Key Points
Expression of the EBV oncogene LMP1 in primary human germinal center B cells, upregulates DDR1, a receptor tyrosine kinase activated by collagen
Primary HRS cells overexpress DDR1, and its activation significantly increases lymphoma cell survival in vitro
Abstract
The malignant Hodgkin and Reed-Sternberg (HRS) cells of Hodgkin lymphoma are surrounded by a tumor microenvironment that is composed of a variety of cell types, as well as noncellular components such as collagen. Although HRS cells harbor oncogenic Epstein-Barr virus (EBV) in approximately 50% of cases, it is not known if the tumor microenvironment contributes to EBV-driven lymphomagenesis. We show that expression of the EBV-encoded latent membrane protein-1 (LMP1) in primary human germinal center B cells, the presumed progenitors of HRS cells, upregulates discoidin domain receptor 1 (DDR1), a receptor tyrosine kinase activated by collagen. We also show that HRS cells intimately associated with collagen frequently overexpress DDR1 and that short-term exposure to collagen is sufficient to activate DDR1 in Hodgkin lymphoma-derived cell lines. The ectopic expression of DDR1 significantly increased the survival of collagen-treated DG75 Burkitt lymphoma cells, following etoposide treatment. Conversely, knockdown of DDR1 significantly decreased the survival of collagen-treated L428 Hodgkin lymphoma cells in the absence of specific apoptotic stimulus, suggesting that DDR1 also influences baseline survival. Our results identify a hitherto unknown function for collagen in protecting Hodgkin lymphoma cells from apoptosis and suggest an important contribution of the tumor microenvironment in promoting the oncogenic effects of EBV.
Introduction
Hodgkin lymphoma is characterized by the presence of a minority of malignant Hodgkin and Reed-Sternberg (HRS) cells surrounded by a prominent tumor microenvironment. This microenvironment is comprised of different cell populations, including T cells, macrophages, fibroblasts, and other cell types. Interactions between HRS cells and these nonmalignant cells are mediated by various receptor-ligand pairs, including for example CD40-CD40L, CD30-CD30L, and RANK-RANKL which contribute to HRS cell proliferation and survival.1,2 However, the Hodgkin lymphoma microenvironment also contains a collagen-rich extracellular matrix. Although collagen is known to contribute to the pathogenesis of other cancer types, including those of the breast, pancreas, and lung,3-5 it is not known if it also contributes to the malignant phenotype of HRS cells.
In approximately 50% of cases, HRS cells harbor the oncogenic Epstein-Barr virus (EBV). EBV-positive HRS cells express a limited number of viral genes which include the Epstein-Barr nuclear antigen 1 (EBNA1) and the latent membrane proteins (LMP1 and -2).6-10 LMP1 is reported to be an oncogene and can constitutively activate several cell signaling pathways known to be aberrantly expressed in Hodgkin lymphoma, including nuclear factor (NF)-κB, JNK, and phosphatidylinositol 3-kinase.11-15 However, LMP1 is also expressed in the germinal center (GC) B cells of normal healthy EBV carriers where it is thought to provide a CD40-like survival signal.16,17
HRS cells show evidence of somatic hypermutation, indicating that they are derived from cells that have been through a GC reaction. Destructive mutations in immunoglobulin (Ig) genes that prevent Ig gene transcription (so called “crippling” mutations) can be detected in around one-quarter of Hodgkin lymphoma suggesting that HRS cells derive from preapoptotic GC B cells. Almost all Hodgkin lymphoma cases with crippling Ig gene mutations are EBV-positive, suggesting that EBV is necessary to override apoptotic signals in GC B cells harboring deleterious Ig gene mutations.18-20
In this study we have sought evidence, linking collagen in the microenvironment of Hodgkin lymphoma to the induction of anti-apoptotic signals by LMP1 in HRS cells. We have focused our studies on the discoidin domain receptor 1 (DDR1), a receptor tyrosine kinase (RTK) that can be activated by collagen.21
The DDR subfamily consists of two members, DDR1 and DDR2, which are activated by triple helical collagens rather than soluble growth factors and which exist as multiple isoforms as a result of alternative splicing around the juxtamembraneous domain. Binding to collagen occurs through the discoidin domains of DDR1 and is integrin independent.22,23 Following binding to collagen, the autophosphorylation of DDR1 triggers the activation of downstream signaling pathways, including phosphatidylinositol 3-kinase/Akt and NF-κB.24,25 Activation of these pathways by DDR1 induces the expression of proinflammatory mediators, as well as matrix-degrading enzymes and is implicated in the development of various inflammatory and fibrotic disorders.26-28 The overexpression of DDR1 has also been reported in several cancers including those of the breast, brain, and lung.29-31 DDR1 has been shown to contribute to oncogenesis by disrupting normal cell-matrix communications and initiating promigratory and proinvasive programs.32-35
In this study, we show that LMP1, but not its physiological homolog CD40, can induce the expression of DDR1 in primary human GC B cells, the presumed progenitors of HRS cells. We also show that the overexpression of DDR1 protects collagen-treated lymphoma cells from cell death. We provide for the first time, evidence of the importance of the microenvironment in regulating the oncogenic effects of LMP1 in B cells.
Materials and methods
Tissue samples and cell lines
Paraffin tissues of 49 cases of pediatric classical Hodgkin lymphoma were obtained from the Children’s Cancer and Leukaemia Group (CCLG; study number 2007-BS-07) and histology verified on review. They included 34 cases of nodular sclerosis subtype, 9 cases of mixed cellularity, 1 case of lymphocyte-rich classical Hodgkin lymphoma and 5 cases defined as “Hodgkin lymphoma not otherwise specified” (see supplemental Table on the Blood Web site). EBV-positive L591 and EBV-negative L428, L1236, KMH2, L540, HDLM2 Hodgkin lymphoma lines, and the EBV-negative DG75 Burkitt lymphoma line were cultured in RPMI 1640 supplemented with 10% fetal calf serum, 2 mM L-glutamine, and 1% penicillin-streptomycin.
