sites compared with the simulations, suggesting that miRNAs play a global role in Myc-mediated repression (Figure 1B). Furthermore, genes repressed >2.5-fold or >5-fold show little or no enrichment over the simulations, suggesting that genes repressed by Myc between 1.5- and 2.5-fold are the genes most frequently targeted by miRNAs (Figure 1C). We next sought to identify which predicted miRNA binding sites are most enriched for in the Myc-repressed gene set. The 2 miRs most significantly enriched for are the miR-17/20 and miR-19a/b families (supplemental Table 1). Also significantly enriched for are the miR-25/92/92ab family and the miR-18ab family. Obtained without prior knowledge of the established link between Myc and the miR-17∼92 cluster, the results of the SigTerms analysis further emphasize the broad role of miR-17∼92 in Myc repression.
The myc-miR-17∼92 axis stimulates BCR signaling
Because the miR-17∼92 cluster is the centerpiece of miRNA-mediated Myc repression, we sought to identify the global effects of miR-17∼92 on the Myc transcriptome. Using the P493-6 system where endogenous miR-17∼92 levels are tightly controlled by Myc, we transfected both MycHIGH and MycLOW cells with exogenous control miRNA mimic or a mixture of 6 miR-17∼92 mimics. This experimental design allowed us to uncouple changes in Myc expression and changes in miR-17∼92 levels (Figure 1D, compare MycHIGH vs MycLOW with control (CTRL) mimics to MycHIGH vs MycLOW with miR-17∼92 mimics). To identify miR-17∼92–dependent Myc targets, we performed mRNA profiling of these 4 cultures followed by GSEA.11 Specifically, we compared the “MycLOW CTRL mimic” condition, in which miR-17∼92 levels are depleted, to the 3 other conditions in which miR-17∼92 miRNAs are more abundant. The GSEA revealed that the differentially expressed genes are selectively enriched in BCR pathway components (Figure 1E). Inspection of the genes found in the core enrichment profile showed that both positive and negative regulators of the BCR signaling pathway were present (supplemental Table 2) and posed the question of whether Myc and miR-17∼92 stimulate or inhibit the BCR signaling.
To characterize the role of Myc in the BCR pathway, increasing amounts of anti-IgM were titrated into the medium to ligate the BCR of P493-6 cells. Phosphorylation of the SYK and the BLNK were used as a gauge for BCR signaling.17 MycHIGH cells exhibited greater levels of SYK and BLNK phosphorylation than MycLOW cells (Figure 1F), supporting a positive role for Myc in BCR signaling. We considered the possibility that low levels of Myc resulted merely in delayed phosphorylation. To explore this scenario, we treated P493-6 cells with anti-IgM and harvested cells at several time points. At each time point tested, there was more phosphorylation of SYK and BLNK when Myc was high compared with when Myc was low; and the kinetics of phosphorylation were independent of Myc status (Figure 1G).
Because of the transient nature of this response, we investigated whether Myc affects the BCR response after ligation with immobilized anti-IgM vs isotype control. This approach enabled examination of long-term (6 and 24 hours) Myc effects. After 6 hours, phosphorylation of BLNK increased more in MycHIGH cells compared with MycLOW cells, and at 24 hours the difference was even greater (Figure 1H). This indicated that Myc stimulates BCR signaling in response to both soluble and immobilized antigens and that the Myc effect is both rapid and sustained.
We next addressed whether miR-17∼92 on its own could stimulate BCR signaling. We transfected exogenous miR-17∼92 mimics into MycLOW P493-6 cells prior to treatment with soluble or immobilized anti-IgM (Figure 2A-B). Upon ligation of the BCR, phosphorylation of both SYK and BLNK were increased in the presence of exogenous miR-17∼92 mimic indicating that miR-17∼92, like Myc, stimulates the BCR response. In contrast, phosphorylation of Lyn (which increased modestly upon ligation of the BCR) was unaffected by exogenous miR-17∼92 mimic treatment (supplemental Figure 1A). We also demonstrate that miR-17∼92 mimic-treated MycHIGH cells responded similarly to control mimic-treated MycHIGH cells (supplemental Figure 1B-C).
To determine whether miR-17∼92 is necessary for Myc to stimulate the BCR response, we transfected MycHIGH cells with either control or miR-17∼92 short-hairpin antisense inhibitors prior to ligation of the BCR. The full Myc-driven BCR response, as measured by phosphorylation of SYK and BLNK, required miR-17∼92, whether soluble or immobilized antibody was used to ligate the BCR (Figure 2C-D).
miR-17∼92 directly targets the transcripts of ITIM-containing proteins CD22 and FCGR2B
We hypothesized that to positively regulate BCR signaling, miR-17∼92 cluster members would need to target its negative regulators and these targets would act upstream of BLNK phosphorylation. Several candidates for this role were present in the core enrichment list of genes (supplemental Table 2). Per TargetScan, 2 of these negative regulators contained predicted miR-17∼92 binding sites in their 3′UTRs, namely cd22 and cd72, both of which encode immunoreceptor tyrosine inhibitory motifs (ITIMs).18,19 Additionally, we searched TargetScan for other ITIM-containing proteins with predicted miR-17∼92 binding sites and identified fcgr2b20 and fcrl4 (also known as IRTA121 ) as putative targets. Quantitative reverse transcription–polymerase chain reaction analysis revealed that Myc repressed each of these gene transcripts (Figure 3A). Furthermore, exogenous miR-17∼92 mimics could completely abolish the effect of Myc withdrawal on cd22 and fcrl4, and partially on fcgr2b and cd72 (Figure 3A). We then performed western blotting to examine changes in protein levels. For FCRL4, FCGR2B, and CD22, MycLOW cells treated with exogenous miR-17∼92 mimics exhibited reduced protein levels compared with those observed in MycHIGH cells (Figure 3B, supplemental Figure 1C). In contrast, CD72 protein levels were mostly unchanged by addition of miR-17∼92 mimic and therefore the regulation of CD72 by Myc is likely to be mostly transcriptional (supplemental Figure 1D). Taken together, these data suggest that miR-17∼92 targets the ITIM-containing protein transcripts of fcrl4, fcgr2b, and cd22.
We then sought to determine whether this regulation was due to direct targeting by miR-17∼92 cluster members or was an indirect effect. To probe for direct binding of miR-92a, -19a, and -18a to predicted binding sites in the fcrl4, cd22, and fcgr2b 3′UTRs respectively, we used dual-luciferase sensor assays. In addition to fragments with wild-type 3′UTRs, we also generated constructs with mutations in the predicted seed sequences to serve as negative controls (Figure 3C-D, supplemental Figure 1E, with mutated nucleotides shaded). While miR-92a is predicted to target fcrl4, we did not observe a decrease in luciferase activity when the miR-92a mimic was cotransfected with the wild-type fcrl4 3′UTR construct, suggesting the regulation of fcrl4 by miR-17∼92 is either indirect or occurs through another, as of yet untested site (supplemental Figure 1E). On the other hand, the cd22 and fcgr2b 3′UTRs are directly targeted by miR-19a and miR-18a (Figure 3C-D). The wild-type cd22 3′UTR exhibited a 25% statistically significant reduction in luciferase activity compared with the mutant 3′UTR construct (Figure 3C). In the fcgr2b experiments, we observed that transfection of miR-18a resulted in decreased luciferase activity for all constructs (Figure 3D). However, the wild-type fcgr2b 3′UTR exhibited a significant additional reduction in luciferase activity compared with the mutant 3′UTR construct.
To confirm their inhibitory role in BCR signaling, we used siRNA-mediated knockdown of CD22 and FCGR2B. MycLOW cells were treated with control, anti-CD22, or anti-FCGR2B siRNAs prior to ligation of the BCR. Knockdown efficiency was moderate, similar to miRNA-mediated effects (compare Figure 3B to 3E). Yet knockdown of either CD22 or FCGR2B led to increased phosphorylation of BLNK upon BCR ligation, with FCGR2B knockdown having a more prominent effect (Figure 3E). Hence, the miR-17∼92–targeted ITIM proteins indeed negatively control the BCR signaling.
miR-17∼92 stimulates Ca2+ signals and stabilizes Myc upon ligation of the BCR
To understand the biological relevance of the miR-17∼92–BCR axis, we sought to identify changes in downstream signaling events. Ligation of the BCR leads to phosphorylation and activation of phospholipase C (PLC)γ2, which generates inositol triphosphate–mediated release of calcium from the endoplasmic reticulum and subsequent activation of calcium entry via plasma membrane Orai channels.22 To explore whether Myc and miR-17∼92 affect this downstream signaling event, we initially treated MycHIGH and MycLOW cells with anti-IgM and probed for phosphorylation of PLCγ2. Like SYK and BLNK, PLCγ2 phosphorylation was increased in MycHIGH cells compared with MycLOW cells (supplemental Figure 2A). To examine the functional consequence of this signaling, we performed real-time single-cell cytosolic calcium measurements to determine how Myc affects cytosolic calcium entry. MycLOW cells exhibit increased basal signaling relative to MycHIGH cells (supplemental Figure 2B). Upon addition of anti-IgM, we observed a sharp increase in the Fura-2 ratio indicative of an increase in cytosolic calcium concentration. When directly comparing MycHIGH cells to MycLOW cells, induced peak Fura-2 ratios were similar; therefore, the response to BCR ligation was greater in MycHIGH cells (supplemental Figure 2B).
We next treated MycLOW cells with miR-17∼92 mimics and MycHIGH cells with miR-17∼92 inhibitors prior to anti-IgM treatment (Figure 4A-B). As expected, the presence of exogenous miR-17∼92 led to increased phosphorylation of PLCγ2 upon ligation of the BCR. In contrast, miR-17∼92 inhibitors limited PLCγ2 phosphorylation. Again, single-cell cytosolic calcium measurements were used to determine the functional effects of this altered signaling. In the MycLOW condition, addition of exogenous miR-17∼92 mimic augmented the calcium signal (Figure 4C). Furthermore, inhibition of miR-17∼92 in MycHIGH cells resulted in an attenuated calcium signal consistent with decreased PLCγ2 activation (Figure 4D). Together, these results mirror the effects on phosphorylation of SYK and BLNK and demonstrate an impact of miR-17∼92 on critical signals downstream of BCR signaling.
In another biologically relevant pathway, ligation of the BCR leads to phosphorylation and activation of the mitogen-activated protein kinase 1 (MAPK1, or Erk) and phosphatidylinositol-3 kinase (PI3K) pathways. Phosphorylation of Erk increased upon ligation of the BCR, and the increase in MycHIGH cells was greater than in MycLOW cells. However, miR-17∼92 mimics had no effect on Erk phosphorylation (supplemental Figure 2E), suggesting that the Myc-Erk axis is not miRNA-dependent. However, when probing for Myc itself, we observed that ligation of the BCR increased Myc levels and that this increase was greater in miR-17∼92–treated MycLOW cells compared with CTRL mimic–treated MycLOW cells (supplemental Figure 2E). To explain this phenomenon, we considered the fact that in a pathway parallel to BLNK phosphorylation, BCR ligation also leads to v-akt murine thymoma viral oncogene homologue (AKT)1 phosphorylation.23 pAKT1 then directly inhibits glycogen synthase kinase 3β (GSK-3β) through phosphorylation of Ser924 and further downstream, GSK-3β targets Myc for degradation by phosphorylating Thr58.25 Consequently, as described by us previously, Akt phosphorylation results in sharply increased Myc levels in B cells.26 Of note, both CD22 and FCGR2B can recruit the SH2 containing inositol phosphatase (SHIP),27,28 which dephosphorylates phosphatidylinositol 3,4,5-triphosphate (PIP3)29 and therefore dampens phosphorylation of Akt in B cells.30 We postulated that miR-17∼92 could increase Akt phosphorylation by inhibiting CD22 and FCGR2B and rendering SHIP less active.
We treated MycLOW P493-6 cells with either control or miR-17∼92 mimic prior to stimulation with soluble anti-IgM and harvested cells 30 and 90 minutes after ligation (Figure 4E). In both control and miR-17∼92 mimic-treated cells, we observed increased phosphorylation of Akt after BCR ligation (Figure 4E). However, this increase was greater in miR-17∼92 mimic-treated cells. miR-17∼92 had been shown to repress phosphotase and tensin homologue (Pten),31 another modulator of Akt phosphorylation; however, in our system, miR-17∼92 treatment did not result in altered Pten levels (Figure 4E), a phenomenon also observed in other lymphoma cell lines.32 Nevertheless, we did observe increased GSK-3β phosphorylation in the presence of exogenous miR-17∼92 mimic (Figure 4E). We then probed for changes in endogenous Myc levels and found that Myc was more strongly upregulated in miR-17∼92–treated cells, consistent with increased GSK-3β inactivation (Figure 4E, supplemental Figure 2E). These results establish that Myc, via miR-17∼92 inhibition of ITIM-containing proteins, stimulates the BCR response and participates in a feed-forward regulatory loop (Figure 4F).
Inhibition of miR-17∼92 blunts the BCR response in DLBCL cell lines
BCR activation has been implicated in several types of lymphomas and is particularly relevant to the pathogenesis of DLBCL. Additionally, very recent evidence suggests that transgenic expression of miR-17∼92 in murine B cells can drive lymphomas resembling DLBCL.33 Thus, we set out to demonstrate that the miR-17∼92-BCR axis is functional in human DLBCL cell lines. We investigated 8 DLBCL cell lines with varying levels of Myc and miR-17∼92. They included 6 BCR subtype and 2 oxidative phosphorylation (OxPhos) subtype per CCC34 and 4 activated B-cell (ABC)-DLBCL and 4 germinal center B-cell (GCB)-DLBCL per cell of origin classification35 (supplemental Figure 3A-C). Due to the genetic diversity, a correlation between Myc and miR-17∼92 levels across this panel was not apparent. Thus, we treated each of these lines with control or miR-17∼92 short-hairpin inhibitors. Consistent with the idea that several ITIM proteins are direct targets of the miR-17∼92 cluster, 6 of 8 DLBCL cell lines express higher levels of CD22 following treatment with the short-hairpin inhibitors (Figure 5A). In the remaining cell lines, HBL-1 and Pfeiffer, we were unable to detect any CD22 protein. Furthermore, 3 of 8 DLBCL cell lines exhibited increased FCGR2B levels. Although we were unable to establish a direct targeting mechanism, FCRL4 was upregulated by the inhibition of miR-17∼92 in 4 of 8 cell lines. Thus, while regulation of the individual ITIM-containing proteins by miR-17∼92 was not universal across genetically diverse cell lines, each cell line exhibits increased expression of at least 1 target upon inhibition of miR-17∼92.
To determine the effects of miR-17∼92 on the overall BCR response, we repeated the miRNA inhibitor treatment in DLBCL cell lines. For each line, SYK and BLNK phosphorylation was measured after ligation of the BCR (except Karpas 422, in which BLNK is not expressed36 ) (supplemental Figure 3D). In 7 of 8 DLBCL cell lines, miR-17∼92 inhibition prior to ligation of the BCR diminished SYK and BLNK phosphorylation, indicating an important role for miR-17∼92 in the BCR response of DLBCLs (Figure 5B). Even in Karpas 422 cells, miR-17∼92 inhibition modestly decreased SYK phosphorylation and corresponded to the minor increase in CD22 levels (Figure 5A-B).
mir17hg is highly expressed in BCR-subtype tumors
To investigate the relationship between Myc and miR-17∼92 in human DLBCLs, we compared the expression of each in the various DLBCL subtypes. Elevated Myc expression is one of the characteristics of the BCR subtype.34 Indeed, we found MYC mRNA expression to be significantly higher in BCR subtype tumors than the OxPhos, host response (HR), and unassigned subtype tumors (Figure 5C). We also observed that mir17hg primary transcript expression was highest in BCR subtype tumors (Figure 5D). In parallel, we were able to validate mir17hg as a suitable surrogate for the mature miRNA levels using paired miRNA/mRNA profiling of DLBCLs12 (supplemental Figure 3E). Additionally, we performed GSEA on these paired samples and found that the mature miR-17∼92 cluster members (averaged) correlated with genes in the Shipp BCR signature (Figure 5E). This correlation approaches significance (P = .078) and fell just short of the 95% CI, possibly due to the small sample size (n = 29).
Discussion
There is a striking similarity between Myc and miRNAs, in that both are capable of exerting modest repression on hundreds of target genes. Using mRNA profiling and TargetScan predictions, we determined that Myc-repressed, but not Myc-stimulated, genes were enriched for predicted miRNA binding sites (Figure 1B). Myc is known to repress several miRNAs7 and targets of these miRNAs are likely upregulated when Myc expression is high; however, our results suggest that repression of miRNAs is not the chief mechanism by which Myc stimulates expression. On the other hand, Myc repression is likely to be mediated in large part through the miRNA pathway. This evidence is further supported by the fact that miR-17∼92 cluster members, a Myc-stimulated cluster, were among the most enriched for miRNAs (supplemental Table 1). We conclude that miRNAs are functionally integral to Myc-mediated repression and that miR-17∼92 members are the dominant effectors.
The 2 proposed classes of miRNA-target interactions are “one miRNA–one robust target” and “one miRNA–a confluence of weak targets.” The former class includes several important miR-17∼92 targets including PTEN, Bim, and p21.31,37-39 We, in contrast, sought to characterize the broad gene expression changes driven by miR-17∼92. GSEA indicated that miR-17∼92 might preferentially target BCR signaling pathway components (Figure 1E). Subsequent experiments designed to explore this scenario revealed that miR-17∼92 stimulated the response to BCR ligation (Figure 2). We hypothesized that in order for miR-17∼92 to have this stimulatory effect, it had to repress negative regulators of BCR signaling. Indeed cd72, fcrl4, fcgr2b, and cd22, all of which are inhibitory of BCR signaling,18,27,40-42 were identified as Myc-repressed genes with putative miR-17∼92 sites. The repression of cd72 appeared to be largely transcriptional and the exact mechanism of fcrl4 regulation remains elusive. In contrast, we confirmed that miR-18a and miR-19a directly target the fcgr2b and cd22 3′UTRs, respectively (Figure 3C-D). Furthermore, knockdown of either target increased the BCR response (Figure 3E). Mechanistically, this is likely to be achieved via recruitment of SHIP or SHP1/2 phosphatases by FCGR2B and CD22 ITIMs, as demonstrated previously.27,28 Thus, miR-17∼92 directly targets ITIM-containing proteins to alter the balance between kinases and phosphatases in favor of BCR signaling. However, not all BCR signaling components are affected, for example, Lyn may be targeted by different phosphatases, and its activation via BCR ligation is impervious to the effects of miR-17∼92.
Ligation of the BCR is known to result in phosphorylation of PLCγ2,43 the opening of plasma membrane calcium channels, and a resultant calcium flux.22 We demonstrate that in MycLOW cells, exogenous miR-17∼92 augments this calcium signal (Figure 4C). Additionally, the loss-of-function approach revealed that inhibition of miR-17∼92 attenuated calcium signaling in MycHIGH cells (Figure 4D). These results establish the functional role for miR-17∼92 in the BCR pathway.
In addition to targeting the SYK-PLCγ2 axis, SHIP dephosphorylates PIP329,44,45 and therefore dampens phosphorylation of Akt in B cells.30,40 Previous work from our laboratory has shown that in B cells, phosphorylation of Akt leads to the phosphorylation/inhibition of GSK-3β and potent stabilization of Myc.26 Consistent with this scenario, in MycLOW cells treated with exogenous miR-17∼92 mimic, we observed increased phosphorylation of Akt and GSK-3β and augmented Myc levels upon ligation of the BCR (Figure 4A). This constitutes a feed-forward loop wherein overexpression of Myc induces miR-17∼92, which amplifies BCR signaling resulting in yet more Myc and more miR-17∼92 (Figure 4F).
Aberrant BCR signaling plays a pivotal role in the pathogenesis of DLBCL. Several studies have demonstrated the importance of the BCR in lymphomagenesis and 2 models of BCR signaling have emerged: tonic and antigen-driven.23,46,47 By definition, antigen-stimulated signaling requires exogenous ligand to trigger the receptor. This has been modeled in Eμ-myc transgenic mice with a modified BCR (BCRHEL) designed to recognize hen egg lysozyme (HEL).48 In the absence of the cognate antigen, coexpression of BCRHEL and the Eμ-myc transgenes already resulted in more rapid tumorigenesis. However, ectopically expressed HEL further increased tumor burden demonstrating that antigen-stimulated BCR signaling cooperates with Myc to drive lymphomagenesis.48 In contrast to antigen-stimulated BCR signaling, tonic BCR signaling does not require exogenous antigen and might be driven by mutations that are presumed (but not proven) to lead to constitutive activity of the receptor.49 For the most part, how tonic signaling can be sustained in vivo remains unclear.
Here, we demonstrate that miR-17∼92 levels are limiting to the BCR response in multiple DLBCL cell lines (Figure 5B), likely because of its propensity to target CD22 and FCGR2B (Figure 5A), although other miR-17∼92 targets could potentially contribute. It follows that lymphomas whose pathogenesis involves aberrant BCR signaling would benefit greatly from miR-17∼92 overexpression. Indeed, Myc and mir17hg expression is highest in the BCR subtype of DLBCL (Figure 5C-D). Additionally, studies investigating copy number variations in DLBCLs have identified the MYC50 and MIR17HG51 loci as frequently amplified (16% and 12%, respectively). This is consistent with the idea that DLBCLs require either upregulation of MYC or amplification of MIR17HG to sustain the BCR response. Of note, the MIR17HG translocations were exclusively found in GCB-DLBCLs, which on average express less MYC than ABC-DLBCLs. However, Lenz et al51 noted that GCB-DLBCLs with MIR17HG amplification have greater MYC mRNA expression than other GCB-DLBCLs and comparable to that seen in ABC-DLBCLs. Because the MIR17HG amplification occurs independently, miR-17∼92 might stimulate MYC transcription in human DLBCLs. Indeed, activation of BCR signaling has been shown to induce MYC mRNA through the Erk52 and Akt53 pathways. The posttranscriptional loop described herein lends additional credence to the importance of the lymphomagenic MYC-BCR axis.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Dr Jonathan Schug (University of Pennsylvania Functional Genomics Core) for help with interpreting microarray analysis. We thank Dr Ari Melnick (Cornell University Medical College) for providing the HBL-1, U2932, TMD8, and SUDHL4 cell lines, Dr Raju Chaganti (MSKCC) for providing the OCI-LY1 and OCI-LY10 cell lines, Dr Subbarao Bondada (University of Kentucky) for providing the Karpas 422 and Pfeiffer cell lines, and Dr Izidore Lossos (University of Miami) for providing the OCI-Ly19 cell line. We thank all members of PIG (Pathology Immunology Group at Penn) and the entire Thomas-Tikhonenko laboratory for their support and critiques, especially Dr Elena Sotillo-Pineiro for reviewing this manuscript.
This work was supported by National Institutes of Health research project grants R01 (National Cancer Institute) CA122334 (A.T.-T.), T32 postdoctoral training grants CA115299 and CA009140 (J.N.P.), and R01 (National Institute of Allergy and Infectious Diseases) AI060921 (P.J.D. and B.D.F.).
Authorship
Contribution: J.N.P. codesigned experiments, generated and analyzed data, and cowrote the manuscript; A.T.-T. codesigned experiments and cowrote the manuscript; P.J.D. and B.D.F. codesigned, generated, and analyzed data, and cowrote the cytosolic calcium measurement parts of the manuscript; and P.R. and A.J.M. performed bioinformatics analyses.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Andrei Thomas-Tikhonenko, 4056 Colket Translational Research Building, 3501 Civic Center Blvd, Philadelphia, PA 19104-4399; e-mail: andreit@mail.med.upenn.edu.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal