Key Points
STAT3 activity is necessary for TEL-AML1 leukemia maintenance.
TEL-AML1 induces STAT3 activation via RAC1 and leading to induction of MYC expression.
Abstract
The t(12;21)(p13;q22) translocation is the most common chromosomal abnormality in pediatric leukemia. Although this rearrangement involves 2 well-characterized transcription factors, TEL and AML1, the molecular pathways affected by the result of the translocation remain largely unknown. Also in light of recent studies showing genetic and functional heterogeneities in cells responsible for cancer clone maintenance and propagation, targeting a single common deregulated pathway may be critical for the success of novel therapies. Here we describe a novel signaling pathway that is essential for oncogenic addiction in TEL-AML1 leukemia. Our data indicate a direct role for TEL-AML1, via increasing the activity of RAC1, in regulating the phosphorylation of signal transducer and activator of transcription 3 (STAT3), which results in transcriptional induction of MYC. We demonstrate that human leukemic cell lines carrying this translocation are highly sensitive to treatment with S3I-201, a specific STAT3 inhibitor, and, more interestingly, that primary human leukemic samples are also responsive to the drug in the same concentration range. Thus, STAT3 inhibition represents a promising possible therapeutic strategy for the treatment of TEL-AML1 leukemia.
Introduction
Childhood acute lymphoblastic leukemia (ALL) is a malignant disorder of lymphoid progenitor cells with a peak prevalence between the ages of 2 and 5 years, and is the most commonly occurring cancer in children.1 The single most frequent chromosomal abnormality associated with childhood ALL is the t(12;21)(p13;q22) rearrangement that creates a fusion gene including the non-DNA binding region of TEL (ETV6) and almost the entire coding region of AML1 (RUNX1).2,3 Clinically, TEL-AML1 positive patients have good prognoses.4 However, relapses are reported to occur in 10% to 20% of children, additional genetic lesions affecting prognosis,5 and the long-term side effects of chemotherapy remain a cause for concern.6
The TEL-AML1 fusion causes perturbations in normal hematopoiesis, enhancing self-renewal of B-cell progenitors and altering hematopoietic stem cell homeostasis.7-10 However, TEL-AML1 has been shown to be capable of generating pre-leukemic clones11 and of being predisposed to hematologic disease,10,12,13 but on its own it does not appear to be sufficient to induce overt leukemia.14 This is consistent with clinical data in which the translocation is already detectable years before the clinical disease, most often in utero,15 and data suggesting that only 1 of 100 newborns with detectable TEL-AML1 transcripts are destined to develop ALL.16 However, alternative models suggesting that the initiating event is as rare as the disease itself, implying that a high proportion (perhaps 100%) of babies born with the TEL-AML1 fusion are likely to develop leukemia,17 would indicate a more central role for this fusion in leukemogenesis. Interestingly, recent studies have shown that ALL is similar to other types of cancer, as it is characterized by the presence of heterogeneous clones with different genetic alterations that are responsible for maintenance and propagation of the disease.18,19 However, in case of TEL-AML1 leukemia, the only alteration shared by all the clones is the fusion.18
Analysis of the function of TEL-AML1 in human leukemic cell lines has demonstrated that its continued expression is required for survival, proliferation, and leukemia engraftment.20-22 However, there is only limited information on the signaling pathways deregulated by the fusion in these cells.22-25 To address this question, we have used small hairpin (sh)RNA-mediated silencing to demonstrate that TEL-AML1 is responsible for inducing signal transducer and activator of transcription 3 (STAT3) activation in human t(12;21) leukemia cells. This activation is mediated via RAC1, results in transcriptional induction of MYC and is critical for the survival of TEL-AML1+ cell lines and TEL-AML1+ primary cells. STAT3 has been shown to be consistently deregulated in several cancers, including other subtypes of ALL, resulting in a drive to develop new more effective inhibitors for cancer therapy.26,27 Our study suggests that t(12;21) ALL would greatly benefit from such novel therapies based on STAT3 inhibition.
Material and methods
Animals
All mice were maintained in the animal facilities of the UCL Institute of Child Health, and experiments were performed according to the United Kingdom Home Office regulations. C57BL/6J mice were purchased from Harlan Laboratories. NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ (NSG) (5- to 10-week-old) mice were used as recipients for transplantation of human cell lines.
Cell culture and proliferation assay
Human leukemic cells (supplemental Materials and methods) were cultured with drugs or dimethylsulfoxide, at the same concentration as that used in the highest drug treatment, for 96 hours. CellTiter 96 Aqueous One Solution Reagent (Promega) was added to each well and cell viability was determined by measuring the absorbance at 490 nm using a 550 BioRad plate-reader (Bio-Rad). S3I-201, Nicotinoyl Hydrazone28 (STAT5in), Stattic, and Inhibitor VII were purchased from Merck Millipore. NSC2376629 was purchased from Tocris Bioscience. Mouse fetal liver hematopoietic progenitor cells (HPC) were isolated, cultured, and transduced, as previously described.7
Primary patient samples
In accordance with the Declaration of Helsinki, informed consent was given by parents or guardians, or both, to use leukemic cell material that remained after diagnostic procedures for research purposes, as approved by the Institutional Review Board of the Erasmus MC. Mononuclear cells were separated by sucrose gradient centrifugation (Lymphoprep, 1.077 g/mL density; Nycomed Pharma) and mononuclear cells were collected for further processing. The percent of leukemic cells was increased toward >90% by eliminating contaminating normal lymphocytes and myeloid progenitor cells using antibody-coated beads, as previously described.30 Cells were exposed to a twofold serial dilution series of STAT3 inhibitor for 96 hours at 37°C in humidified air containing 5% CO2. Next, cells were incubated with 0.33 µg/µL tetrazolium salt 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS; Promega) and 7.5 µg/mL of the electron coupling reagent phenazine methosulfate (Sigma) for 3 hours. The amount of soluble formazan product formed by viable cells was quantified at 490 nm absorbance on a VersaMax microplate reader.
Retroviral and lentiviral transduction of mouse primary cells and human cell lines
Retroviral TEL-AML1 supernatants were produced and used to transduce mouse HPC, as previously described.7 Transduced HPC were used for further experiments after 48, 72, or 96 hours of transduction. Lentiviral MISSION shRNA constructs (supplemental Materials and methods) were purchased from Sigma-Aldrich. The TEL-specific shRNA was used to specifically silence the TEL-AML1 fusion gene expression in Reh cells, because this cell line has deleted the wild-type TEL allele. For transplantation experiments, viable scramble, and shSTAT3, shRNA-transduced Reh cells were purified 5 days after transduction using the Dead Cell Removal kit (Miltenyi Biotec). Mice were sublethally irradiated with 3 Gy using a 137Cs γ-irradiator, and they were intravenously injected with equivalent numbers (1 × 105) of viable transduced cells per mouse. Mice were sacrificed when they developed clinical signs of disease.
Colony-forming assays
Colony formation of human cell lines was assessed 14 days after plating out 1 × 104 cells in HSC002 methylcellulose medium (R&D Systems). Colonies were stained with p-iodonitrotetrazolium.
Western blot analysis
Western blot analysis (supplemental Materials and methods) was performed according to manufacturer’s instructions. RAC guanosine triphosphate (GTP) pull down and RAC1 detection was performed with the Active Rac1 Pull-Down and Detection Kit, according to the manufacturer’s instructions (Thermo Scientific).
Flow cytometry
Cells were prepared as previously reported31 (supplemental Materials and methods). For intracellular staining, human leukemic cells were lysed and fixed in a single step using BD™ Phosflow Lyse/Fix buffer (BD Phosflow), permeabilized with BD™ Phosflow Perm Buffer III (BD Phosflow) and stained with PE-conjugated anti-STAT3 (pY705) according to manufacturer’s instructions (BD Biosciences). Apoptosis was detected using the Annexin V Apoptosis Detection Kit I (eBioscience). Cell cycle analysis was performed using the Click-iT EdU Alexa Flour 647 Flow Cytometry Assay Kit (Invitrogen). Apoptosis and cell cycle analysis of shSTAT3 and shTEL-AML1 (shTA)-transduced cells was performed 48 hours after treatment with the Dead Cell Removal kit (Miltenyi Biotec).
Quantitative RT-PCR analysis
Total RNA was isolated from cells using the RNeasy Mini Kit (Qiagen) and was converted into complementary DNA (cDNA) using the High Capacity RNA-to-cDNA Kit (Applied Biosystems), according to the manufacturer’s instructions. Quantitative reverse-transcription polymerase chain reaction was performed on isolated mRNA using TaqMan probe based chemistry and an ABI Prism 7900HT fast Sequence Detection System (Life Technologies) (supplemental Materials and methods).
Statistical analysis
Statistical analysis of survival curves was performed using the Mantel-Cox log-rank test and the standard deviation by using the Student t test.
Results
Inhibition of STAT3 in TEL-AML1 leukemic cells blocks proliferation and induces apoptosis
Recent analyses of aberrant signaling pathways operating in leukemia have identified critical roles for deregulated activation of signal transducer and activator of transcription (STAT) proteins, in particular STAT3 and STAT5.27 To determine the role of STAT activation in TEL-AML1 leukemia, we examined the proliferation and survival of different human leukemia cell lines after treatment with specific inhibitors of STAT3 and STAT5. We analyzed 2 different cell lines carrying the t(12;21) translocation (Reh, At-2), 2 with BCR-ABL rearrangement, representing a known STAT3 dependent oncogenic rearrangement32 (Tom-I, Sup-B15), and 3 with different genetic alterations (697, Bel-I, Semk) (supplemental Table 1). Blocking STAT3 activity with the specific inhibitor S3I-20133 resulted in inhibited proliferation of the TEL-AML1 cell lines to an even greater extent than that of the BCR-ABL cells (Figure 1A). Similar inhibition was observed with 2 distinct STAT3 inhibitors (supplemental Figure 1). Although STAT5 has been previously associated with TEL-AML1 leukemia, downstream of erythropoietin receptor upregulation,25 its inhibition did not have a significant effect on the proliferation of Reh and At-2 cells (Figure 1B). To examine the mechanism of the proliferation block induced by STAT3 suppression in Reh and At-2 cells, we measured the percentage of apoptosis and the cell cycle profile after 24 hours exposure to 50 μM of S3I-201. The results showed a significant increase of apoptosis (Figure 1C; supplemental Figure 2) and cell cycle arrest (Figure 1D; supplemental Figure 3) correlating with upregulation of p21 and p27 expression and induction of caspase 3 cleavage (supplemental Figure 4A-B). Next, molecularly, we investigated the inhibition of STAT3. For this purpose, 2 different shRNA targeting STAT3 were used (supplemental Figure 5). Both shRNA resulted in similar induction of apoptosis and inhibition of cell cycle progression (Figure 1E-F; supplemental Figure 6). Together, the sensitivity to S3I-201 and STAT3 knock-down establish an important role for STAT3 in the survival of TEL-AML1 cell lines.
Pharmacological and shRNA-mediated inhibition of STAT3 in TEL-AML1 leukemic cells blocks proliferation and induces apoptosis. (A-B) Proliferation of leukemic cell lines as measured by MTS assay after 96 hours culture with the indicated concentrations of the STAT3 inhibitor, S3I-301 (A), and STAT5 inhibitor (STAT5in) (B). Results are normalized to the proliferation of dimethylsulfoxide (DMSO)-treated cells. (C) Percentage of apoptotic Annexin V+PI- cells 24 hours after DMSO or S3I-301 treatment. (D) Percentage of cells in the S-phase of the cell-cycle 24 hours after DMSO or S3I-201 exposure. Cells were pulsed with 10 µM 5-ethynyl-2´-deoxyuridine (Edu) for 1 hour. (E) Percentage of apoptotic Annexin V+PI- 6 days after transduction with control scramble or 2 different STAT3 (shSTAT3) shRNA. (F) Percentage of cells in the S-phase of the cell-cycle 6 days after transduction with control scramble or 2 different STAT3 (shSTAT3) shRNA. Cells were pulsed with 10 µM 5-ethynyl-2´-deoxyuridine (Edu) for 1 hour. All data are representative of 3 independent experiments and show mean ± standard deviation of triplicate measurements. *P < .05, **P < .01, ***P < .005 compared with control (Student’s unpaired t test).
Pharmacological and shRNA-mediated inhibition of STAT3 in TEL-AML1 leukemic cells blocks proliferation and induces apoptosis. (A-B) Proliferation of leukemic cell lines as measured by MTS assay after 96 hours culture with the indicated concentrations of the STAT3 inhibitor, S3I-301 (A), and STAT5 inhibitor (STAT5in) (B). Results are normalized to the proliferation of dimethylsulfoxide (DMSO)-treated cells. (C) Percentage of apoptotic Annexin V+PI- cells 24 hours after DMSO or S3I-301 treatment. (D) Percentage of cells in the S-phase of the cell-cycle 24 hours after DMSO or S3I-201 exposure. Cells were pulsed with 10 µM 5-ethynyl-2´-deoxyuridine (Edu) for 1 hour. (E) Percentage of apoptotic Annexin V+PI- 6 days after transduction with control scramble or 2 different STAT3 (shSTAT3) shRNA. (F) Percentage of cells in the S-phase of the cell-cycle 6 days after transduction with control scramble or 2 different STAT3 (shSTAT3) shRNA. Cells were pulsed with 10 µM 5-ethynyl-2´-deoxyuridine (Edu) for 1 hour. All data are representative of 3 independent experiments and show mean ± standard deviation of triplicate measurements. *P < .05, **P < .01, ***P < .005 compared with control (Student’s unpaired t test).
TEL-AML1 induces phosphorylation of STAT3
To further investigate whether TEL-AML1 plays a role in STAT3 activation, we expressed the fusion cDNA in mouse fetal liver HPC. In this model, TEL-AML1 has been previously shown to promote B-lymphocyte development, enhance self-renewal of B-cell precursors, and lead to the establishment of long-term growth factor–dependent pre-B-cell lines.7 Here, we show that the transduction of mouse HPC with TEL-AML1 results in increased levels of STAT3 phosphorylation, at both Ser727 and Tyr705, the increase correlating with the levels of TEL-AML1 expressed (Figure 2A; supplemental Figure 7). A recent report used shRNA-mediated silencing of TEL-AML1 in human leukemic cell lines to demonstrate cell cycle inhibition and impaired engraftment in a xenotransplant mouse model.22 In line with this observation, we show that silencing TEL-AML1 expression with shRNA targeting the 5′ region of TEL, still present in the fusion gene transcript, causes a block in G1/S cell cycle transition in Reh cells (supplemental Figure 8). Moreover, this is accompanied by reduced levels of Tyr705 STAT3 phosphorylation (Figure 2B).
TEL-AML1 induces phosphorylation of STAT3. (A) Western blot analysis of p-S727 and p-Y705 STAT3 and total STAT3 in mouse c-kit+Ter119- fetal liver HPC, 72 hours after transduction with murine stem cell virus TEL-AML1 expressing (T/A) or empty (MSCV CD2) retroviral vectors. Numbers represent densitometric quantitation of phospho-STAT3 bands normalized to total STAT3. (B) Flow cytometric analysis of p-Y705 STAT3 in Reh 6 days after transduction with control scramble (mean fluorescence intensity [MFI] = 21.04) or shTA shRNA (MFI = 12.01) shRNA. Isotype control staining (MFI = 10.23) is also shown. All data are representative of 3 independent experiments.
TEL-AML1 induces phosphorylation of STAT3. (A) Western blot analysis of p-S727 and p-Y705 STAT3 and total STAT3 in mouse c-kit+Ter119- fetal liver HPC, 72 hours after transduction with murine stem cell virus TEL-AML1 expressing (T/A) or empty (MSCV CD2) retroviral vectors. Numbers represent densitometric quantitation of phospho-STAT3 bands normalized to total STAT3. (B) Flow cytometric analysis of p-Y705 STAT3 in Reh 6 days after transduction with control scramble (mean fluorescence intensity [MFI] = 21.04) or shTA shRNA (MFI = 12.01) shRNA. Isotype control staining (MFI = 10.23) is also shown. All data are representative of 3 independent experiments.
RAC1 is the main STAT3 activator in TEL-AML1 leukemia
RAC1 is the best characterized member of the ρ GTP-binding protein family, a group of proteins that regulate diverse cellular functions34 and have been recently shown to play critical roles in normal hematopoiesis and leukemia.35 Interestingly, RAC1 has been shown to be capable of increasing the levels of STAT3 phosphorylation through different mechanisms.36 For this reason, we decided to examine whether the activation of STAT3 observed in TEL-AML1 leukemia could be mediated by RAC1. The level of active RAC1-GTP was measured after transduction of HPC with TEL-AML1, and shRNA-mediated knock-down of the fusion gene in Reh cells. Expression of TEL-AML1 in mouse HPC resulted in a marked increase in active RAC1 (Figure 3A; supplemental Figure 9A), whereas silencing of the fusion in human leukemic cells led to reduced RAC1 activity (Figure 3B). To examine whether TEL-AML1-mediated RAC1 activation was responsible for elevated STAT3 activity, we treated Reh and At-2 cells with the RAC specific inhibitor NSC23766. RAC inhibition led to reduced levels of STAT3 phosphorylation (Figure 3C; supplemental Figure 9B) and consequent leukemic cell apoptosis (Figure 3D).
RAC1 is the main STAT3 activator in TEL-AML1 leukemia. (A-B) Western blot analysis of GTP-Rac1 pull-downs and total Rac1 from mouse HPC (A), 72 hours after transduction with MSCV T/A or MSCV CD2, and Reh cells (B), 5 days after transduction with control scramble or shTA shRNA. Numbers represent densitometric quantitation of GTP-Rac1 normalized to total RAC1 (A), and GTP-RAC1, and total RAC1 normalized to HSP90 (B). (C) Flow cytometric analysis of p-Y705 STAT3 in Reh, 5 hours after treatment with 50 μM (MFI = 12.99) and 100 μM (MFI = 11.66) NSC23766 or dimethylsulfoxide (MFI = 15.57). (D) Percentage of Reh and At-2 apoptotic Annexin V+PI- cells 24 hours after dimethylsulfoxide or 50 µM NSC23766 treatment. Data are representative of 2 (B) and 3 (A, C, and D) independent experiments. *P < .05, **P < .01, ***P < .005 compared with control (Student unpaired t test). GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
RAC1 is the main STAT3 activator in TEL-AML1 leukemia. (A-B) Western blot analysis of GTP-Rac1 pull-downs and total Rac1 from mouse HPC (A), 72 hours after transduction with MSCV T/A or MSCV CD2, and Reh cells (B), 5 days after transduction with control scramble or shTA shRNA. Numbers represent densitometric quantitation of GTP-Rac1 normalized to total RAC1 (A), and GTP-RAC1, and total RAC1 normalized to HSP90 (B). (C) Flow cytometric analysis of p-Y705 STAT3 in Reh, 5 hours after treatment with 50 μM (MFI = 12.99) and 100 μM (MFI = 11.66) NSC23766 or dimethylsulfoxide (MFI = 15.57). (D) Percentage of Reh and At-2 apoptotic Annexin V+PI- cells 24 hours after dimethylsulfoxide or 50 µM NSC23766 treatment. Data are representative of 2 (B) and 3 (A, C, and D) independent experiments. *P < .05, **P < .01, ***P < .005 compared with control (Student unpaired t test). GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
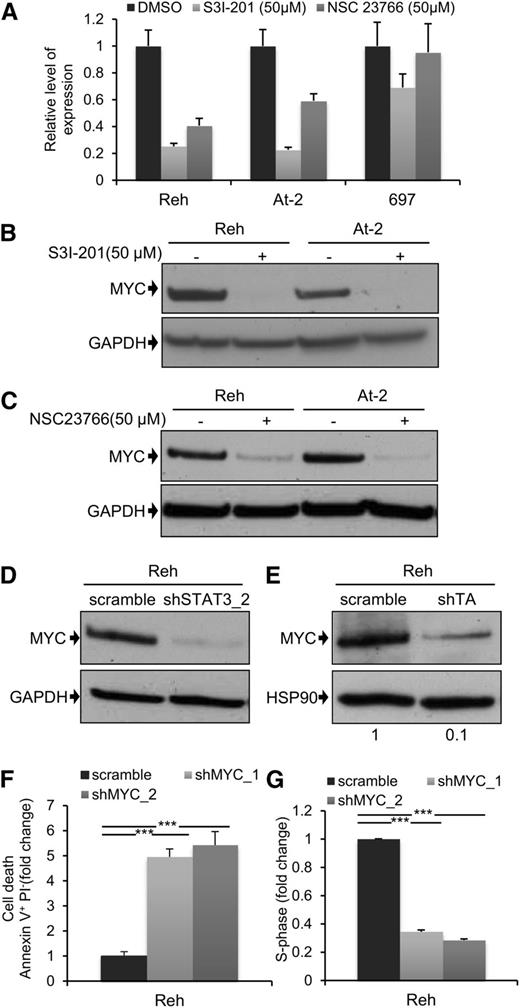
MYC expression is dependent on TEL-AML1mediated STAT3 activity
Next, we decided to examine the downstream mechanism for the leukemogenic activity of STAT3. MYC and BIRC5 are 2 of the best characterized STAT3 transcriptional target genes,36 and they play major roles in normal and malignant hematopoiesis.37,38 For these reasons, we analyzed the effect of STAT3, and RAC1, suppression on MYC and BIRC5 expression. Inhibition of STAT3 activity by S3I-201 led to a marked reduction in mRNA expression of both genes 6 hours after drug exposure (Figure 4A; supplemental Figure 10A). There was also a complete loss of MYC protein (Figure 4B), and partial suppression of BIRC5 protein (supplemental Figure 10B), after 24 hours. Interestingly, although there was no detectable apoptosis 4 hours after treatment of Reh and At-2 cells with S3I-201 (supplemental Figure 10C), at this time point MYC expression was already significantly reduced (supplemental Figure 10D). This indicates that reduced MYC expression was not a consequence of apoptosis. Similar results were obtained using the RAC1 inhibitor NSC23766 (Figure 4C), and this was confirmed after STAT3 silencing (Figure 4D). We also examined the effect of S3I-201 and NSC23766 on MYC and BIRC5 mRNA and protein expression in the non-TEL-AML1 cell line 697 (Figure 4A; supplemental Figure 10A-E). Although S3I-201, but not NSC23766, caused a modest reduction in MYC and BIRC5 expression, neither drug had a comparable inhibitory effect to that observed in the TEL-AML1 cells. Furthermore, knock-down of TEL-AML1 in Reh cells resulted in inhibition of MYC protein expression, confirming the role of this fusion in regulating STAT3- dependent–MYC induction (Figure 4E). Because MYC may thus play an important role in TEL-AML1 leukemogenesis, we decided to analyze the effect of silencing MYC in Reh cells (supplemental Figure 10F). This resulted in a similar induction of apoptosis and inhibition of cell cycle progression, as observed after STAT3 knock-down (Figure 4F-G).
MYC expression is dependent on TEL-AML1–mediated STAT3 activity. (A) Quantitative reverse-transcription polymerase chain reaction analysis of MYC mRNA expression 6 hours after treatment with 50 µM S3I-201, 50 µM NSC23766, or dimethylsulfoxide in Reh, At-2, and 697 cells. Columns represent mean ± standard deviation of quadruplicate measurements. (B-E) Western blot analysis of MYC protein expression in Reh and At-2 cells 24 hours after treatment with 50 µM S3I-201 (B), or 50 µM NSC23766 (C), or 6 days after transduction with control scramble, STAT3 (shSTAT3) shRNA (D), or shTA shRNA (E). Numbers represent densitometric quantitation of MYC bands normalized to GAPDH (B-D) and HSP90 (E). Percentage of apoptotic Annexin V+PI- cells (F) and S-phase cells (G) 6 days after transduction with scramble or 2 different MYC (shMYC) shRNA. Data are representative of 2 (D) and 3 (A-G) independent experiments and show mean ± standard deviation of triplicate measurements. *P < .05, **P < .01, ***P < .005 compared with control (Student unpaired t test). GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
MYC expression is dependent on TEL-AML1–mediated STAT3 activity. (A) Quantitative reverse-transcription polymerase chain reaction analysis of MYC mRNA expression 6 hours after treatment with 50 µM S3I-201, 50 µM NSC23766, or dimethylsulfoxide in Reh, At-2, and 697 cells. Columns represent mean ± standard deviation of quadruplicate measurements. (B-E) Western blot analysis of MYC protein expression in Reh and At-2 cells 24 hours after treatment with 50 µM S3I-201 (B), or 50 µM NSC23766 (C), or 6 days after transduction with control scramble, STAT3 (shSTAT3) shRNA (D), or shTA shRNA (E). Numbers represent densitometric quantitation of MYC bands normalized to GAPDH (B-D) and HSP90 (E). Percentage of apoptotic Annexin V+PI- cells (F) and S-phase cells (G) 6 days after transduction with scramble or 2 different MYC (shMYC) shRNA. Data are representative of 2 (D) and 3 (A-G) independent experiments and show mean ± standard deviation of triplicate measurements. *P < .05, **P < .01, ***P < .005 compared with control (Student unpaired t test). GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
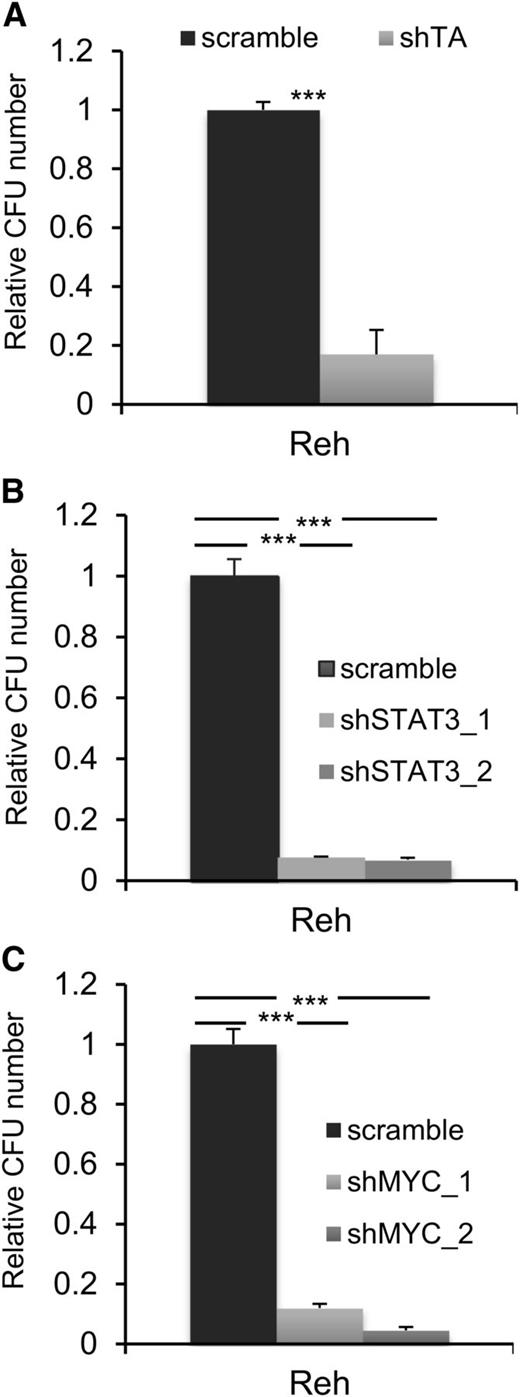
Activation of STAT3 is necessary for the self-renewal and leukemogenic activity of TEL-AML1
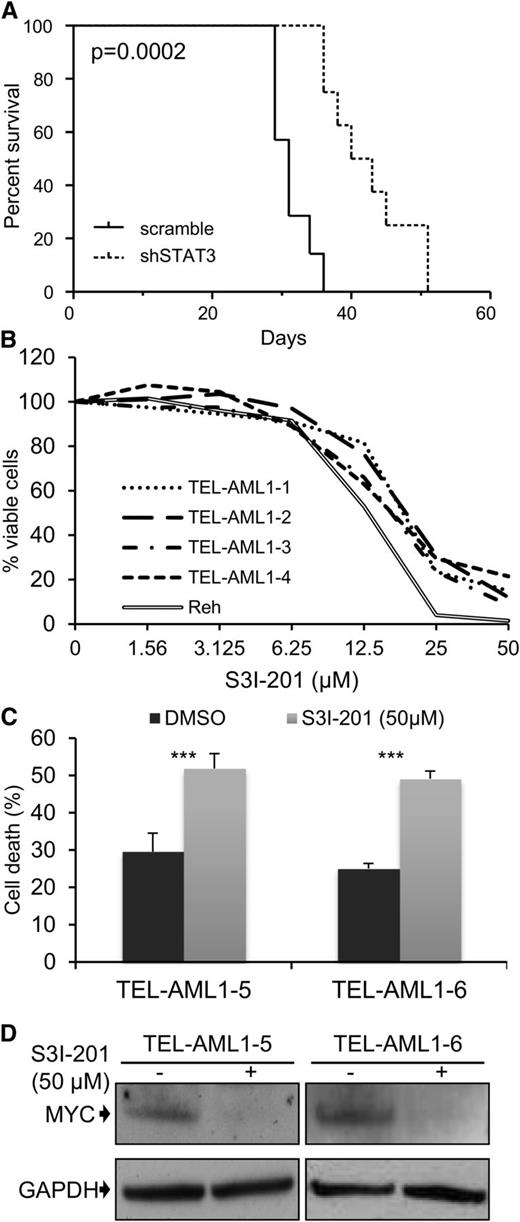
A common feature of all cancers, including leukemia, is the capacity for unlimited malignant cell self-renewal. To analyze the importance of TEL-AML1 in this process, we examined the effect of silencing fusion gene expression on leukemic cell colony formation. TEL-AML1 knock-down significantly impaired the ability of Reh cells to form colonies in methylcellulose (Figure 5A; supplemental Figure 11A). STAT3 silencing also caused a marked reduction in the number of colonies formed by Reh and At-2 cells (Figure 5B; supplemental Figure 11B-C). Furthermore, MYC silencing resulted in comparable inhibition of colony formation to that observed with STAT3 knock-down (Figure 5C; supplemental Figure 11D). Colony formation by TEL-AML1 leukemic cells was also specifically inhibited by S3I-201 treatment when compared with that of non-TEL-AML1 cells (supplemental Figure 12). To address the importance of STAT3 in leukemia progression in vivo, next we transplanted immunocompromised NSG mice with Reh cells after STAT3 silencing (supplemental Figure 13). Although all transplanted mice succumbed to leukemia, STAT3 knock-down resulted in a significant delay in leukemia progression (Figure 6A). Furthermore, Reh cells isolated from the latter showed elevated levels of STAT3 expression in comparison with those observed in cells prior to transplantation, demonstrating in vivo selection against leukemic cells with low STAT3 expression (supplemental Figure 14A). Similar selection against cells expressing low levels of STAT3 was also seen in long-term, in vitro culture of transduced Reh cells (supplemental Figure 14B-C). To extend our study to primary leukemia, next we tested the effect of STAT3 inhibition on the viability of 4 independent TEL-AML1–positive patient samples (supplemental Table 2). Treatment with S3I-201 impaired the viability of all 5 primary leukemia samples, the IC50 of the responses being equivalent to that of Reh cells (Figure 6B; supplemental Figure 15). Moreover treatment with the inhibitor induced high levels of apoptosis (Figure 6C) and completely abolished MYC protein expression in the primary ALL cells (Figure 6D), consistent with the data obtained in Reh cells.
STAT3 is necessary for colony formation by TEL-AML1+ cells. (A-C) Colony formation in methylcellulose by Reh cells 6 days after transduction with control scramble or shTA (A), shSTAT3 (B), or shMYC (C) shRNA. Plots show the number of colonies formed relative to control cultures, columns representing mean ± standard deviation values of triplicate cultures. Data are representative of 3 independent experiments. **P < .01, ***P < .005 compared with control (Student unpaired t test). CFU, colony-forming unit.
STAT3 is necessary for colony formation by TEL-AML1+ cells. (A-C) Colony formation in methylcellulose by Reh cells 6 days after transduction with control scramble or shTA (A), shSTAT3 (B), or shMYC (C) shRNA. Plots show the number of colonies formed relative to control cultures, columns representing mean ± standard deviation values of triplicate cultures. Data are representative of 3 independent experiments. **P < .01, ***P < .005 compared with control (Student unpaired t test). CFU, colony-forming unit.
Leukemia progression of TEL-AML1+ cell lines and survival of t(12;21) primary leukemia cells is impaired by STAT3 inhibition. Leukemia progression of TEL-AML1+ cell lines and survival of t(12;21) primary leukemia cells is impaired by STAT3 inhibition. (A) Kaplan-Meier survival curve for NSG mice transplanted with control scramble (n = 7) or shSTAT3 (n = 8) transduced Reh cells. P = .0002 versus scramble control (Mantel-Cox log-rank test). (B) Viability as measured by MTS assay of 4 independent TEL-AML1+ primary human leukemic samples after treatment with different S3I-201 concentrations for 96 hours. The response of Reh cells is included as a reference (C). Percentage apoptosis of 2 different primary human leukemic samples after 24-hour exposure to 50 µM S3I-201. (D) Western blot analysis of MYC protein expression in primary cells 24 hours after treatment with 50 µM S3I-201. **P < .01, ***P < .005 compared with control (Student unpaired t test). DMSO, dimethylsulfoxide; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Leukemia progression of TEL-AML1+ cell lines and survival of t(12;21) primary leukemia cells is impaired by STAT3 inhibition. Leukemia progression of TEL-AML1+ cell lines and survival of t(12;21) primary leukemia cells is impaired by STAT3 inhibition. (A) Kaplan-Meier survival curve for NSG mice transplanted with control scramble (n = 7) or shSTAT3 (n = 8) transduced Reh cells. P = .0002 versus scramble control (Mantel-Cox log-rank test). (B) Viability as measured by MTS assay of 4 independent TEL-AML1+ primary human leukemic samples after treatment with different S3I-201 concentrations for 96 hours. The response of Reh cells is included as a reference (C). Percentage apoptosis of 2 different primary human leukemic samples after 24-hour exposure to 50 µM S3I-201. (D) Western blot analysis of MYC protein expression in primary cells 24 hours after treatment with 50 µM S3I-201. **P < .01, ***P < .005 compared with control (Student unpaired t test). DMSO, dimethylsulfoxide; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Discussion
In this study, we describe a new signaling pathway operating in t(12;21) leukemia, actively regulated by TEL-AML1 and for the first time susceptible to pharmacological inhibition in primary leukemia cells. In this pathway, TEL-AML1 activates RAC1, in turn, that induces STAT3 activation. STAT3 activation is necessary for the survival, proliferation, and self-renewal of TEL-AML1+ leukemia through transcriptional induction of MYC expression (Figure 7).
Model of TEL-AML1–induced leukemogenesis. TEL-AML1 induces RAC1 activation that, in turn, promotes STAT3 phosphorylation and consequently the induction of MYC transcription. Active STAT3 is necessary for the survival, proliferation, and self-renewal of TEL-AML1expressing ALL cells.
Model of TEL-AML1–induced leukemogenesis. TEL-AML1 induces RAC1 activation that, in turn, promotes STAT3 phosphorylation and consequently the induction of MYC transcription. Active STAT3 is necessary for the survival, proliferation, and self-renewal of TEL-AML1expressing ALL cells.
STAT proteins are a family of 7 different transcription factors responsible for transducing signals from activated cytokine receptors to the nucleus. Binding of cytokine receptors by their ligands results in activation of Janus kinases (JAKs), which then phosphorylate and induce Src homology 2-dependent dimerization of STAT proteins. This leads to their translocation into and accumulation in the nucleus, and transcriptional activation of target genes.27 STATs are involved in a variety of biological processes, including cellular growth, differentiation, apoptosis, and immune responses. Several studies suggest a direct role for STAT proteins in regulating the development, survival, and proliferation of early B-cell progenitors. For instance, STAT3 and STAT5 play major roles in B development due to their importance in IL-7–mediated B lymphopoieisis.39,40 STAT3 is also frequently activated in cancer and plays important roles in oncogenesis and leukemogenesis.27 In leukemia, activation of STAT3 has been demonstrated mainly in acute myeloid leukemia, whereas alteration of STAT5 is more frequent in ALL.27 However, our data indicate that STAT3 is activated in all of the ALL cell lines examined (supplemental Figure 16A) and that TEL-AML1+ cells are sensitive to STAT3 inhibition, but not to inhibition of STAT5.
The principal kinase-mediating cytokine–receptor-induced phosphorylation of STAT3 is JAK2. Chromosomal translocations generating fusion proteins with constitutively active JAK2 moieties have been reported to occur in leukemia41 and activating JAK2 mutations are associated with high-risk pediatric ALL.42 However, our data suggest that STAT3 activity is regulated by a distinct mechanism in TEL-AML1+ leukemia. In our working model, STAT3 activity is necessary for proliferation, survival, clonogenicity, and leukemic engraftment of TEL-AML1+ cells, but activation of STAT3 appears to be independent of JAK2. Thus, the JAK2 inhibitor AG490 does not have any effect on the proliferation and survival of TEL-AML1+ cells (supplemental Figure 16B), whereas the BCR-ABL+ TOM-1 cells exhibit sensitivity to JAK2 inhibition, as previously reported.43 Instead, the TEL-AML1 fusion protein itself is responsible for maintaining STAT3 activity, via induction of RAC1. A precedent for JAK2-independent STAT3 activation in hematologic disease has recently been set by the discovery of activating STAT3 mutations in 40% of T-cell large granular lymphocytic leukemia.44 Our study describes an alternative mechanism operating in TEL-AML1+ leukemia, ensuring constitutive STAT3 activation that is uncoupled from normal regulatory pathways.
Activation of STAT3 by RAC1 has been previously reported by a number of different studies. However, the mechanisms by which this is achieved remain unclear. Two studies demonstrate that RAC1 activates STAT3 directly, but evidence also exists for indirect induction via interleukin 6 signaling.36 In the present study, the insensitivity of the TEL-AML1+ cells to JAK2 inhibition suggest that activation of STAT3 by RAC1 in these cells is likely to occur through a direct intracellular mechanism. It is also not clear whether TEL-AML1 causes direct activation of RAC1 at the protein level or whether this is achieved by transcriptional induction or repression of regulatory proteins. RAC1 is a ubiquitously expressed member of the ρ family of small GTPAses. These molecules shuttle between an inactive GDP-bound and an active GTP-bound state, with the transition being controlled by 2 sets of regulatory proteins. Guanine nucleotide exchange factors (GEFs) activate ρ GTPases by stimulating release of GDP and its replacement with GTP. The hydrolysis of GTP is stimulated by GTPase-activating proteins (GAPs), which then promote the transition of active ρ GTPases to the inactive state. A further level of regulation is achieved by guanine nucleotide dissociation inhibitors (GDIs) that inhibit ρ GTPases by binding them and preventing nucleotide release.34 Both GEFs and GAPs are regulated by complex mechanisms that control their subcellular translocation to regions of ρ GTPase proximity, release from autoinhibition and allosteric changes in their catalytic domains. However, their relative activities will also be determined by their basal levels of expression. In this context, analysis of global gene expression studies on patient samples and ALL cell lines indicate that elevated expression levels of particular GEFs are indeed associated with TEL-AML1 leukemia.45,46 For example, although not a direct transcriptional target of TEL-AML1, the ASEF gene is expressed at elevated levels in t(12;21) ALL and can cooperate with TEL-AML1 in promoting mouse B-cell progenitor colony formation.46
One of the most important transcriptional targets of STAT3 is the MYC gene.47 This gene encodes a transcription factor that is of fundamental importance to cell growth, proliferation, and stem cell maintenance, such that its homozygous deletion results in embryonic lethality.37 MYC is also important in B-cell development, constitutive MYC expression resulting in elevated numbers of B progenitors, while compound deletion of Myc and N-Myc family members causing a block in pro-B to pre-B transition.37 As a result of its central role in controlling numerous functions in normal cell biology, MYC is deregulated in most cancers, including many hematologic malignancies.37,48 Our data demonstrate an absolute requirement for STAT3 activity in the maintenance of MYC expression at both RNA and protein levels in TEL-AML1+ ALL cells. Interestingly, although all of the ALL cell lines we analyzed exhibited comparable levels of active STAT3 (data not shown), there was a marked difference in their sensitivity to STAT3 inhibition. Further analysis showed that STAT3 inhibition in the insensitive 697 ALL cells resulted in only partial MYC repression. Therefore, the differential sensitivity of the different ALL subtypes are likely due to the relative dependence of MYC expression upon STAT3 activity.
Despite the excellent prognosis of t(12,21) ALL as a whole,4 subgroups with less favorable prognosis exist, and long-term consequences of intensive chemotherapy can be significant.5,6 For these reasons, understanding the molecular basis underlying the development of TEL-AML1 leukemia could help identify novel therapeutic targets and develop new treatments for this disease. Recent studies have found genetic and functional heterogeneities in the cells responsible for leukemia clone maintenance and propagation in TEL-AML1+ and BCR-ABL+ ALL.18,19 In particular, some patients were found to harbor complex patterns of leukemic TEL-AML1+ subclones, patterns that were maintained in serial transplantations of immunodeficient mice.18 Interestingly, the only genetic abnormality that was common to all of these subclones was the fusion gene itself. This indicates that novel therapies targeting the one common genetic abnormality, the TEL-AML fusion, or essential downstream mediators, such as the STAT3 signaling pathway, would be particularly effective at eliminating TEL-AML1+ leukemic cells, which may prove to have lower toxicities and fewer unwanted side effects.
In brief, our findings represent yet another example of aberrant STAT3 activation in leukemia. Our data also show that in common with 70% of all cancers, MYC plays a fundamental role in TEL-AML1+ leukemia. The prevalence of MYC deregulation in cancer has caused it to be an attractive pharmacologic target for cancer treatment for many years. However, due its importance in normal cell function and stem cell maintenance, and the practical difficulties in designing MYC inhibitory drugs, more attention has been recently focused on the inhibition of downstream or upstream signaling pathways. Because STAT3 is dispensable for homeostatic hematopoietic stem cell function49 and can be targeted pharmacologically without any short-term toxicity,50 our data establish STAT3 inhibition as a means of achieving safe inhibition of MYC, thereby constituting a promising novel therapeutic strategy for the treatment of TEL-AML1 leukemia.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Ayad Eddaoudi, UCL Institute of Child Health Flow Cytometry Facility, for providing assistance with flow cytometry, all the staff of the UCL Institute of Child Health Western Laboratories for excellent animal husbandry, Alberto Bardelli, Federica Di Nicolantonio (University of Torino, Italy) and John Anderson (UCL Institute of Child Health, United Kingdom) for critically reading the manuscript, Didier Trono for lentiviral packaging constructs, and Renate Panzer-Grümayer and Ronald Stam for human cell lines.
This work was supported by grants from the Great Ormond Street Hospital Children’s Charity (ICH22 and W1003) (M.M. and V.W.-V.), the Leukaemia and Lymphoma Research (10032 and 07047) (J.d.B. and O.W., respectively).
Authorship
Contribution: M.M. and O.W. designed research; M.M., J.d.B., V.W.-V., R.P., and M.L.d.B. performed experiments; M.M., J.d.B., and V.W.-V. interpreted data; R.P. and M.L.d.B. analyzed experiments with human primary samples; M.M. and O.W. wrote the manuscript; O.W. supervised the study; and M.M., J.d.B., V.W.-V., R.P., M.L.d.B., and O.W. discussed and commented on the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Owen Williams, Molecular Haematology and Cancer Biology Unit, UCL Institute of Child Health, London WC1N 1EH, UK; e-mail: owen.williams@ucl.ac.uk.


![Figure 2. TEL-AML1 induces phosphorylation of STAT3. (A) Western blot analysis of p-S727 and p-Y705 STAT3 and total STAT3 in mouse c-kit+Ter119- fetal liver HPC, 72 hours after transduction with murine stem cell virus TEL-AML1 expressing (T/A) or empty (MSCV CD2) retroviral vectors. Numbers represent densitometric quantitation of phospho-STAT3 bands normalized to total STAT3. (B) Flow cytometric analysis of p-Y705 STAT3 in Reh 6 days after transduction with control scramble (mean fluorescence intensity [MFI] = 21.04) or shTA shRNA (MFI = 12.01) shRNA. Isotype control staining (MFI = 10.23) is also shown. All data are representative of 3 independent experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/122/4/10.1182_blood-2012-11-465252/4/m_542f2.jpeg?Expires=1767710584&Signature=Zm2Sm~TRneKjbJcPS2ykSUm~2xWpPOupr3pVaipke0KIK59g8I4YfGZqZ4KGCHLsLM5Wwam6HtpKUH6inSQMKAjcg5WlBTccYkEPzwgzwVgmHh1gFa8-IZzwh2q6paYCnP86UWe8jHGtYbqvR8i4sLYUVB3fKtOaHaVMv8NjjcFZMT4PgDe95SfYlQGQZe~iYK9WMfmfbwtRvgiexzY0PhXknVqYNDKnhg7ICQ8l29QtN89fNi26~sDTjoL5OILZJtkAti2bdbR2XqX0049mOpExQTBA6D9dOHgf49KHMf~TSJFIds205B37uRBmBVBU~jP18PiWx~nv6MybxR5FRQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)