Isolation and treatment of GC B cells
GC B cells were isolated from fresh tonsils by magnetic separation with α-CD10-phycoerythrin (Becton Dickinson) and α-PE microbeads as described previously, and with informed consent in accordance with the Declaration of Helsinki and under local ethics committee approval (ref no. 06/Q2702/50).36 A total of 4 × 106 CD10+ cells were treated with 200 ng/mL soluble CD40L (Autogen Bioclear) or nucleofected with 7 μg pSG5 or pSG5-LMP1 expression vector together with 3 μg of pMACSLNGFR in B-cell solution using program U15 on the Nucleofector I. After 16 hours cultivation, transfected cells were stained with α-low-affinity nerve growth factor receptor-allophycocyanin (LNGFR-APC) (Miltenyi Biotec) and propidium iodide (PI). APC-labeled and PI-negative cells were collected on a MoFlo sorter (Beckman-Coulter). For analysis of DDR1 expression, transfected cells were washed in AutoMACS (Miltenyi Biotec) containing EDTA for gentle permeabilization, then stained with anti-DDR1 antibody and fluorescein isothiocyanate (FITC)-conjugated anti-rabbit antibody, and gated on morphology and PI positivity using the following controls to set up compensation and quadrants: unstained cells, cells positive for CD10+PE, cells positive for NGFR-APC, and cells stained with each antibody alone.
Transfection of cell lines
A total of 3 × 106 L428 or 2 × 106 L591 cells were nucleofected (using L or T solution, respectively) with 2 µg (L591) or 3 µg (L428) of ON-TARGETplus SMARTpool siRNA (Dharmacon; [containing siRNA sequences: 5′-UGGUUACUCUUCAGCGAAA-3′, 5′-GGAGCUACCGGCUGCGUUA-3′, 5′-GCGUGUGUGUGCAGGACGA-3′, and 5′-ACGAGCAGGUCAUCGAGAA-3′]), or with scrambled siRNA as a control. We chose to use the Dharmacon ON-TARGETplus SMARTpool system designed to reduce off-target effects in two ways. Firstly, the sense strand is modified to prevent uptake by RNA-induced silencing complex and favor antisense strand loading. Secondly, the antisense strand is modified to destabilize off-target activity and enhance target specificity.37,38 A total of 2 × 106 DG75 cells were transfected in V nucleofection solution with the pIRES2-EGFP vector (Clontech Laboratories) containing DDR1a or no insert as a control (gift of Dr Simon Johnson, University of Nottingham, UK). Transfection was validated using quantitative reverse-transcription polymerase chain reaction (Q-RT-PCR) and immunoblotting for each experiment.
Cytotoxicity assays
Transfected BL-derived DG75 cells or HL-derived L428 cells were seeded at 1 × 105/mL and 100 µg/mL soluble type I collagen (Sigma-Aldrich) added, and where indicated treated with either 50 µM etoposide or dimethylsulfoxide as control. Cells were stained with Annexin V-FITC Apoptosis Detection kit as recommended by the manufacturer (BD Pharmigen) and with PI before analysis on a flow cytometer.
Q-RT-PCR
Q-RT-PCR was performed with commercially available primers for DDR1 (either Hs00233612_m1 or Hs01058424_g1), intercellular adhesion molecule 1 (ICAM1) (Hs00164932), or TNF alpha-induced protein 3 (TNFAIP3) (Hs00234713). DDR1a and DDR1b were amplified using the following primers: DDR1a- 5′-CCCCAATGGCTCTGCCTA-3′ and 5′-AACAATGTCAGCCTCGGCATA-3′; DDR1b- 5′-GGCCAAACCCACCAACAC-3′ and 5′-AACAATGTCAGCCTCGGCATA-3′); and probe (6-FAM-ACTATATGGAGCCTGAGAAG-MGB). The 2−ΔΔ CT method for quantifying expression relative to the housekeeping control was used which was either B2M or glyceraldehyde-3-phosphate dehydrogenase; normalized values were expressed relative to one sample set to a value of 1. Results were reported as the mean of 3 replicates.
Immunoblotting and immunoprecipitation
Cells were lysed in RIPA buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1 mM EDTA, and 1% Nonidet P-40) with 1 mM sodium vanadate and protease inhibitors (Complete Protease Inhibitor Cocktail Tablets, Roche Diagnostics). Bio-Rad DC Protein Assay Kit quantified the protein. Laemmli sample buffer was added before SDS-PAGE. Membranes were incubated at 4°C overnight with primary antibody diluted in 5% nonfat milk powder in TBS-Tween (Sigma-Aldrich). Antibodies used were directed to DDR1 (C-20; 1:250, Santa Cruz Biotechnology), pan-phospho-tyrosine (4G10; 1:1000, Millipore), β-actin (c-2; 1:1000, Santa Cruz Biotechnology), and MCM-7 (DCS-141; 1:2000, Sigma Aldrich), and both were used as a loading control for immunoblotting. After a TBS-Tween-20 (0.1%) wash, blots were incubated for 1 hour with appropriate secondary HRP-conjugated goat anti-mouse IgG or anti-rabbit (Dako). Detection was with enhanced chemiluminescence (GE Healthcare).
Immunoprecipitation was performed using 50% Protein G agarose bead slurry (Millipore). Five-hundred micrograms of protein was incubated with 2 µg of primary DDR1 antibody or a duplicate control lysate with an irrelevant rabbit immunoglobulin (instead of DDR1 antibody) as a negative control. Lysates were precleared by adding 50 µL of agarose bead-RIPA slurry, and then incubated with antibody overnight. One-hundred microliters of 50% agarose bead slurry was added to each sample and incubated for 2 hours on a rotating wheel at 4°C. After the final spin, the beads were re-suspended in SDS buffer, boiled for 5 minutes at 95°C to elute immune complexes, which were subjected to immunoblotting.
Tissue staining
Tissue sections were stained using immunohistochemistry after low-temperature antigen retrieval as previously described.39 Primary antibody diluted in PBS-Tween-20 (0.1%) was applied for 1 hour. Antibodies used were directed to LMP1 (CS1–4; 1:25, Dako) and DDR1 (C-20; 1:80, Santa Cruz Biotechnology). Detection was performed with HRP-conjugated goat anti-rabbit or goat anti-mouse secondary IgG (Dako) for 30 minutes, followed by visualization in DAB. Cases were recorded as DDR1-positive if ≥10% of HRS cells were positive; positive cases were further subdivided into 4 categories on the basis of the percentage of positive HRS cells: 1 = 10% to 25%; 2 = 26% to 50%; 3 = 51% to 75%; and 4 = >75%. The intensity of staining in HRS cells was also recorded compared with blood vessels which served as an internal positive control: 1 = staining in HRS cells weaker than blood vessels in the same tissue section; 2 = staining in HRS cells of same intensity as blood vessels; and 3 = staining stronger than in blood vessels. For van Gieson’s stain, sections were treated with 4% iron alum, washed in tap water, stained in Mayer’s hematoxylin, and then van Gieson’s solution for 10 minutes.
Polymerase chain reaction (PCR) sequencing
RNA was harvested from cell lines using Qiagen RNeasy minikit as per manufacturer’s instructions. Extracted RNA was reverse transcribed to complementary DNA (cDNA) using SuperScript III first-strand synthesis system (Invitrogen). cDNA was sequenced using BigDye sequencing kit (Applied Biosystems). Ten nanograms of either directly purified or gel extracted PCR product was added to the sequencing mix which consisted of 1 µL of BigDye, 3.5 µL of 5X buffer, 1 µL of either forward or reverse sequencing primer (10 pmol/µL). The following primers were used; pair 1: forward 5′-CCTTAGGCCCGAGGGATC-3′ reverse 5′-TGCCTGAGATCACCTCCTGA-3′; pair 2: forward 5′-AGCTACCGGCTGCGTTACTC-3′ reverse 5′-ACGCCGGAAGCGACATTCCA-3′; pair 3: forward 5′-CCAGGCTATGCAGGTCCACT-3′ reverse 5′-AGCACCTGGGCGGTTGTTGA-3′; pair 4: forward 5′-ACGGAGGGTGTTGGAAGAGG-3′ reverse 5′-GATCTTGAGGGCTGTCGACC-3′; pair 5 forward 5′-CTCGACTCCGCTTCAAGGAG-3′ reverse 5′-CAACTAGGCAGTTCCGCGTG-3′; pair 6 forward 5′-CAATGCTGCTGCATGATGTGGCAG-3′ reverse 5′-TGGCTTCCCCTGGATCGCTC-3′, and juxtamembraneous domain primer forward 5′-GAGCTGACGGTTCACCTCTC-3′ reverse 5′-AGCAGCATTGGGTAGCTGAT-3′. Samples were denatured at 96°C for 1 minute followed by 25 cycles of rapid thermal ramp to 96°C for 10 seconds, 50°C for 5 seconds, and 60°C for 4 minutes. After PCR amplification, the cDNA was precipitated by the addition of 96% ethanol with EDTA (0.25M). The cDNA was pelleted by centrifugation at 16 000 rcf for 20 minutes, and then washed twice with 70% ethanol. Samples were air-dried for 15 minutes and resuspended in 10 µL Hi-Di formamide, then subjected to automated sequence analysis.
Statistical analysis
Data points were reported as experimental averages and error bars represented standard deviation. Comparisons between experimental groups used the 2-sided Student t test, with P < .05 considered statistically significant.
Results
Upregulation of DDR1 following the expression of LMP1 in primary human GC B cells
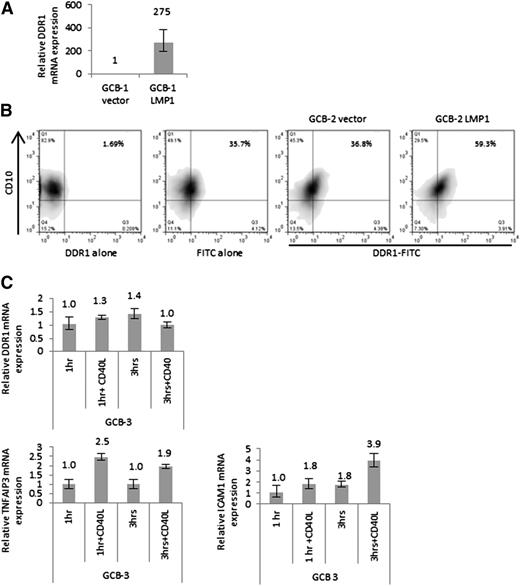
We first investigated the influence of LMP1 on DDR1 expression in primary human GC B cells, the presumed progenitors of HRS cells. CD10-expressing GC B cells were isolated from fresh tonsils and co-transfected with either LMP1 expression vector or empty vector, together with a vector encoding a truncated NGFR gene. Transfected cells were enriched by fluorescence-activated cell (FACS) sorting of NGFR-expressing cells. We confirmed the ability of LMP1 to regulate known target genes in isolated GC B cells, including the NF-κB target gene, ICAM1 (supplemental Figure 1). Q-RT-PCR analysis then revealed that while DDR1 expression was undetectable (Ct value >40) in 11 of 12 vector-transfected GC B cell samples, a robust increase in DDR1 expression was observed in all 12 LMP1-transfected GC B cell samples (mean Ct value = 30.73) (Figure 1A). In a separate experiment, we used flow cytometry to measure the proportion of GC B cells expressing surface DDR1 protein. Figure 1B shows that control transfected cells did not express detectable DDR1 surface protein, whereas DDR1 protein expression was detectable in 22.5% of LMP1-transfected cells which is consistent with the relatively inefficient expression of transgenes we have previously observed in transfected and sorted primary GC B cells.40 Next, we treated isolated GC B cells with CD40L to activate the cellular CD40 receptor, the physiological homolog of LMP1. We investigated the effects of CD40 ligation on DDR1 expression at both 1 and 3 hours, time points that our preliminary experiments had revealed consistently upregulated known CD40 targets including TNFAIP3 and ICAM1. Figure 1C shows while CD40 ligation led to a robust increase in the expression of TNFAIP3 and ICAM1, it did not induce DDR1 mRNA expression.
Upregulation of DDR1 following the expression of LMP1 in primary human GC B cells. (A) Q-RT-PCR analysis of DDR1 mRNA levels following the expression of LMP1 in primary human GC B cells compared with cells transfected with an empty vector as a control. LMP1 expression was followed by the robust upregulation of DDR1 mRNA. Shown is 1 of 4 experiments performed in triplicate on different donor GC B cells. Normalized values were expressed relative to empty vector set to a value of 1. (B) FACS analysis of DDR1 expression in GC B cells transfected with LMP1 or an empty vector. Left panels show cells stained with only primary DDR1 antibody or rabbit secondary antibody and used as negative controls. Right panels show staining of cells with both antibodies following transfection with empty vector or LMP1. Cells transfected with empty vector show no change in DDR1 expression compared with cells stained with rabbit secondary antibody alone, indicating that DDR1 is not expressed in empty vector transfected GC B cells. LMP1 expression led to detectable DDR1 protein expression in almost a quarter of GC B cells. (C) DDR1 mRNA expression following the treatment of primary human GC B cells with CD40L (200 ng/mL) for 1 and 3 hours in comparison with unstimulated GC B cells for the same corresponding time points. Treatment with CD40L did not induce the upregulation of DDR1. In contrast, there was a robust increase in the expression of the known CD40 targets TNFAIP3 and ICAM1. Data shown is representative of 3 replicates with normalized values expressed relative to control set to a value of 1.
Upregulation of DDR1 following the expression of LMP1 in primary human GC B cells. (A) Q-RT-PCR analysis of DDR1 mRNA levels following the expression of LMP1 in primary human GC B cells compared with cells transfected with an empty vector as a control. LMP1 expression was followed by the robust upregulation of DDR1 mRNA. Shown is 1 of 4 experiments performed in triplicate on different donor GC B cells. Normalized values were expressed relative to empty vector set to a value of 1. (B) FACS analysis of DDR1 expression in GC B cells transfected with LMP1 or an empty vector. Left panels show cells stained with only primary DDR1 antibody or rabbit secondary antibody and used as negative controls. Right panels show staining of cells with both antibodies following transfection with empty vector or LMP1. Cells transfected with empty vector show no change in DDR1 expression compared with cells stained with rabbit secondary antibody alone, indicating that DDR1 is not expressed in empty vector transfected GC B cells. LMP1 expression led to detectable DDR1 protein expression in almost a quarter of GC B cells. (C) DDR1 mRNA expression following the treatment of primary human GC B cells with CD40L (200 ng/mL) for 1 and 3 hours in comparison with unstimulated GC B cells for the same corresponding time points. Treatment with CD40L did not induce the upregulation of DDR1. In contrast, there was a robust increase in the expression of the known CD40 targets TNFAIP3 and ICAM1. Data shown is representative of 3 replicates with normalized values expressed relative to control set to a value of 1.
HRS cells overexpressing DDR1 are intimately associated with collagen
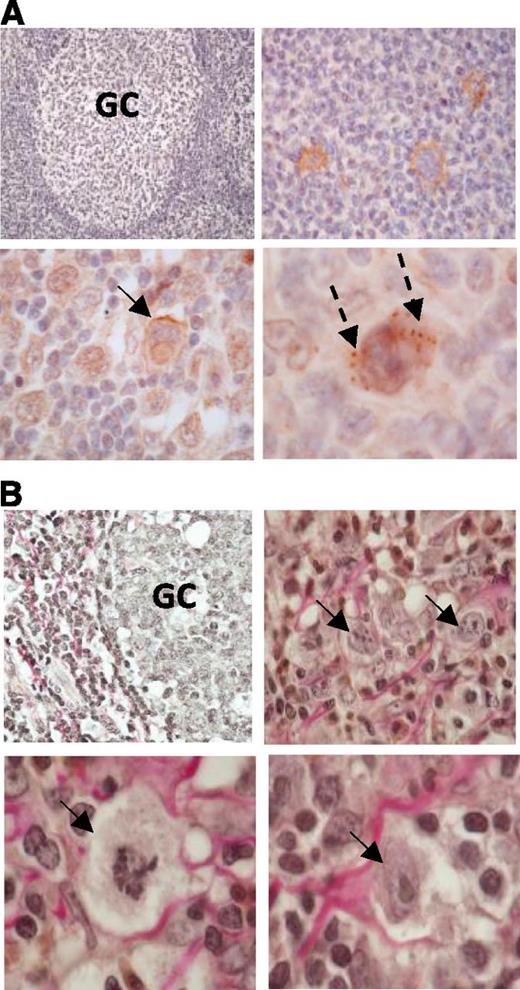
Having established that LMP1 can upregulate DDR1 in human GC B cells, we next studied DDR1 expression in primary tissues. LMP1 staining was used to classify 28 of 49 cases as EBV-positive. Immunohistochemistry revealed that while DDR1 was not expressed in the normal GC B cells of reactive tonsils, it was expressed in HRS cells in 22 of 28 (78.6%) cases of EBV-positive Hodgkin lymphoma, as well as in 13 of 21 (61.9%) cases of EBV-negative Hodgkin lymphoma. There were no significant differences in DDR1 expression (either in intensity of staining or proportion of positive cells staining) and histological subtype or EBV status (supplemental Table). In DDR1-positive cases, expression was localized to the cell membrane and as focal aggregates in the cytoplasm (Figure 2A). We observed collagen deposition in all cases, including both DDR1-positive and DDR1-negative samples. Furthermore, in all cases, collagen was found to be closely associated with HRS cells; although the extent to which this was observed varied from case to case and was dependent upon the frequency of HRS cells, as well as on the amount of collagen present within the tumor (Figure 2B).
DDR1 overexpressing primary HRS cells intimately associate with collagen. (A) Immunohistochemistry stain for DDR1 expression in Hodgkin lymphoma. DDR1 was either absent or only weakly expressed in normal GC cells of a nonmalignant tonsil (left, upper panel). Original magnification ×200. Remaining panels show representative examples of DDR1 expression in primary HRS cells. DDR1 expression in HRS cells was localized to the cell membrane (arrows) or as focal aggregates in the cytoplasm (broken arrows). Original magnifications: right upper panel ×400; left lower panel ×600 oil immersion lens; right lower panel ×1000 oil immersion lens. (B) van Gieson’s stain of a nonmalignant tonsil shows limited collagen deposition within the GC (left, upper panel; original magnification ×400) and by comparison DDR1-positive HRS cells (arrows) are frequently intimately associated with collagen (red staining). Original magnifications: right upper panel ×600 oil immersion lens; lower panels ×1000 oil immersion lens. GC, germinal center.
DDR1 overexpressing primary HRS cells intimately associate with collagen. (A) Immunohistochemistry stain for DDR1 expression in Hodgkin lymphoma. DDR1 was either absent or only weakly expressed in normal GC cells of a nonmalignant tonsil (left, upper panel). Original magnification ×200. Remaining panels show representative examples of DDR1 expression in primary HRS cells. DDR1 expression in HRS cells was localized to the cell membrane (arrows) or as focal aggregates in the cytoplasm (broken arrows). Original magnifications: right upper panel ×400; left lower panel ×600 oil immersion lens; right lower panel ×1000 oil immersion lens. (B) van Gieson’s stain of a nonmalignant tonsil shows limited collagen deposition within the GC (left, upper panel; original magnification ×400) and by comparison DDR1-positive HRS cells (arrows) are frequently intimately associated with collagen (red staining). Original magnifications: right upper panel ×600 oil immersion lens; lower panels ×1000 oil immersion lens. GC, germinal center.
DDR1 is overexpressed in Hodgkin lymphoma cell lines
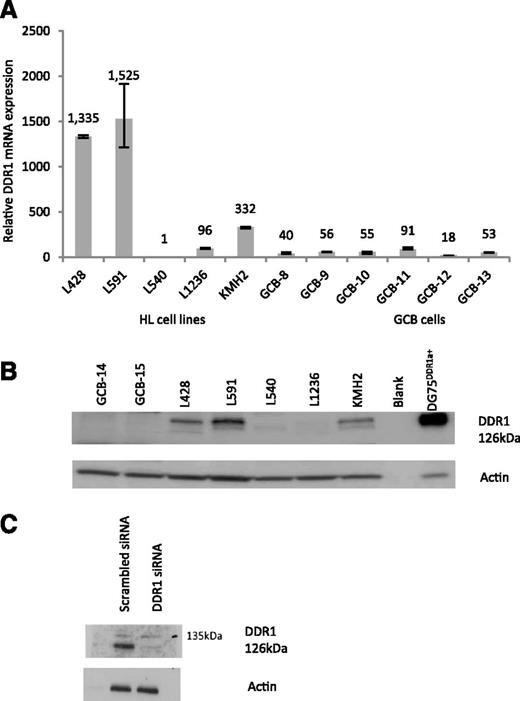
We next explored the expression of DDR1 in Hodgkin lymphoma-derived cell lines. To do this, we amplified an mRNA sequence within the discoidin domain of DDR1 present in all known isoforms. Figure 3A shows that compared with GC B cells isolated from tonsils, DDR1 mRNA levels were higher in 4 of 5 Hodgkin lymphoma-derived cell lines. We then studied DDR1 protein expression in the Hodgkin lymphoma cell lines using immunoblotting. Figure 3B shows the presence of a 126KDa protein at the expected molecular weight of full length DDR1 and which matched the DDR1 mRNA data. To confirm the specificity of this antibody, we knocked down DDR1 in both L428 and L591 cell lines using a pool of siRNAs, which recognize all known isoforms. Figure 3C shows that the 126KDa band was almost entirely depleted following inhibition of DDR1 by RNA interference. The observation that DDR1 expression is also detectable in some EBV-negative HL cell lines in addition to the EBV-positive L591 line is consistent with the detection of DDR1 in EBV-negative primary HRS cells described above.
DDR1 is overexpressed in Hodgkin lymphoma cell lines. (A) A Q-RT-PCR assay which amplifies a sequence within the discoidin domain of DDR1 present in all known isoforms revealed that compared with GC B cells isolated from tonsils, DDR1 mRNA levels were higher in 4 of 5 Hodgkin lymphoma-derived cell lines. Normalized values were expressed relative to L540 set to a value of 1. (B) Immunoblotting for DDR1 in the same panel of cell lines showed the presence of a 126KDa protein at the expected weight of full length DDR1 and matched the levels of total DDR1 mRNA. Two representative GC B samples were DDR1 negative. DG75 cells transfected with DDR1 expression vector were used as a positive control. (C) Treatment of L591 cells with a pool of siRNAs, which recognizes all DDR1 isoforms, showed that the 126KDa band was decreased following inhibition by DDR1 RNA interference.
DDR1 is overexpressed in Hodgkin lymphoma cell lines. (A) A Q-RT-PCR assay which amplifies a sequence within the discoidin domain of DDR1 present in all known isoforms revealed that compared with GC B cells isolated from tonsils, DDR1 mRNA levels were higher in 4 of 5 Hodgkin lymphoma-derived cell lines. Normalized values were expressed relative to L540 set to a value of 1. (B) Immunoblotting for DDR1 in the same panel of cell lines showed the presence of a 126KDa protein at the expected weight of full length DDR1 and matched the levels of total DDR1 mRNA. Two representative GC B samples were DDR1 negative. DG75 cells transfected with DDR1 expression vector were used as a positive control. (C) Treatment of L591 cells with a pool of siRNAs, which recognizes all DDR1 isoforms, showed that the 126KDa band was decreased following inhibition by DDR1 RNA interference.
DDR1a is the major isoform expressed in Hodgkin lymphoma cell lines and is regulated by LMP1 in GC B cells
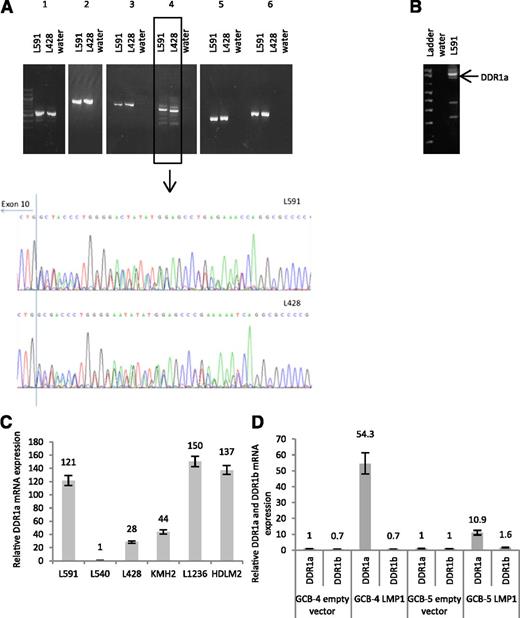
We then used RT-PCR to amplify transcripts spanning the entire coding region of DDR1 from two of the cell lines with high levels of DDR1 expression (L591 and L428). Amplification of the juxtamembraneous domain revealed multiple bands. Sequencing of this PCR product revealed two overlapping sequences consistent with the presence of the DDR1a and the DDR1d isoforms (Figure 4A). We then used a second set of primers optimized to amplify the juxtamembraneous domain, subjected the amplified product to gel electrophoresis and sequenced individual bands from the gel. The strongest band present was found to correspond to the kinase active DDR1a isoform (Figure 4B). Of the other two kinase active forms, DDR1b was also present but at much lower levels and DDR1c was undetectable. Transcripts for the kinase inactive form, DDR1d, as well as those encoding several potential novel isoforms were also present (supplemental Figure 3). We confirmed the expression of DDR1a in 5 of 6 Hodgkin lymphoma cell lines by Q-RT-PCR (Figure 4C).
DDR1a is the major isoform expressed in Hodgkin lymphoma cell lines and is regulated by LMP1 in GC B cells. (A) PCR of cDNA from 2 representative Hodgkin lymphoma cell lines, L428 and L591 revealed the presence of a single band for 5 of 6 primer pairs spanning the coding region of DDR1. Multiple bands were detected by primer pair 4 spanning the entire juxtamembraneous domain. Sequencing of the PCR product of primer pair 4 revealed two overlapping sequences consistent with the presence of the DDR1a isoform and the DDR1d isoform. (B) To confirm these observations, a second set of primers was optimized to amplify only the juxtamembraneous domain. The amplified product was subjected to gel electrophoresis and individual bands sequenced from the gel. The strongest band present was found to correspond to the kinase-active DDR1a isoform. Transcripts for the kinase inactive form, DDR1d, as well as those encoding several potential novel isoforms were also present (supplemental Figure 3). Shown here are data from L591 cells, although similar results were obtained for L428 cells. (C) An isoform-specific Q-RT-PCR assay confirmed the expression of the DDR1a isoform in Hodgkin lymphoma cell lines. Normalized values were expressed relative to L540 set to a value of 1. (D) Q-RT-PCR analysis revealed that LMP1 increased the expression of DDR1a, but not the other kinase-active isoform, DDR1b, in human GC B cells. Normalized values were expressed relative to empty vector set to a value of 1.
DDR1a is the major isoform expressed in Hodgkin lymphoma cell lines and is regulated by LMP1 in GC B cells. (A) PCR of cDNA from 2 representative Hodgkin lymphoma cell lines, L428 and L591 revealed the presence of a single band for 5 of 6 primer pairs spanning the coding region of DDR1. Multiple bands were detected by primer pair 4 spanning the entire juxtamembraneous domain. Sequencing of the PCR product of primer pair 4 revealed two overlapping sequences consistent with the presence of the DDR1a isoform and the DDR1d isoform. (B) To confirm these observations, a second set of primers was optimized to amplify only the juxtamembraneous domain. The amplified product was subjected to gel electrophoresis and individual bands sequenced from the gel. The strongest band present was found to correspond to the kinase-active DDR1a isoform. Transcripts for the kinase inactive form, DDR1d, as well as those encoding several potential novel isoforms were also present (supplemental Figure 3). Shown here are data from L591 cells, although similar results were obtained for L428 cells. (C) An isoform-specific Q-RT-PCR assay confirmed the expression of the DDR1a isoform in Hodgkin lymphoma cell lines. Normalized values were expressed relative to L540 set to a value of 1. (D) Q-RT-PCR analysis revealed that LMP1 increased the expression of DDR1a, but not the other kinase-active isoform, DDR1b, in human GC B cells. Normalized values were expressed relative to empty vector set to a value of 1.
Having shown that DDR1a is the predominant isoform in Hodgkin lymphoma cells, we then used Q-RT-PCR assays to measure its expression in LMP1-expressing primary human GC B cells. Figure 4D shows that LMP1 increased the expression of DDR1a in two separate donor GC B cells. We conclude that DDR1a is the predominant kinase-active isoform present in Hodgkin lymphoma cells and can also be induced by LMP1 expression in GC B cells.
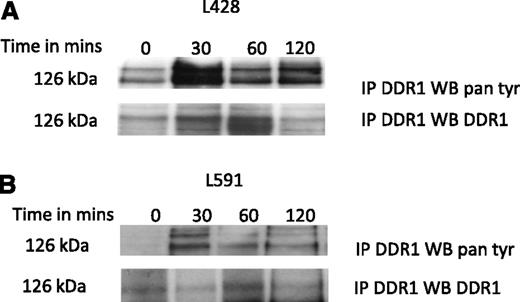
Collagen induces the phosphorylation and activation of DDR1 in HRS cells
We next studied if collagen can activate DDR1 in HRS cells. To do this, L428 and L591 cells were serum starved and then re-suspended in media supplemented with collagen. The phosphorylation of DDR1 was measured using immunoprecipitation with a DDR1 antibody, followed by immunoblotting with a pan-phospho-tyrosine antibody. We found that the addition of collagen induced the robust phosphorylation of DDR1 in both L428 and L591 cells (Figure 5).
Collagen induces the phosphorylation and activation of DDR1 in HRS cells. (A) Detection of DDR1 phosphorylation in L428 cells, and (B) L591 cells with anti-phosphotyrosine antibody (4G10). The addition of collagen induced the robust phosphorylation of DDR1 in cells peaking at 30 minutes. Cell lysates were immunoprecipitated with anti-DDR1, and then probed with 4G10 or reprobed with anti-DDR1 antibody.
Collagen induces the phosphorylation and activation of DDR1 in HRS cells. (A) Detection of DDR1 phosphorylation in L428 cells, and (B) L591 cells with anti-phosphotyrosine antibody (4G10). The addition of collagen induced the robust phosphorylation of DDR1 in cells peaking at 30 minutes. Cell lysates were immunoprecipitated with anti-DDR1, and then probed with 4G10 or reprobed with anti-DDR1 antibody.
Collagen-induced activation of endogenous DDR1 confers resistance to cell death in L428 Hodgkin lymphoma cells
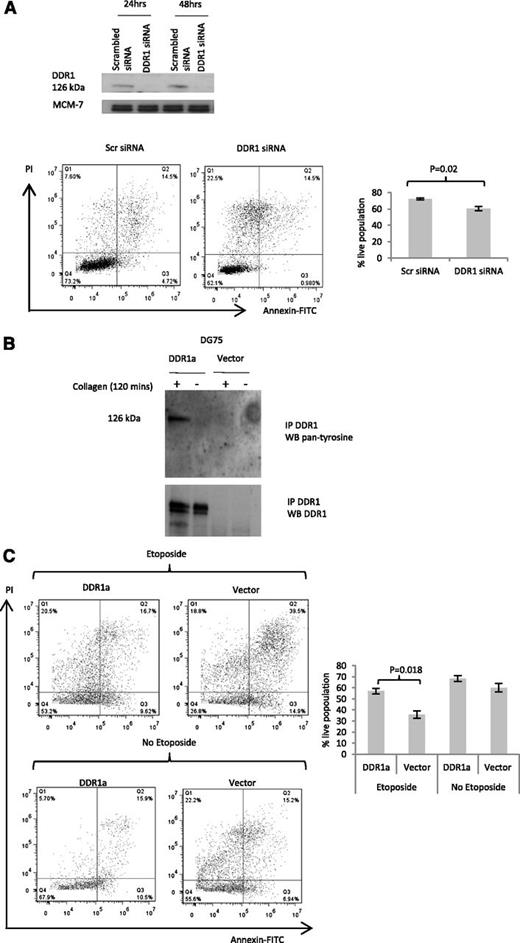
To explore if endogenously expressed DDR1 could influence cell death in lymphoma cells, we inhibited DDR1 expression by RNA interference in collagen-treated L428 Hodgkin lymphoma cells. After confirming the successful inhibition by RNA interference of DDR1 in these cells, we used flow cytometry to measure apoptosis 48 hours after treatment with siRNA. We found that the inhibition of endogenous DDR1 significantly and reproducibly increased baseline levels of cell death in collagen-treated L428 cells (Figure 6A). We conclude that the overexpression of DDR1 can protect collagen-treated lymphoma cells from cell death.
Collagen-induced activation of DDR1 confers resistance to cell death. (A) siRNA-mediated depletion of DDR1 protein shown by immunoblotting (upper panel) increased cell death in collagen-treated L428 Hodgkin lymphoma cells (lower panels). The differences between live populations, defined as Annexin-negative and PI-negative, were significant across 3 experimental replicates (P = .02). (B) Collagen induced the phosphorylation of ectopically expressed DDR1 in DG75 cells. As expected, no phosphorylation of DDR1 was observed following the cultivation of these cells in the absence of collagen. (C) Representative flow cytometry data showed that the addition of collagen to DG75 cells overexpressing DDR1 protected them from etoposide-induced death. The lower panel shows that the differences between live populations, defined as Annexin-negative and PI-negative, were significant across 3 experimental replicates (P = .018).
Collagen-induced activation of DDR1 confers resistance to cell death. (A) siRNA-mediated depletion of DDR1 protein shown by immunoblotting (upper panel) increased cell death in collagen-treated L428 Hodgkin lymphoma cells (lower panels). The differences between live populations, defined as Annexin-negative and PI-negative, were significant across 3 experimental replicates (P = .02). (B) Collagen induced the phosphorylation of ectopically expressed DDR1 in DG75 cells. As expected, no phosphorylation of DDR1 was observed following the cultivation of these cells in the absence of collagen. (C) Representative flow cytometry data showed that the addition of collagen to DG75 cells overexpressing DDR1 protected them from etoposide-induced death. The lower panel shows that the differences between live populations, defined as Annexin-negative and PI-negative, were significant across 3 experimental replicates (P = .018).
Collagen protects DG75 lymphoma cells from etoposide-induced cell death
Finally, we explored the possibility that DDR1 might also protect lymphoma cells from chemotherapy induced death. To do this, we overexpressed DDR1a in the BL-derived cell line, DG75. We found that collagen induced the phosphorylation of ectopically expressed DDR1 in DG75 cells (Figure 6B). As expected, no phosphorylation of DDR1 was observed following the cultivation of these cells in the absence of collagen. Figure 6C shows that the overexpression of DDR1 significantly decreased etoposide-induced cell death in collagen-treated DG75 cells as measured by flow cytometry staining with Annexin V-FITC. DDR1 overexpression had no significant effect on cell death in DG75 cells in the absence of etoposide. This is perhaps not unexpected given that DG75 cells do not normally express DDR1 and presumably do not require this receptor under normal growth conditions. These results suggest that DDR1 expression in lymphoma cells can confer resistance to chemotherapeutic drugs.
Discussion
DDR1 is an RTK which has previously been implicated in the pathogenesis of several common cancers including those of the breast, lung, and brain.29-31 In this study, we have shown that the EBV gene, LMP1, can induce the expression of DDR1 in primary human GC B cells, the presumed progenitors of HRS cells. Furthermore, we have shown that DDR1 is overexpressed in HRS cells in the majority of cases of Hodgkin lymphoma. We also showed that DDR1 could be activated by its ligand, collagen, in Hodgkin lymphoma-derived cell lines. Importantly, although we observed as expected, the extensive deposition of collagen in the tissues of patients with nodular sclerosis HL, we also found that collagen was present in all cases of mixed cellularity and lymphocyte rich classical HL.
The activation of DDR1 by collagen appears to be important for the HRS cell phenotype since we showed that the depletion of DDR1 in collagen-treated L428 HL cells increased cell death in the absence of a specific apoptotic stimulus. Furthermore, etoposide-induced cell death decreased in collagen-treated DG75 BL cells overexpressing DDR1; observations that are consistent with a previous study showing that DDR1 can confer resistance to chemotherapy in breast cancer.24 Our observations not only point to an important function for collagen in mediating the DDR1-dependent survival of lymphoma cells, but also suggest that strategies to deplete or inhibit DDR1 in tumor cells might enhance chemotherapy responses in lymphoma patients.
Although these experiments are able to reveal the impact of DDR1 on the phenotype of established lymphoma cells, they are unable to provide any information on the contribution of DDR1 to lymphoma development, a process that is already complete in cell lines. These functional cell line models are also limited in so far as they cannot reproduce the complexity of the Hodgkin lymphoma tumor microenvironment. Therefore, we cannot exclude the possibility that the effects of the collagen-DDR1 interaction will be different in the presence of this complex microenvironment.
In almost all individuals, primary infection with EBV is followed by asymptomatic persistence of the virus in memory B cells where EBV gene expression is probably completely silenced.41 The mechanism by which EBV gains access to the memory B cell pool is controversial; the most widely held model proposes that EBV-infected memory B cells are generated via a GC reaction in which LMP1 provides a surrogate CD40 signal and the other latent membrane protein, LMP2, a BCR-like stimulus.42-44 Our previous observations that primary human GC B cells are permissive for the expression of both LMP1 and LMP2 have provided some support for this model.36,45 However, in rare instances, EBV can also give rise to various forms of lymphoma, most of which are derived from GC B cells. The present study goes some way to explaining this apparent paradox by suggesting that the oncogenic activities of LMP1 in GC B cells may, at least in part, be dependent on cues from the microenvironment. It is interesting to speculate whether such signals from the microenvironment may be initiating factors in the pathogenesis of EBV-driven lymphomas.
EBV-positive and EBV-negative HRS cells show deregulated activation of numerous signaling pathways and transcription factors that have been shown to have essential roles in the pathophysiology of these cells.1 Among these, CD40 has been shown to be important for the proliferation and survival of HRS cells; observations which have suggested a pathogenic role for CD40L-expressing T cells in the HL microenvironment.46-48 However, we did not observe the upregulation of DDR1 following the stimulation of the CD40 receptor on GC B cells, suggesting that LMP1 mediates the overexpression of DDR1 through signaling functions which are different to those activated by CD40. Our observations are consistent with previous reports of the existence of potentially oncogenic functions of LMP1 in B cells that do not overlap those of CD40.49,50 Therefore, the DDR1 pathway described here is likely to be an alternative or an addition to CD40 signaling in the pathogenesis of Hodgkin lymphoma. Furthermore, the observation that DDR1 overexpression is not restricted to EBV-positive cases is consistent with the existence of nonviral mechanisms capable of promoting DDR1 expression in the absence of EBV.
DDR1 is known to exist in multiple isoforms generated by differential splicing around the juxtamembraneous domain.21 We showed that LMP1 upregulated the DDR1a isoform which was also the major isoform expressed in Hodgkin lymphoma cell lines. DDR1a is one of the three known kinase active isoforms. DDR1a has already been shown to be expressed in leukocytes and to promote cell adhesion, migration, and macrophage differentiation.34 cDNA sequencing also revealed that Hodgkin lymphoma cell lines express DDR1d, a kinase inactive isoform, the function of which is poorly understood, as well as transcripts potentially coding for novel DDR1 isoforms. It remains to be established if DDR1d or these novel transcripts are involved in the pathogenesis of Hodgkin lymphoma.
Conventional treatments that include chemotherapy and radiotherapy have dramatically improved survival for many Hodgkin lymphoma patients. However, clinical outcomes for a subgroup of Hodgkin lymphoma patients who have refractory disease or who relapse early is particularly poor.51 New treatments are required for this subset of patients with poorer outcomes, as well as to reduce the acute toxicity and long-term sequelae for those patients who will be cured. It has recently been shown that several RTK inhibitors including dasatinib and nilotinib, can suppress DDR1 activation.52 We believe our data support the use of RTK inhibitors in the treatment of patients with Hodgkin lymphoma, and in particular, the possibility that the suppression of DDR1 might enhance chemotherapy response.
The online version of this article contains a data supplement.
There is an Inside Blood commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by Leukaemia and Lymphoma Research (United Kingdom). The authors wish to thank the Children’s Cancer and Leukaemia Group Tissue Bank (United Kingdom) for providing patient samples, and the Experimental Cancer Medicine Centre (Birmingham, United Kingdom) for infrastructure funding.
Authorship
Contribution: F.Z.C. designed and performed the research, analyzed the data, created the figures, and wrote the manuscript; M.V. performed GC B transfection, planned and supervised experimental work, and reviewed the manuscript; S.B. supervised experimental work and assisted with sequencing data; E.N. performed CD40 stimulation; M.-A.B. analyzed immunohistochemistry data; P.R.K. contributed to the design of the study, supervised the research, and helped write the manuscript; and P.G.M. planned and implemented the research, supervised the experimental work and data analyses, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Fathima Zumla Cader, School of Cancer Sciences, University of Birmingham, Vincent Drive, Birmingham, B15 2TT, United Kingdom; e-mail: f.z.cader@bham.ac.uk.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal