Key Points
UDS demonstrated that BCR-ABL KD mutations detectable with conventional methods may just be the tip of the iceberg.
The information provided by conventional Sanger sequencing may not always be sufficient to predict responsiveness to a given TKI.
Abstract
In chronic myeloid leukemia and Philadelphia chromosome–positive acute lymphoblastic leukemia, tyrosine kinase inhibitor (TKI) therapy may select for drug-resistant BCR-ABL mutants. We used an ultra-deep sequencing (UDS) approach to resolve qualitatively and quantitatively the complexity of mutated populations surviving TKIs and to investigate their clonal structure and evolution over time in relation to therapeutic intervention. To this purpose, we performed a longitudinal analysis of 106 samples from 33 patients who had received sequential treatment with multiple TKIs and had experienced sequential relapses accompanied by selection of 1 or more TKI-resistant mutations. We found that conventional Sanger sequencing had misclassified or underestimated BCR-ABL mutation status in 55% of the samples, where mutations with 1% to 15% abundance were detected. A complex clonal texture was uncovered by clonal analysis of samples harboring multiple mutations and up to 13 different mutated populations were identified. The landscape of these mutated populations was found to be highly dynamic. The high degree of complexity uncovered by UDS indicates that conventional Sanger sequencing might be an inadequate tool to assess BCR-ABL kinase domain mutation status, which currently represents an important component of the therapeutic decision algorithms. Further evaluation of the clinical usefulness of UDS-based approaches is warranted.
Introduction
The first Bcr-Abl tyrosine kinase inhibitor (TKI), imatinib, was introduced in the treatment protocols of chronic myeloid leukemias (CMLs) and Philadelphia chromosome–positive (Ph+) acute lymphoblastic leukemias (ALLs) more than a decade ago.1 Soon after, however, it was observed that BCR-ABL kinase domain (KD) mutated forms with reduced or no sensitivity to imatinib could be selected.2 Second-(dasatinib/nilotinib/bosutinib) and third-generation (ponatinib) TKIs with much fewer insensitive mutations are now already approved or pending approval (Table 13-10 ). Sequential switch from a TKI to another may rescue response, although further gain of mutations by the same (compound mutations) or different (polyclonal mutations) Ph+ clones is possible.11
Summary of the BCR-ABL KD amino acid substitutions identified in clinical samples from patients reported to be resistant to the currently approved TKIs
| Imatinib . | Nilotinib . | Dasatinib . | Bosutinib . | Ponatinib . | |||||
|---|---|---|---|---|---|---|---|---|---|
| M237V | L273M | F311L | E355D/G | V379I | A397P | Y253F/H* | V299L† | V299L† | ? |
| M244V | E275K/Q | T315I‡ | F359V/I/C* | A380T | S417F/Y | E255K/V* | T315I‡ | T315I‡ | |
| L248R | D276G | F317L/V/I/C† | D363Y | F382L | I418S/V | T315I‡ | F317L/V/I/C† | ? | |
| G250E/R | T277A | F359V/I/C | L364I | L384M | S438C | F359V/I/C* | |||
| Q252R/H | E279K | Y342H | A365V | L387M/F | E453G/K | ||||
| Y253F/H* | V280A/I | M343T | L370P | M388L | E459K/V | ||||
| E255K/V* | V289A | A344V | V371A | Y393C | P480L | ||||
| E258D | V299L† | M351T | E373K | H396R/P | F486S | ||||
| Imatinib . | Nilotinib . | Dasatinib . | Bosutinib . | Ponatinib . | |||||
|---|---|---|---|---|---|---|---|---|---|
| M237V | L273M | F311L | E355D/G | V379I | A397P | Y253F/H* | V299L† | V299L† | ? |
| M244V | E275K/Q | T315I‡ | F359V/I/C* | A380T | S417F/Y | E255K/V* | T315I‡ | T315I‡ | |
| L248R | D276G | F317L/V/I/C† | D363Y | F382L | I418S/V | T315I‡ | F317L/V/I/C† | ? | |
| G250E/R | T277A | F359V/I/C | L364I | L384M | S438C | F359V/I/C* | |||
| Q252R/H | E279K | Y342H | A365V | L387M/F | E453G/K | ||||
| Y253F/H* | V280A/I | M343T | L370P | M388L | E459K/V | ||||
| E255K/V* | V289A | A344V | V371A | Y393C | P480L | ||||
| E258D | V299L† | M351T | E373K | H396R/P | F486S | ||||
Imatinib, dasatinib, and nilotinib are approved both by the Food and Drug Administration (FDA) and European Medicines Agency (EMA) for first- or subsequent-line use. Busutinib and ponatinib have recently been approved by the FDA for patients with resistance (or intolerance) to prior TKI therapy. Amino acid substitutions reported to be capable to survive imatinib therapy are almost 50.3 For patients harboring T315I, pharmacologic options include the recently FDA-approved ponatinib (for CML and Ph+ ALL patients with resistance to a prior TKI therapy)4 or omacetaxine mepesuccinate,5 an alkaloid with a mechanism of action other than Bcr-Abl kinase inhibition (for CP or AP CML patients with resistance to 2 or more TKIs).
? indicates that bosutinib-resistant mutations other than T315I and ponatinib-resistant mutations, if any, still need to be assessed.
V299L and F317L/V/I/C retain insensitivity also to dasatinib.7-9
T315I is a pan-resistant mutation retaining insensitivity to dasatinib, nilotinib, and bosutinib.10
Capillary Sanger sequencing (SS) is the most widely adopted method to assess BCR-ABL KD mutation status3 despite its multiple technical limitations: it cannot robustly identify mutated populations <10% to 15%, it provides only rough estimates of mutated clone abundance, and it cannot discriminate between polyclonal and compound mutations, unless it is preceded by a cumbersome step of cloning. Before the advent of next-generation sequencing (NGS) technologies, however, no method was available that could improve upon these limitations while allowing for scanning of any sequence variant at any position within the KD. NGS carries out thousands to millions of picoliter-scale sequencing reactions simultaneously yielding thousands to millions of sequence reads, each one corresponding to a single, clonally amplified, DNA molecule.12 The use of this approach to sequence a nucleotide position multiple times, thus achieving high sensitivity, is defined as ultra-deep sequencing (UDS). Among NGS technologies commercially available, the Roche 454 Life Sciences is particularly suitable for projects of targeted amplicon resequencing, the average read length being 400 to 600 bp.13 Its application for the UDS of the BCR-ABL KD would allow for: (1) full characterization of the spectrum of minor (<10%-15%) mutated variants; (2) the ability to follow the dynamics of resistant mutations over time; and (3) reconstruction of the clonal architecture of mutated populations in the case of multiple mutations occurring within the same amplicon.
We thus decided to take advantage of a UDS-based approach in order to resolve qualitatively and quantitatively the complexity of mutated Ph+ populations surviving TKIs and to investigate their clonal structure and evolution over time in relation to treatment.
Materials and methods
Patients
We retrospectively selected 33 CML or Ph+ ALL patients who had received sequential treatment with multiple TKIs (2-4 TKIs among imatinib, dasatinib, nilotinib, ponatinib) and had experienced sequential relapses accompanied by selection of 1 or more TKI-resistant mutations. Their main characteristics are presented in Table 2. Written informed consent had been obtained, in accordance with the Declaration of Helsinki. Up to 10 samples were analyzed for each patient, for a total of 106 samples.
Features of the patients included in the present study
| Features . | n . |
|---|---|
| Patients, total | 33 |
| Median age, y (range) | 52 (18-79) |
| Male to female ratio | 19:14 |
| Disease phase/type | |
| CML | 18 |
| Chronic phase | 9 |
| Accelerated phase | 1 |
| Myeloid blast crisis | 3 |
| Lymphoid blast crisis | 5 |
| Ph+ ALL | 15 |
| No. of lines of TKI therapy received | |
| 2 | 22 |
| 3 | 8 |
| 4 or more | 3 |
| Features . | n . |
|---|---|
| Patients, total | 33 |
| Median age, y (range) | 52 (18-79) |
| Male to female ratio | 19:14 |
| Disease phase/type | |
| CML | 18 |
| Chronic phase | 9 |
| Accelerated phase | 1 |
| Myeloid blast crisis | 3 |
| Lymphoid blast crisis | 5 |
| Ph+ ALL | 15 |
| No. of lines of TKI therapy received | |
| 2 | 22 |
| 3 | 8 |
| 4 or more | 3 |
We also randomly selected and analyzed 15 CML patients who had achieved an optimal response to imatinib (according to the European Leukemia Net [ELN] definitions)14 for comparison.
This study was approved by the review boards of the S. Orsola-Malpighi Hospital (Bologna) and of the Institute of Hematology and Blood Transfusion (Prague); it was conducted in accordance with the Declaration of Helsinki.
SS of the BCR-ABL KD
SS of the BCR-ABL KD was performed on an ABI PRISM 3730 (Applied Biosystems) as previously described.15
UDS of the BCR-ABL KD
RNA was converted to complementary DNA (cDNA) with the Transcriptor High-Fidelity cDNA Synthesis kit (Roche Applied Science). To select for the translocated ABL allele, a first step of amplification was performed by polymerase chain reaction (PCR) with a forward primer either on BCR exon 1a (in case of e1a2 BCR-ABL fusion) or on the border of BCR exons 12-13 (in case of b2a2 or b3a2 BCR-ABL fusions) and a reverse primer on ABL, exon 10. A second amplification step was then performed to generate 4 partly overlapping amplicons covering the KD of ABL (supplemental Figure 1, available on the Blood website), tagged with a 10-base “barcode” sequence (multiplex identifier) for sample pooling. Amplifications were done using the FastStart High-Fidelity PCR System kit (Roche Applied Science). UDS was performed on a Roche GS Junior (454-Life Sciences) according to the manufacturer’s instructions. Primer sequences, PCR, and sequencing protocols are detailed in the supplemental Methods. Amplicon Variant Analyzer (version 2.7; 454-Life Sciences) and Sequence Pilot (version 4.0.1; JSI-Medical Systems) were used to align reads to the reference ABL sequence (GenBank accession no. X16416.1) and to calculate variant frequencies. The presence of all relevant mutations was also manually verified by inspection of individual flowgrams at the corresponding positions, with particular attention to homopolymeric regions. Samples harboring multiple mutations were analyzed to gain further insights into the clonal architecture of mutated populations. To this purpose, 2 further amplicons were designed generating clonal reads for cases harboring a P-loop mutation plus a threonine 315 or phenylalanine 317 mutation, and for cases harboring a threonine 315 or phenylalanine 317 mutation plus a methionine 315, phenylalanine 359 or A-loop mutation: the most frequent mutation combinations (supplemental Figure 1). UDS runs were designed to enable high-sensitivity mutation calling, the target sequence coverage ranging from 3438 to 9976 independent reads for each nucleotide position. The sensitivity and reproducibility of an ultra-deep amplicon sequencing approach based on the 454 technology had been previously explored in the framework of the IRON (Interlaboratory Robustness of Next-generation sequencing)16 consortium. More recently, robustness and clinical utility were addressed in an even greater detail by Grossmann et al.17 Moreover, we further explored and confirmed the sensitivity and reproducibility of our UDS-based BCR-ABL KD mutation screening assay in a series of additional experiments detailed in the supplemental Methods.
Because (1) several studies concordantly demonstrated high reliability and reproducibility of 454 technology for variants ≥1%,18-20 and (2) serial dilution experiments (see supplemental Methods for details) confirmed the ability of our assay to detect mutations as low as 1% (whereas lower levels were not investigated), we decided to reduce the likelihood of false-positive results by filtering-out variants with <1% abundance and to exploit high coverage only for haplotype and clonal evolution analyses.
Phylogenetic analysis
Multiple read alignment and clustering was done with Jalview (version 2.8; www.jalview.org). Phylogenetic reconstruction was performed using the bioneighbor joining method with the Kimura 2-parameter substitution model with 1000 bootstrap replicates as implemented in the program SEAVIEW.21,22 To minimize the impact of sequencing errors, only sequences that represent >0.5% of the reads sequenced for at least 1 time point were considered for phylogenetic analyses.
Results
BCR-ABL KD mutation status may be more complex than SS shows
All the nucleotide substitutions that had been detected by SS (“major” mutations) were also detected by UDS, with fairly good concordance between the percentage of variant reads assessed by UDS and the mutation abundance estimated from the relative peak height in the SS chromatogram (Table 3; response definitions in references 14 and 23). In 58 of 106 (55%) samples, however, UDS revealed that mutations undetectable by SS (from now on referred to as “minor” mutations; abundance between 1% and 15%) were also present (Table 3). The type of minor mutations detected by UDS could frequently be accounted for by TKI exposure history because half (57 of 111, 51%) could be recognized to be poorly sensitive either to the TKI being administered or to the previous TKI received, or both (Table 1 reports the mutations know to be resistant to imatinib, dasatinib, nilotinib). In 4 patients (MBC-11, ALL-23, ALL-32, ALL-33), different Ph+ populations were found to have acquired different nucleotide substitutions leading to the same TKI-resistant amino acid change (eg, g>c and g>t at position 903, both resulting in a Q252H mutation; t>c at 1096 and c>a at 1098, both resulting in an F317L mutation). In the remaining cases, minor mutations were either silent (20 of 111, 18%) or never reported in association with TKI resistance (34 of 111, 28%).
Comparison between mutations detected by SS and mutations detected by UDS and estimated clonal composition of the samples harboring multiple mutations as assessed by UDS
| Code . | Date . | TKI . | Line . | Mutations by SS . | Mutations by UDS* . | Estimated mutated populations by UDS† . | Disease status and response . |
|---|---|---|---|---|---|---|---|
| CP-01-01 | 2/29/2012 | DAS | 2 | H396R (∼50), F317L (∼30) | H396R (55.05), F317L (28.23) | H396R (43.99), F317L (17.17), H396R+F317L (11.06) | Complete cytogenetic response but no molecular response after 6 mo on DAS |
| CP-01-02 | 5/2/2012 | DAS | 2 | F317L (∼70), H396R (∼20) | F317L (63.07), H396R (15.74), T315I (5.42) | F317L (55.47),H396R (7.60), H396R+F317L (7.38), T315I (4.44), H396R+T315I (0.76), F317L+T315I (0.22) | Complete hematologic response, cytogenetic response not assessed |
| CP-01-03 | 7/7/2012 | NIL | 3 | T315I (∼100) | T315I (99.28) | T315I (99.28) | Complete hematologic response, no cytogenetic response |
| CP-02-01 | 3/4/2008 | IM | 1 | F359V (∼20) | F359V (17.33) | F359V (17.33) | Loss of complete hematologic response after 5 mo on IM |
| CP-02-02 | 4/2/2008 | DAS | 2 | T315I (∼100) | T315I (94.80) | T315I (94.80) | Progression to LBC |
| CP-03-01 | 3/7/2005 | IM | 1 | G250E (∼100) | G250E (93.72), F317L (1.78) | G250E (92.20), G250E+F317L (1.52), F317L (0.26) | Minor cytogenetic response after 12 mo on IM |
| CP-03-02 | 9/14/2005 | DAS | 2 | G250E (∼70), F317L (∼20) | G250E (74.71), F317L (22.51) | G250E (62.00), G250E+F317L (12.71), F317L (9.80) | Minor cytogenetic response |
| CP-03-03 | 11/17/2005 | DAS | 2 | G250E (∼70), F317L (∼30) | G250E (60.73), F317L (27.06) | G250E (46.44), G250E+F317L (14.29), F317L (12.77) | Not available |
| CP-03-04 | 2/13/2006 | DAS | 2 | G250E (∼50), F317L (∼40) | G250E (45.47), F317L (37.49), H295H (4.91), C330C (1.48) | G250E (30.46), F317L (20.40), G250E+F317L (12.47), F317L+H295H (2.14), G250E+F317L+H295H (1.19), H295H (0.89), F317L+C330C (0.82), G250E+H295H (0.69), G250E+F317L+C330C (0.47), G250E+C330C (0.19) | Complete hematologic response, no cytogenetic response |
| CP-03-05 | 5/15/2006 | NIL | 3 | G250E (∼100), E255E (∼100) | G250E (87.17), E255E (85.78), F317L (10.44) | G250E+E255E (77.90), G250E+F317L+E255E (7.66), F317L (2.56), G250E (1.61), F317L+E255E (0.22) | Complete hematologic response, no cytogenetic response |
| CP-04-01 | 10/20/2005 | IM | 1 | L384M (∼100) | L384M (87.04), E255V (15.14) | N.A. | Loss of complete cytogenetic response after 24 mo on IM |
| CP-04-02 | 12/22/2005 | NIL | 2 | L384M (∼70), E255V (∼30) | L384M (68.33), E255V (32.02), M351I (2.64) | N.A. | Loss of complete hematologic response |
| CP-04-03 | 1/24/2006 | NIL | 2 | E255V (∼100) | E255V (80.71), L384M (14.40) | N.A. | No hematologic response |
| CP-05-01 | 1/20/2005 | IM | 1 | G250E (∼100) | G250E (99.51) | G250E (99.51) | Loss of complete cytogenetic response after 36 mo on IM |
| CP-05-02 | 3/23/2005 | DAS | 2 | F317L (∼30), G250E (∼30) | F317L (24.93), G250E (22.89), C305C (11.70), K274E (10.38) | G250E (16.52), F317L (15.06), C305C (9.30), G250E+F317L (4.30), K274E (4.09) F317L+K274E (3.70), G250E+K274E (1.04), C305C+F317L (0.97), C305+CK274E (0.91), G250E+F317L+K274E (0.51), G250E+C305C+F317L (0.39), G250E+C305C+K274E (0.13) | Not available |
| CP-05-03 | 4/20/2005 | DAS | 2 | F317L (∼100) | F317L (99.50) | F317L (99.50) | Complete hematologic response, no cytogenetic response |
| CP-06-01 | 4/19/2005 | IM | 1 | H396R (∼100) | H396R (99.63), A413A (1.71), K247N (1.11) | N.A. | Partial cytogenetic response after 18 mo on IM |
| CP-06-02 | 5/17/2005 | DAS | 2 | None | H396R (16.07), F317L (7.43) | H396R (16.07), F317L (7.43) | Complete cytogenetic response (= 0/200 Ph+ by FISH) |
| CP-06-03 | 6/14/2005 | DAS | 2 | F317L (∼20) | F317L (20.86), H396R (3.54) | F317L (20.20), H396R (2.88), F317L+H396R (0.66) | Loss of complete cytogenetic response (= 30/200 Ph+ by FISH) |
| CP-07-01 | 3/22/2007 | IM | 1 | None | A433A (1.48), P408P (1.42), K378R (1.32) | A433A (1.48), P408P (1.42), K378R (1.32) | No cytogenetic response after 12 mo on IM |
| CP-07-02 | 9/8/2009 | NIL | 2 | T315I (∼30) | T315I (25.99), M351T (5.93), T345T (6.37), R332R (5.91) | T315I (20.47), T345T (6.15), R332R (5.75), T315I+M351T (5.30), M351T (0.47), T315I+T345T (0.22), M351T+R332R (0.16) | No cytogenetic response |
| CP-07-03 | 3/4/2010 | NIL | 2 | T315I (∼70) | T315I (65.85), Y253H (16.65), W235R (3.77), F497L (3.14), T406I (2.43), Q477Q (2.37), F486S (2.28), D363N (1.47) | N.A. | No cytogenetic response; transplanted 6 mo later |
| CP-08-01 | 6/22/2010 | IM | 1 | M351T (∼100), E499E (∼100) | M351T (99.84), E499E (99.84) | N.A. | No cytogenetic response after 12 mo on IM |
| CP-08-02 | 4/26/2012 | NIL | 2 | Y253H (∼100), E499E (∼100) | Y253H (95.64), E499E (99.67), M351T (3.20) | N.A. | No cytogenetic response |
| CP-09-01 | 6/22/2004 | IM | 1 | M244V (∼20) | M244V (19.4), H396R (2.88), L298V (1.91), L364I (1.64) | N.A. | No cytogenetic response after 15 mo on IM |
| CP-09-02 | 7/14/2005 | IM | 1 | M244V (∼80) | M244V (79.31), H396R (6.75), L298V (3.75), L364I (3.28) | N.A. | Loss of complete hematologic response |
| CP-09-03 | 9/3/2007 | DAS | 2 | M244V (∼70), T315A (∼60) | M244V (73.36), T315A (57.53), F425S (2.18), T406I (1.75) | N.A. | Complete hematologic response but no cytogenetic response |
| AP-10-01 | 3/7/2005 | IM | 1 | E355G (∼50) | E355G (43.75), L341P (21.75), F496L (19.63), L428L (11.01), T315I (10.24), Y456Y (4.81) | N.A. | Progression from CP to AP after 9 mo on IM |
| AP-10-02 | 4/11/2005 | DAS | 2 | T315I (∼50) | T315I (42.60), F317L (1.25) | T315I (42.60), F317L (1.25) | Progression to MBC |
| MBC-11-01 | 4/12/2010 | IM | 1 | M351T (∼100), L248V (∼30) | N.D. | N.D. | Loss of complete hematologic response after 6 mo on IM |
| MBC-11-02 | 8/23/2010 | DAS | 2 | M351T (∼100), F317L (∼70), L248V (∼20) | M351T (100.00), F317L(ttc>tta)(65.52), L248V (19.45), del(248-274) (9.52), F317L(ttc>ctc)(8.52), V299L (1.99) | M351T+F317L(ttc>tta) (52.60), M351T (10.30), M351T+L248V (9.29), M351T+F317L(ttc>tta)+L248V (8.78), M351T+F317L(ttc>ctc) (6.33), M351T+del(248-274)(4.77), M351T+F317L(ttc>tta)+del(248-274)(4.14), M351T+V299L (1.41), M351T+L248V+F317L(ttc>ctc) (1.19), M351T+del(248-274)+F317L(ttc>ctc) (0.61), M351T+F317L(ttc>ctc)+V299L (0.39), M351T+L248V+V299L (0.19) | No hematologic response |
| MBC-12-01 | 3/5/2012 | IM | 1 | F359V (∼70) | F359V (63.24), L387M (4.18), M351T (3.42), V379I (1.62) | F359V (60.26), L387M (2.47), M351T (2.45), F359V+L387M (1.61), V379I (1.12), F359V+M351T (0.87), F359V+V379I (0.50), M351T+L387M (0.10) | Loss of complete cytogenetic response after 12 mo on IM |
| MBC-12-02 | 9/24/2012 | DAS | 2 | L387M (∼60), T315A (∼50), F359V (∼20) | L387M (57.43), T315A (57.19), F359V (16.24), T315I (8.22), F317V (3.98), F317L (1.15) | L387M+T315A (55.99), F359V+T315I (8.06), F359V (4.37), F359V+F317V (3.63), T315A (1.20), L387M+F317L (0.97), F317V (0.19), F359V+F317L (0.18), L387M+T315I (0.16), L387M+F317V (0.16), L387M (0.15) | Loss of complete hematologic response |
| MBC-13-01 | 5/28/2010 | DAS | 2 | T315A (∼100) | T315A (73.75), V299L (8.19) | T315A (72.40), V299L (6.84), T315A +V299L (1.35) | Loss of complete hematologic response after 6 mo on DAS |
| MBC-13-02 | 12/9/2010 | NIL | 3 | T315A (∼100), E255V (∼70), G250E (∼15) | T315A (92.87), E255V (69.74), G250E (12.22), E255K (1.03) | T315A+E255V (65.07), T315A (15.72), T315A+G250E (10.44), E255V (3.73), T315A+E255V+G250E (0.94), G250E (0.84), T315A+E255K (0.70), E255K (0.33) | No hematologic response |
| LBC-14-01 | 12/20/2011 | IM | 1 | G250E (∼40), E255V (∼20) | G250E (34.28), E255V (15.05), E255K (2.34), Y253F (1.10) | G250E (32.91), E255V (14.09), E255K (2.13), Y253F (0.90), G250E+E255V (0.96), G250E+E255K (0.21), G250E+Y253F (0.20) | Progression to lymphoid bast crisis after 32 mo on IM |
| LBC-14-02 | 2/8/2012 | DAS | 2 | T315I (∼30) | T315I (28.92) | T315I (28.92) | No hematologic response |
| LBC-15-01 | 5/26/2011 | IM | 1 | Y253H (∼100) | Y253H (99.88) | Y253H (99.88) | Hematologic relapse |
| LBC-15-02 | 12/5/2011 | DAS | 2 | Y253H (∼50), F317L (∼50) | Y253H (54.90), F317L (54.40) | Y253H+F317L (43.00), Y253H (11.90), F317L (11.40) | Hematologic relapse |
| LBC-16-01 | 3/14/2005 | IM | 1 | E255K (∼100) | E255K (98.84) | E255K (98.84) | Progression to LBC after 9 mo on IM |
| LBC-16-02 | 4/19/2005 | DAS | 2 | E255K (∼100), T315I (∼100) | E255K (99.84), T315I (99.48), L273S (1.30) | E255K+T315I (97.83), E255K (0.71), E255K+T315I+L273S (1.30), T315I (0.35) | No hematologic response |
| LBC-16-03 | 5/23/2005 | DAS | 2 | E255K (∼100), T315I (∼100) | E255K (99.53), T315I (99.53) | E255K+T315I (99.53) | No hematologic response |
| LBC-17-01 | 11/29/2010 | IM | 1 | L387M (∼30) | L387M (34.12) | L387M (28.18) | Loss of complete hematologic response after 6 mo on IM |
| LBC-17-02 | 2/27/2012 | DAS | 2 | L387M (∼100), T315I (∼50), M318V (∼50), F317L (∼50), Y320N (∼50) | L387M (96.33), T315I (51.46), M318V (51.19), F317L (45.21), Y320N (44.79) | L387M+T315I+M318V (47.90), L387M+F317L+Y320N (42.21), L387M (2.63), T315I+M318V (1.82), F317L+Y320N (1.37), L387M+F317L (1.10), L387M+T315I+M318V+Y320N (0.80), L387M+M318V (0.50), L387M+T315I+F317L (0.36), L387M+Y320N (0.25), L387M+T315I (0.25), L387M+T315I+M318V+F317L (0.17), L387M+T315I+Y320N (0.16) | Loss of cytogenetic and hematologic response |
| LBC-18-01 | 11/11/2007 | IM | 1 | F359V (∼60) | F359V (57.89) | F359V (57.89) | Progression to LBC after 3 mo on IM |
| LBC-18-02 | 1/17/2008 | DAS | 2 | None | F317L (8.48), F317I (1.02), F359V (1.02) | F317L (8.48), F317I+F359V (1.02) | Complete hematologic response, partial cytogenetic response |
| LBC-18-03 | 2/12/2008 | DAS | 2 | F359V (∼100), F317I (∼100) | F317I (92.35), F359V (90.11), F317L (4.25) | F359V+F317I (88.01), F317I (4.34), F317L (3.12), F359V+F317L (1.13), F359V (0.97) | Loss of complete hematologic response |
| ALL-19-01 | 9/27/2005 | IM | 1 | Y253H (∼100) | Y253H (99.79) | Y253H (99.79) | Hematologic relapse after 6 mo on IM |
| ALL-19-02 | 1/3/2006 | DAS | 2 | None | P465L (2.78), I432I (1.60), T277I (1.56), E352V (1.41), A474A (1.15) | N.A. | Complete hematologic and cytogenetic response, molecularly detectable disease |
| ALL-19-03 | 3/2/2006 | DAS | 2 | None | A399V (1.58) | A399V (1.58) | Complete hematologic and cytogenetic response, molecularly detectable disease |
| ALL-19-04 | 6/15/2006 | DAS | 2 | Y253H (∼100), T315I (∼50) | Y253H (99.70), T315I (40.29), N336S (5.39), W405R (1.89) | N.A. | Hematologic relapse |
| ALL-20-01 | 1/11/2005 | IM | 1 | M351T (∼100) | M351T (99.91) | M351T (99.91) | Hematologic relapse after 12 mo on IM |
| ALL-20-02 | 3/12/2005 | DAS | 2 | None | L370L (5.45), I432I (2.65) | N.A. | Complete hematologic and cytogenetic response, molecularly detectable disease |
| ALL-20-03 | 1/16/2006 | DAS | 2 | M351T (∼100), F317L (∼50) | M351T (99.77), F317L (53.27) | M351T (46.50), M351T+F317L (53.27) | Hematologic relapse |
| ALL-20-04 | 4/7/2006 | DAS | 2 | M351T (∼100), F317L (∼100) | M351T (95.46), F317L (85.32), A399T (5.64), Y353H (1.00) | M351T+F317L (85.32), M351T+A399T (5.64), M351T (3.50), M351T+F317L+Y353H (1.00) | Progressive disease |
| ALL-21-01 | 5/30/2005 | DAS | 2 | F317L (∼100) | F317L (99.99), M237I (2.03) | F317L (97.96), F317L+M237I (2.03) | Hematologic relapse after 3 mo on DAS |
| ALL-21-02 | 10/27/2005 | NIL | 3 | F317L (∼100), Y253H (∼20) | F317L (98.59), Y253H (17.35) | F317L (81.24), F317L+Y253H (17.35) | Complete hematologic response but no cytogenetic response |
| ALL-21-03 | 1/16/2006 | NIL | 3 | F317L (∼100), Y253H (∼20) | F317L (100.00), Y253H (18.77) | F317L (81.23), F317L+Y253H (18.77) | Hematologic relapse |
| ALL-21-04 | 3/17/2006 | NIL | 3 | F317L (∼100), Y253H (∼100) | F317L (100.00), Y253H (100.00) | F317L+Y253H (100.00) | Progressive disease |
| ALL-22-01 | 12/23/2005 | IM | 1 | F359V (∼100) | F359V (99.76), M237T (2.11) | N.A. | Hematologic relapse after 26 mo on IM |
| ALL-22-02 | 1/29/2005 | DAS | 2 | F359V (∼100), T315I (∼100) | F359V (99.30), T315I (98.77), Y449Y (10.50) | N.A. | Progressive disease |
| ALL-23-01 | 9/14/2011 | DAS | 2 | E255K (∼30), T315I (∼20) | E255K (24.04), T315I (19.37), G303G (1.51) | E255K (20.88), T315I (16.21), E255K+T315I (3.16), G303G (1.51) | Hematologic relapse after 9 mo on DAS |
| ALL-23-02 | 10/27/2011 | PON | 3 | T315I (∼100) | T315I (99.78), Y312C (1.22) | T315I (98.28), T315I+Y312C (1.22) | Complete hematologic response |
| ALL-23-03 | 11/3/2011 | PON | 3 | T315I (∼100) | T315I (99.83) | T315I (99.83) | Complete hematologic response |
| ALL-23-04 | 12/15/2011 | POST-SCT, NONE | / | None | None | None | Complete hematologic and cytogenetic response, molecularly detectable disease |
| ALL-23-05 | 1/11/2012 | POST-SCT, IM | 4 | E255K (∼100) | E255K (99.88) | E255K (99.88) | Complete hematologic response but loss of cytogenetic response |
| ALL-23-06 | 1/25/2012 | PON | 5 | E255K (∼100) | E255K (99.36), S417S (2.21) | N.A. | Hematologic relapse |
| ALL-23-07 | 2/8/2012 | PON | 5 | E255K (∼30) | E255K (22.49) | E255K (22.49) | Complete hematologic response |
| ALL-23-08 | 2/15/2012 | PON | 5 | E255K (∼100) | E255K (99.88) | E255K (99.88) | Partial hematologic response |
| ALL-23-09 | 3/12/2012 | DAS | 6 | E255K (∼70), T315I (∼50) | E255K (76.10), T315I (57.35), Q252H (cag>cac)(14.19), Q252H (cag>cat)(7.49), G250E (1.05) | E255K+T315I (51.60), E255K+Q252H (cag>cac) (13.94), E255K+Q252H (cag>cat) (7.34),T315I (5.75), E255K (2.52), E255K+ G250E (0.70), Q252H (cag>cac) (0.25), G250E (0.35), Q252H (cag>cat) (0.15) | Progressive disease |
| ALL-23-10 | 3/28/2012 | PON | 7 | E255K (∼100), T315I (∼100) | E255K (99.64), T315I (98.56) | E255K+T315I (98.20), E255K (1.44), T315I (0.36) | Progressive disease |
| ALL-24-01 | 6/15/2011 | NIL | 1 | Y253H (∼30) | Y253H (27.35), P465L (1.55) | N.A. | Complete hematologic and cytogenetic response, molecularly detectable disease |
| ALL-24-02 | 7/28/2011 | IM | 2 | Y253H (∼100) | Y253H (99.99), R367L (3.62) | Y253H (96.37), Y253H+R367L (3.62) | Complete hematologic and cytogenetic response, but 1-log increase in BCR-ABL transcript levels |
| ALL-24-03 | 3/1/2012 | DAS | 3 | None | Y302S (1.62) | Y302S (1.62) | Complete hematologic and cytogenetic response, no molecular assessment performed |
| ALL-24-04 | 3/29/2012 | DAS | 3 | Y253H (∼100), T315I (∼100) | Y253H (99.10), T315I (99.10), L273L (1.09) | Y253H+T315I (98.01); Y253H+T315I+L273L (1.09) | Hematologic relapse |
| ALL-25-01 | 12/2/2011 | IM | 1 | Y253H (∼20) | Y253H (18.38) | Y253H (18.38) | Complete hematologic and cytogenetic response, molecularly detectable disease after 18 mo on IM |
| ALL-25-02 | 1/18/2012 | DAS | 2 | Y253H (∼100), T315I (∼100) | Y253H (87.69), T315I (86.16) | Y253H+T315I (80.53), Y253H (7.16), T315I (5.63) | Hematologic relapse |
| ALL-26-01 | 1/11/2012 | IM | 1 | E255K (∼60) | E255K (54.75), F359V (13.81), T315I (3.84) | N.A. | Hematologic relapse after 6 mo on IM |
| ALL-26-02 | 6/16/2012 | DAS | 2 | T315I (∼100) | T315I (99.42) | T315I (99.42) | Progressive disease |
| ALL-27-01 | 1/14/2011 | IM | 1 | T315I (∼100) | T315I (99.87) | T315I (99.87) | Complete hematologic but not cytogenetic response after 3 mo on IM |
| ALL-27-02 | 1/31/2011 | IM | 1 | T315I (∼100) | T315I (99.74) | T315I (99.74) | Stable disease |
| ALL-27-03 | 2/7/2011 | NIL | 2 | T315I (∼80) | T315I (75.55), E255K (9.70), E255V (1.53) | T315I (70.47), E255K (5.10), T315I+E255K (4.60), E255V (1.05), T315I+E255V (0.48) | Stable disease |
| ALL-27-04 | 2/21/2011 | NIL | 2 | T315I (∼60), E255K (∼30) | T315I (53.80), E255K (25.63), E255V (1.33) | T315I (46.10), E255K (18.26), T315I+E255K (7.37), E255V (1.00), T315I+E255V (0.33) | Hematologic relapse |
| ALL-28-01 | 12/5/2007 | IM | 1 | Y253H (∼50), E255K (∼20) | Y253H (56.01), E255K (14.92), T315I (8.33), Q252H (6.32), A269T (3.57), Y253F (1.37) | Y253H (52.30), E255K (14.70), T315I (5.29), Q252H (3.92), Y253H+T315I (2.93), Q252H+A269T (2.40), Y253F (1.26), Y253H+269T (0.63), A269T (0.54), Y253H+E255K (0.15), Y253F+T315I (0.11) | Hematologic relapse after 9 mo on IM |
| ALL-28-02 | 3/26/2008 | DAS | 2 | T315I (∼100) | T315I (90.96) | T315I (90.96) | Progressive disease |
| ALL-29-01 | 6/6/2012 | IM POST-SCT | 2 | T315? | T315M (30.20) | T315M (30.20) | Complete hematologic response, molecularly detectable disease |
| ALL-29-02 | 6/20/2012 | IM | 2 | T315? | T315M (22.32) | T315M (22.32) | Hematologic relapse |
| ALL-29-03 | 7/10/2012 | IM | 2 | T315? | T315M (30.28) | T315M (30.28) | Progressive disease |
| ALL-29-04 | 8/1/2012 | IM | 2 | T315? | T315M (57.28), T315I (1.09) | T315M (57.28), T315I (1.09) | Stable disease |
| ALL-29-05 | 8/22/2012 | PON | 3 | T315?, E255V (∼20) | T315M (28.61), T315I (22.93), E255V (12.40) | T315M (25.20), T315I (20.01), E255V (5.03), T315M+E255V (3.41), T315I+E255V (2.92) | Progressive disease |
| ALL-30-01 | 3/30/2010 | NIL | 1 | E255V (∼40) | E255V (34.70) | E255V (34.70) | Complete hematologic response, molecularly detectable disease after 1 mo on IM |
| ALL-30-02 | 5/12/2010 | IM | 2 | E255V (∼100) | E255V (91.90), V304A (3.91), L302R (1.61), G303W (1.61) | See Figure 3 | Complete hematologic response, molecularly detectable disease with 1-log increase in BCR-ABL transcript levels |
| ALL-30-03 | 7/19/2010 | IM | 4 | E255V (∼50), Y253H (∼50) | E255V (46.50), Y253H (39.30), E255K (1.02) | See Figure 3 | Hematologic relapse |
| ALL-30-04 | 9/7/2010 | DAS | 5 | E255V (∼100) | E255V (91.60), T315I (2.00), E255K (1.80), K262R (1.00) | See Figure 3 | Stable disease |
| ALL-30-05 | 10/5/2010 | DAS | 5 | E255V (∼50), T315I (∼30) | E255V (53.00), T315I (23.00), Q252E (14.30), E255K (2.10) | See Figure 3 | Progressive disease |
| ALL-31-01 | 4/4/2006 | IM | 1 | F317L (∼100) | F317L (99.64) | F317L (99.64) | Hematologic relapse after 5 mo on IM |
| ALL-31-02 | 6/6/2006 | DAS | 2 | D276G (∼50), F317L (∼50), T315A (∼50) | T315A (51.12), F317L (45.81), D276G (44.86) | F317L (33.45), T315A+D276G (32.00), T315A (17.87), F317L+D276G (11.11), D276G (1.25), T315A+F317L (0.75), T315A+F317L+D276G (0.50) | Hematologic relapse after transient hematologic improvement |
| ALL-31-03 | 7/14/2006 | IM | 3 | F317L (∼100) | F317L (99.76), R332R (4.17) | F317L (95.59), F317L+R332R (4.17) | Stable disease |
| ALL-31-04 | 8/23/2006 | IM | 3 | T315A (∼100), G250E (∼100) | T315A (100.00), G250E (90.48) | T315A+G250E (90.48), T315A (9.52) | Progressive disease |
| ALL-31-05 | 9/26/2006 | NIL | 4 | T315A (∼100), G250E (∼100) | T315A (99.83), G250E (99.83) | T315A+G250E (99.83) | Progressive disease |
| ALL-31-06 | 10/24/2006 | NIL | 4 | T315A (∼100), G250E (∼60), D276G (∼30), Y253H (∼30) | T315A (99.96), G250E (67.93), D276G (28.89), Y253H (27.21), Q252E (1.49) | T315A+G250E (65.82), T315A+Y253H+D276G (26.02), T315A (3.76), T315A+G250E+D276G (2.11), T315A+Q252E (0.78), T315A+Y253H (0.48), T315A+Y253H+D276G+Q252E (0.48), T315A+D276G (0.28), T315A+Y253H+Q252E (0.23) | Progressive disease |
| ALL-32-01 | 1/6/2012 | IM | 2 | Y253H (∼100) | Y253H (99.79) | Y253H (99.79) | Hematologic relapse |
| ALL-32-02 | 4/12/2012 | DAS | 3 | None | None | None | Complete hematologic and cytogenetic response, molecularly detectable disease |
| ALL-32-03 | 8/8/2012 | DAS | 3 | None | Y253H (2.49), T315I (1.19) | Y253H (1.30), Y253H+T315I (1.19) | Complete hematologic response, 2-log increase in BCR-ABL transcript levels |
| ALL-32-04 | 9/17/2012 | DAS | 3 | Y253H (∼100), T315I (∼60), F317L(ttc>tta) (∼20) | Y253H (100.00), T315I (79.11), F317L(ttc>tta)(15.70), F317L(ttc>ctc)(4.04) | Y253H+T315I ((78.77), Y253H+ F317L(ttc>tta)(15.36), Y253H+ F317L(ttc>ctc)(4.04), Y253H (1.49), Y253H+T315I+F317L(ttc>tta)(0.34) | Loss of cytogenetic response |
| ALL-33-01 | 3/27/2005 | IM | 1 | G250E (∼100) | G250E (99.50) | G250E (99.50) | Hematologic relapse |
| ALL-33-02 | 9/5/2005 | DAS | 2 | G250E (∼100), F317L (∼70) | N.D. | N.D. | Hematologic relapse after a 5-mo complete hematologic response |
| ALL-33-03 | 12/1/2005 | NIL | 3 | G250E (∼100), F317L (∼50), Y253H (∼30) | G250E (99.99), F317L(ttc>tta)(43.63), Y253H (26.36), V299L (4.84), L248R (1.89), F317L(ttc>ctc)(1.01) | G250E (35.54), G250E+F317L(ttc>tta) (31.90), G250E+Y253H (14.86), G250E+F317L(ttc>tta)+Y253H (9.84), G250E+V299L (2.72), G250E+L248R (1.13), G250E+Y253H+V299L (0.99), G250E+F317L(ttc>tta)+V299L (0.77), G250E+F317L(ttc>tta)+L248R (0.76), G250E+F317L(ttc>ctc) (0.70), G250E+F317L(ttc>tta)+V299L+Y253H (0.36), G250E+F317L(ttc>ctc)+Y253H (0.31) | Progressive disease |
| Code . | Date . | TKI . | Line . | Mutations by SS . | Mutations by UDS* . | Estimated mutated populations by UDS† . | Disease status and response . |
|---|---|---|---|---|---|---|---|
| CP-01-01 | 2/29/2012 | DAS | 2 | H396R (∼50), F317L (∼30) | H396R (55.05), F317L (28.23) | H396R (43.99), F317L (17.17), H396R+F317L (11.06) | Complete cytogenetic response but no molecular response after 6 mo on DAS |
| CP-01-02 | 5/2/2012 | DAS | 2 | F317L (∼70), H396R (∼20) | F317L (63.07), H396R (15.74), T315I (5.42) | F317L (55.47),H396R (7.60), H396R+F317L (7.38), T315I (4.44), H396R+T315I (0.76), F317L+T315I (0.22) | Complete hematologic response, cytogenetic response not assessed |
| CP-01-03 | 7/7/2012 | NIL | 3 | T315I (∼100) | T315I (99.28) | T315I (99.28) | Complete hematologic response, no cytogenetic response |
| CP-02-01 | 3/4/2008 | IM | 1 | F359V (∼20) | F359V (17.33) | F359V (17.33) | Loss of complete hematologic response after 5 mo on IM |
| CP-02-02 | 4/2/2008 | DAS | 2 | T315I (∼100) | T315I (94.80) | T315I (94.80) | Progression to LBC |
| CP-03-01 | 3/7/2005 | IM | 1 | G250E (∼100) | G250E (93.72), F317L (1.78) | G250E (92.20), G250E+F317L (1.52), F317L (0.26) | Minor cytogenetic response after 12 mo on IM |
| CP-03-02 | 9/14/2005 | DAS | 2 | G250E (∼70), F317L (∼20) | G250E (74.71), F317L (22.51) | G250E (62.00), G250E+F317L (12.71), F317L (9.80) | Minor cytogenetic response |
| CP-03-03 | 11/17/2005 | DAS | 2 | G250E (∼70), F317L (∼30) | G250E (60.73), F317L (27.06) | G250E (46.44), G250E+F317L (14.29), F317L (12.77) | Not available |
| CP-03-04 | 2/13/2006 | DAS | 2 | G250E (∼50), F317L (∼40) | G250E (45.47), F317L (37.49), H295H (4.91), C330C (1.48) | G250E (30.46), F317L (20.40), G250E+F317L (12.47), F317L+H295H (2.14), G250E+F317L+H295H (1.19), H295H (0.89), F317L+C330C (0.82), G250E+H295H (0.69), G250E+F317L+C330C (0.47), G250E+C330C (0.19) | Complete hematologic response, no cytogenetic response |
| CP-03-05 | 5/15/2006 | NIL | 3 | G250E (∼100), E255E (∼100) | G250E (87.17), E255E (85.78), F317L (10.44) | G250E+E255E (77.90), G250E+F317L+E255E (7.66), F317L (2.56), G250E (1.61), F317L+E255E (0.22) | Complete hematologic response, no cytogenetic response |
| CP-04-01 | 10/20/2005 | IM | 1 | L384M (∼100) | L384M (87.04), E255V (15.14) | N.A. | Loss of complete cytogenetic response after 24 mo on IM |
| CP-04-02 | 12/22/2005 | NIL | 2 | L384M (∼70), E255V (∼30) | L384M (68.33), E255V (32.02), M351I (2.64) | N.A. | Loss of complete hematologic response |
| CP-04-03 | 1/24/2006 | NIL | 2 | E255V (∼100) | E255V (80.71), L384M (14.40) | N.A. | No hematologic response |
| CP-05-01 | 1/20/2005 | IM | 1 | G250E (∼100) | G250E (99.51) | G250E (99.51) | Loss of complete cytogenetic response after 36 mo on IM |
| CP-05-02 | 3/23/2005 | DAS | 2 | F317L (∼30), G250E (∼30) | F317L (24.93), G250E (22.89), C305C (11.70), K274E (10.38) | G250E (16.52), F317L (15.06), C305C (9.30), G250E+F317L (4.30), K274E (4.09) F317L+K274E (3.70), G250E+K274E (1.04), C305C+F317L (0.97), C305+CK274E (0.91), G250E+F317L+K274E (0.51), G250E+C305C+F317L (0.39), G250E+C305C+K274E (0.13) | Not available |
| CP-05-03 | 4/20/2005 | DAS | 2 | F317L (∼100) | F317L (99.50) | F317L (99.50) | Complete hematologic response, no cytogenetic response |
| CP-06-01 | 4/19/2005 | IM | 1 | H396R (∼100) | H396R (99.63), A413A (1.71), K247N (1.11) | N.A. | Partial cytogenetic response after 18 mo on IM |
| CP-06-02 | 5/17/2005 | DAS | 2 | None | H396R (16.07), F317L (7.43) | H396R (16.07), F317L (7.43) | Complete cytogenetic response (= 0/200 Ph+ by FISH) |
| CP-06-03 | 6/14/2005 | DAS | 2 | F317L (∼20) | F317L (20.86), H396R (3.54) | F317L (20.20), H396R (2.88), F317L+H396R (0.66) | Loss of complete cytogenetic response (= 30/200 Ph+ by FISH) |
| CP-07-01 | 3/22/2007 | IM | 1 | None | A433A (1.48), P408P (1.42), K378R (1.32) | A433A (1.48), P408P (1.42), K378R (1.32) | No cytogenetic response after 12 mo on IM |
| CP-07-02 | 9/8/2009 | NIL | 2 | T315I (∼30) | T315I (25.99), M351T (5.93), T345T (6.37), R332R (5.91) | T315I (20.47), T345T (6.15), R332R (5.75), T315I+M351T (5.30), M351T (0.47), T315I+T345T (0.22), M351T+R332R (0.16) | No cytogenetic response |
| CP-07-03 | 3/4/2010 | NIL | 2 | T315I (∼70) | T315I (65.85), Y253H (16.65), W235R (3.77), F497L (3.14), T406I (2.43), Q477Q (2.37), F486S (2.28), D363N (1.47) | N.A. | No cytogenetic response; transplanted 6 mo later |
| CP-08-01 | 6/22/2010 | IM | 1 | M351T (∼100), E499E (∼100) | M351T (99.84), E499E (99.84) | N.A. | No cytogenetic response after 12 mo on IM |
| CP-08-02 | 4/26/2012 | NIL | 2 | Y253H (∼100), E499E (∼100) | Y253H (95.64), E499E (99.67), M351T (3.20) | N.A. | No cytogenetic response |
| CP-09-01 | 6/22/2004 | IM | 1 | M244V (∼20) | M244V (19.4), H396R (2.88), L298V (1.91), L364I (1.64) | N.A. | No cytogenetic response after 15 mo on IM |
| CP-09-02 | 7/14/2005 | IM | 1 | M244V (∼80) | M244V (79.31), H396R (6.75), L298V (3.75), L364I (3.28) | N.A. | Loss of complete hematologic response |
| CP-09-03 | 9/3/2007 | DAS | 2 | M244V (∼70), T315A (∼60) | M244V (73.36), T315A (57.53), F425S (2.18), T406I (1.75) | N.A. | Complete hematologic response but no cytogenetic response |
| AP-10-01 | 3/7/2005 | IM | 1 | E355G (∼50) | E355G (43.75), L341P (21.75), F496L (19.63), L428L (11.01), T315I (10.24), Y456Y (4.81) | N.A. | Progression from CP to AP after 9 mo on IM |
| AP-10-02 | 4/11/2005 | DAS | 2 | T315I (∼50) | T315I (42.60), F317L (1.25) | T315I (42.60), F317L (1.25) | Progression to MBC |
| MBC-11-01 | 4/12/2010 | IM | 1 | M351T (∼100), L248V (∼30) | N.D. | N.D. | Loss of complete hematologic response after 6 mo on IM |
| MBC-11-02 | 8/23/2010 | DAS | 2 | M351T (∼100), F317L (∼70), L248V (∼20) | M351T (100.00), F317L(ttc>tta)(65.52), L248V (19.45), del(248-274) (9.52), F317L(ttc>ctc)(8.52), V299L (1.99) | M351T+F317L(ttc>tta) (52.60), M351T (10.30), M351T+L248V (9.29), M351T+F317L(ttc>tta)+L248V (8.78), M351T+F317L(ttc>ctc) (6.33), M351T+del(248-274)(4.77), M351T+F317L(ttc>tta)+del(248-274)(4.14), M351T+V299L (1.41), M351T+L248V+F317L(ttc>ctc) (1.19), M351T+del(248-274)+F317L(ttc>ctc) (0.61), M351T+F317L(ttc>ctc)+V299L (0.39), M351T+L248V+V299L (0.19) | No hematologic response |
| MBC-12-01 | 3/5/2012 | IM | 1 | F359V (∼70) | F359V (63.24), L387M (4.18), M351T (3.42), V379I (1.62) | F359V (60.26), L387M (2.47), M351T (2.45), F359V+L387M (1.61), V379I (1.12), F359V+M351T (0.87), F359V+V379I (0.50), M351T+L387M (0.10) | Loss of complete cytogenetic response after 12 mo on IM |
| MBC-12-02 | 9/24/2012 | DAS | 2 | L387M (∼60), T315A (∼50), F359V (∼20) | L387M (57.43), T315A (57.19), F359V (16.24), T315I (8.22), F317V (3.98), F317L (1.15) | L387M+T315A (55.99), F359V+T315I (8.06), F359V (4.37), F359V+F317V (3.63), T315A (1.20), L387M+F317L (0.97), F317V (0.19), F359V+F317L (0.18), L387M+T315I (0.16), L387M+F317V (0.16), L387M (0.15) | Loss of complete hematologic response |
| MBC-13-01 | 5/28/2010 | DAS | 2 | T315A (∼100) | T315A (73.75), V299L (8.19) | T315A (72.40), V299L (6.84), T315A +V299L (1.35) | Loss of complete hematologic response after 6 mo on DAS |
| MBC-13-02 | 12/9/2010 | NIL | 3 | T315A (∼100), E255V (∼70), G250E (∼15) | T315A (92.87), E255V (69.74), G250E (12.22), E255K (1.03) | T315A+E255V (65.07), T315A (15.72), T315A+G250E (10.44), E255V (3.73), T315A+E255V+G250E (0.94), G250E (0.84), T315A+E255K (0.70), E255K (0.33) | No hematologic response |
| LBC-14-01 | 12/20/2011 | IM | 1 | G250E (∼40), E255V (∼20) | G250E (34.28), E255V (15.05), E255K (2.34), Y253F (1.10) | G250E (32.91), E255V (14.09), E255K (2.13), Y253F (0.90), G250E+E255V (0.96), G250E+E255K (0.21), G250E+Y253F (0.20) | Progression to lymphoid bast crisis after 32 mo on IM |
| LBC-14-02 | 2/8/2012 | DAS | 2 | T315I (∼30) | T315I (28.92) | T315I (28.92) | No hematologic response |
| LBC-15-01 | 5/26/2011 | IM | 1 | Y253H (∼100) | Y253H (99.88) | Y253H (99.88) | Hematologic relapse |
| LBC-15-02 | 12/5/2011 | DAS | 2 | Y253H (∼50), F317L (∼50) | Y253H (54.90), F317L (54.40) | Y253H+F317L (43.00), Y253H (11.90), F317L (11.40) | Hematologic relapse |
| LBC-16-01 | 3/14/2005 | IM | 1 | E255K (∼100) | E255K (98.84) | E255K (98.84) | Progression to LBC after 9 mo on IM |
| LBC-16-02 | 4/19/2005 | DAS | 2 | E255K (∼100), T315I (∼100) | E255K (99.84), T315I (99.48), L273S (1.30) | E255K+T315I (97.83), E255K (0.71), E255K+T315I+L273S (1.30), T315I (0.35) | No hematologic response |
| LBC-16-03 | 5/23/2005 | DAS | 2 | E255K (∼100), T315I (∼100) | E255K (99.53), T315I (99.53) | E255K+T315I (99.53) | No hematologic response |
| LBC-17-01 | 11/29/2010 | IM | 1 | L387M (∼30) | L387M (34.12) | L387M (28.18) | Loss of complete hematologic response after 6 mo on IM |
| LBC-17-02 | 2/27/2012 | DAS | 2 | L387M (∼100), T315I (∼50), M318V (∼50), F317L (∼50), Y320N (∼50) | L387M (96.33), T315I (51.46), M318V (51.19), F317L (45.21), Y320N (44.79) | L387M+T315I+M318V (47.90), L387M+F317L+Y320N (42.21), L387M (2.63), T315I+M318V (1.82), F317L+Y320N (1.37), L387M+F317L (1.10), L387M+T315I+M318V+Y320N (0.80), L387M+M318V (0.50), L387M+T315I+F317L (0.36), L387M+Y320N (0.25), L387M+T315I (0.25), L387M+T315I+M318V+F317L (0.17), L387M+T315I+Y320N (0.16) | Loss of cytogenetic and hematologic response |
| LBC-18-01 | 11/11/2007 | IM | 1 | F359V (∼60) | F359V (57.89) | F359V (57.89) | Progression to LBC after 3 mo on IM |
| LBC-18-02 | 1/17/2008 | DAS | 2 | None | F317L (8.48), F317I (1.02), F359V (1.02) | F317L (8.48), F317I+F359V (1.02) | Complete hematologic response, partial cytogenetic response |
| LBC-18-03 | 2/12/2008 | DAS | 2 | F359V (∼100), F317I (∼100) | F317I (92.35), F359V (90.11), F317L (4.25) | F359V+F317I (88.01), F317I (4.34), F317L (3.12), F359V+F317L (1.13), F359V (0.97) | Loss of complete hematologic response |
| ALL-19-01 | 9/27/2005 | IM | 1 | Y253H (∼100) | Y253H (99.79) | Y253H (99.79) | Hematologic relapse after 6 mo on IM |
| ALL-19-02 | 1/3/2006 | DAS | 2 | None | P465L (2.78), I432I (1.60), T277I (1.56), E352V (1.41), A474A (1.15) | N.A. | Complete hematologic and cytogenetic response, molecularly detectable disease |
| ALL-19-03 | 3/2/2006 | DAS | 2 | None | A399V (1.58) | A399V (1.58) | Complete hematologic and cytogenetic response, molecularly detectable disease |
| ALL-19-04 | 6/15/2006 | DAS | 2 | Y253H (∼100), T315I (∼50) | Y253H (99.70), T315I (40.29), N336S (5.39), W405R (1.89) | N.A. | Hematologic relapse |
| ALL-20-01 | 1/11/2005 | IM | 1 | M351T (∼100) | M351T (99.91) | M351T (99.91) | Hematologic relapse after 12 mo on IM |
| ALL-20-02 | 3/12/2005 | DAS | 2 | None | L370L (5.45), I432I (2.65) | N.A. | Complete hematologic and cytogenetic response, molecularly detectable disease |
| ALL-20-03 | 1/16/2006 | DAS | 2 | M351T (∼100), F317L (∼50) | M351T (99.77), F317L (53.27) | M351T (46.50), M351T+F317L (53.27) | Hematologic relapse |
| ALL-20-04 | 4/7/2006 | DAS | 2 | M351T (∼100), F317L (∼100) | M351T (95.46), F317L (85.32), A399T (5.64), Y353H (1.00) | M351T+F317L (85.32), M351T+A399T (5.64), M351T (3.50), M351T+F317L+Y353H (1.00) | Progressive disease |
| ALL-21-01 | 5/30/2005 | DAS | 2 | F317L (∼100) | F317L (99.99), M237I (2.03) | F317L (97.96), F317L+M237I (2.03) | Hematologic relapse after 3 mo on DAS |
| ALL-21-02 | 10/27/2005 | NIL | 3 | F317L (∼100), Y253H (∼20) | F317L (98.59), Y253H (17.35) | F317L (81.24), F317L+Y253H (17.35) | Complete hematologic response but no cytogenetic response |
| ALL-21-03 | 1/16/2006 | NIL | 3 | F317L (∼100), Y253H (∼20) | F317L (100.00), Y253H (18.77) | F317L (81.23), F317L+Y253H (18.77) | Hematologic relapse |
| ALL-21-04 | 3/17/2006 | NIL | 3 | F317L (∼100), Y253H (∼100) | F317L (100.00), Y253H (100.00) | F317L+Y253H (100.00) | Progressive disease |
| ALL-22-01 | 12/23/2005 | IM | 1 | F359V (∼100) | F359V (99.76), M237T (2.11) | N.A. | Hematologic relapse after 26 mo on IM |
| ALL-22-02 | 1/29/2005 | DAS | 2 | F359V (∼100), T315I (∼100) | F359V (99.30), T315I (98.77), Y449Y (10.50) | N.A. | Progressive disease |
| ALL-23-01 | 9/14/2011 | DAS | 2 | E255K (∼30), T315I (∼20) | E255K (24.04), T315I (19.37), G303G (1.51) | E255K (20.88), T315I (16.21), E255K+T315I (3.16), G303G (1.51) | Hematologic relapse after 9 mo on DAS |
| ALL-23-02 | 10/27/2011 | PON | 3 | T315I (∼100) | T315I (99.78), Y312C (1.22) | T315I (98.28), T315I+Y312C (1.22) | Complete hematologic response |
| ALL-23-03 | 11/3/2011 | PON | 3 | T315I (∼100) | T315I (99.83) | T315I (99.83) | Complete hematologic response |
| ALL-23-04 | 12/15/2011 | POST-SCT, NONE | / | None | None | None | Complete hematologic and cytogenetic response, molecularly detectable disease |
| ALL-23-05 | 1/11/2012 | POST-SCT, IM | 4 | E255K (∼100) | E255K (99.88) | E255K (99.88) | Complete hematologic response but loss of cytogenetic response |
| ALL-23-06 | 1/25/2012 | PON | 5 | E255K (∼100) | E255K (99.36), S417S (2.21) | N.A. | Hematologic relapse |
| ALL-23-07 | 2/8/2012 | PON | 5 | E255K (∼30) | E255K (22.49) | E255K (22.49) | Complete hematologic response |
| ALL-23-08 | 2/15/2012 | PON | 5 | E255K (∼100) | E255K (99.88) | E255K (99.88) | Partial hematologic response |
| ALL-23-09 | 3/12/2012 | DAS | 6 | E255K (∼70), T315I (∼50) | E255K (76.10), T315I (57.35), Q252H (cag>cac)(14.19), Q252H (cag>cat)(7.49), G250E (1.05) | E255K+T315I (51.60), E255K+Q252H (cag>cac) (13.94), E255K+Q252H (cag>cat) (7.34),T315I (5.75), E255K (2.52), E255K+ G250E (0.70), Q252H (cag>cac) (0.25), G250E (0.35), Q252H (cag>cat) (0.15) | Progressive disease |
| ALL-23-10 | 3/28/2012 | PON | 7 | E255K (∼100), T315I (∼100) | E255K (99.64), T315I (98.56) | E255K+T315I (98.20), E255K (1.44), T315I (0.36) | Progressive disease |
| ALL-24-01 | 6/15/2011 | NIL | 1 | Y253H (∼30) | Y253H (27.35), P465L (1.55) | N.A. | Complete hematologic and cytogenetic response, molecularly detectable disease |
| ALL-24-02 | 7/28/2011 | IM | 2 | Y253H (∼100) | Y253H (99.99), R367L (3.62) | Y253H (96.37), Y253H+R367L (3.62) | Complete hematologic and cytogenetic response, but 1-log increase in BCR-ABL transcript levels |
| ALL-24-03 | 3/1/2012 | DAS | 3 | None | Y302S (1.62) | Y302S (1.62) | Complete hematologic and cytogenetic response, no molecular assessment performed |
| ALL-24-04 | 3/29/2012 | DAS | 3 | Y253H (∼100), T315I (∼100) | Y253H (99.10), T315I (99.10), L273L (1.09) | Y253H+T315I (98.01); Y253H+T315I+L273L (1.09) | Hematologic relapse |
| ALL-25-01 | 12/2/2011 | IM | 1 | Y253H (∼20) | Y253H (18.38) | Y253H (18.38) | Complete hematologic and cytogenetic response, molecularly detectable disease after 18 mo on IM |
| ALL-25-02 | 1/18/2012 | DAS | 2 | Y253H (∼100), T315I (∼100) | Y253H (87.69), T315I (86.16) | Y253H+T315I (80.53), Y253H (7.16), T315I (5.63) | Hematologic relapse |
| ALL-26-01 | 1/11/2012 | IM | 1 | E255K (∼60) | E255K (54.75), F359V (13.81), T315I (3.84) | N.A. | Hematologic relapse after 6 mo on IM |
| ALL-26-02 | 6/16/2012 | DAS | 2 | T315I (∼100) | T315I (99.42) | T315I (99.42) | Progressive disease |
| ALL-27-01 | 1/14/2011 | IM | 1 | T315I (∼100) | T315I (99.87) | T315I (99.87) | Complete hematologic but not cytogenetic response after 3 mo on IM |
| ALL-27-02 | 1/31/2011 | IM | 1 | T315I (∼100) | T315I (99.74) | T315I (99.74) | Stable disease |
| ALL-27-03 | 2/7/2011 | NIL | 2 | T315I (∼80) | T315I (75.55), E255K (9.70), E255V (1.53) | T315I (70.47), E255K (5.10), T315I+E255K (4.60), E255V (1.05), T315I+E255V (0.48) | Stable disease |
| ALL-27-04 | 2/21/2011 | NIL | 2 | T315I (∼60), E255K (∼30) | T315I (53.80), E255K (25.63), E255V (1.33) | T315I (46.10), E255K (18.26), T315I+E255K (7.37), E255V (1.00), T315I+E255V (0.33) | Hematologic relapse |
| ALL-28-01 | 12/5/2007 | IM | 1 | Y253H (∼50), E255K (∼20) | Y253H (56.01), E255K (14.92), T315I (8.33), Q252H (6.32), A269T (3.57), Y253F (1.37) | Y253H (52.30), E255K (14.70), T315I (5.29), Q252H (3.92), Y253H+T315I (2.93), Q252H+A269T (2.40), Y253F (1.26), Y253H+269T (0.63), A269T (0.54), Y253H+E255K (0.15), Y253F+T315I (0.11) | Hematologic relapse after 9 mo on IM |
| ALL-28-02 | 3/26/2008 | DAS | 2 | T315I (∼100) | T315I (90.96) | T315I (90.96) | Progressive disease |
| ALL-29-01 | 6/6/2012 | IM POST-SCT | 2 | T315? | T315M (30.20) | T315M (30.20) | Complete hematologic response, molecularly detectable disease |
| ALL-29-02 | 6/20/2012 | IM | 2 | T315? | T315M (22.32) | T315M (22.32) | Hematologic relapse |
| ALL-29-03 | 7/10/2012 | IM | 2 | T315? | T315M (30.28) | T315M (30.28) | Progressive disease |
| ALL-29-04 | 8/1/2012 | IM | 2 | T315? | T315M (57.28), T315I (1.09) | T315M (57.28), T315I (1.09) | Stable disease |
| ALL-29-05 | 8/22/2012 | PON | 3 | T315?, E255V (∼20) | T315M (28.61), T315I (22.93), E255V (12.40) | T315M (25.20), T315I (20.01), E255V (5.03), T315M+E255V (3.41), T315I+E255V (2.92) | Progressive disease |
| ALL-30-01 | 3/30/2010 | NIL | 1 | E255V (∼40) | E255V (34.70) | E255V (34.70) | Complete hematologic response, molecularly detectable disease after 1 mo on IM |
| ALL-30-02 | 5/12/2010 | IM | 2 | E255V (∼100) | E255V (91.90), V304A (3.91), L302R (1.61), G303W (1.61) | See Figure 3 | Complete hematologic response, molecularly detectable disease with 1-log increase in BCR-ABL transcript levels |
| ALL-30-03 | 7/19/2010 | IM | 4 | E255V (∼50), Y253H (∼50) | E255V (46.50), Y253H (39.30), E255K (1.02) | See Figure 3 | Hematologic relapse |
| ALL-30-04 | 9/7/2010 | DAS | 5 | E255V (∼100) | E255V (91.60), T315I (2.00), E255K (1.80), K262R (1.00) | See Figure 3 | Stable disease |
| ALL-30-05 | 10/5/2010 | DAS | 5 | E255V (∼50), T315I (∼30) | E255V (53.00), T315I (23.00), Q252E (14.30), E255K (2.10) | See Figure 3 | Progressive disease |
| ALL-31-01 | 4/4/2006 | IM | 1 | F317L (∼100) | F317L (99.64) | F317L (99.64) | Hematologic relapse after 5 mo on IM |
| ALL-31-02 | 6/6/2006 | DAS | 2 | D276G (∼50), F317L (∼50), T315A (∼50) | T315A (51.12), F317L (45.81), D276G (44.86) | F317L (33.45), T315A+D276G (32.00), T315A (17.87), F317L+D276G (11.11), D276G (1.25), T315A+F317L (0.75), T315A+F317L+D276G (0.50) | Hematologic relapse after transient hematologic improvement |
| ALL-31-03 | 7/14/2006 | IM | 3 | F317L (∼100) | F317L (99.76), R332R (4.17) | F317L (95.59), F317L+R332R (4.17) | Stable disease |
| ALL-31-04 | 8/23/2006 | IM | 3 | T315A (∼100), G250E (∼100) | T315A (100.00), G250E (90.48) | T315A+G250E (90.48), T315A (9.52) | Progressive disease |
| ALL-31-05 | 9/26/2006 | NIL | 4 | T315A (∼100), G250E (∼100) | T315A (99.83), G250E (99.83) | T315A+G250E (99.83) | Progressive disease |
| ALL-31-06 | 10/24/2006 | NIL | 4 | T315A (∼100), G250E (∼60), D276G (∼30), Y253H (∼30) | T315A (99.96), G250E (67.93), D276G (28.89), Y253H (27.21), Q252E (1.49) | T315A+G250E (65.82), T315A+Y253H+D276G (26.02), T315A (3.76), T315A+G250E+D276G (2.11), T315A+Q252E (0.78), T315A+Y253H (0.48), T315A+Y253H+D276G+Q252E (0.48), T315A+D276G (0.28), T315A+Y253H+Q252E (0.23) | Progressive disease |
| ALL-32-01 | 1/6/2012 | IM | 2 | Y253H (∼100) | Y253H (99.79) | Y253H (99.79) | Hematologic relapse |
| ALL-32-02 | 4/12/2012 | DAS | 3 | None | None | None | Complete hematologic and cytogenetic response, molecularly detectable disease |
| ALL-32-03 | 8/8/2012 | DAS | 3 | None | Y253H (2.49), T315I (1.19) | Y253H (1.30), Y253H+T315I (1.19) | Complete hematologic response, 2-log increase in BCR-ABL transcript levels |
| ALL-32-04 | 9/17/2012 | DAS | 3 | Y253H (∼100), T315I (∼60), F317L(ttc>tta) (∼20) | Y253H (100.00), T315I (79.11), F317L(ttc>tta)(15.70), F317L(ttc>ctc)(4.04) | Y253H+T315I ((78.77), Y253H+ F317L(ttc>tta)(15.36), Y253H+ F317L(ttc>ctc)(4.04), Y253H (1.49), Y253H+T315I+F317L(ttc>tta)(0.34) | Loss of cytogenetic response |
| ALL-33-01 | 3/27/2005 | IM | 1 | G250E (∼100) | G250E (99.50) | G250E (99.50) | Hematologic relapse |
| ALL-33-02 | 9/5/2005 | DAS | 2 | G250E (∼100), F317L (∼70) | N.D. | N.D. | Hematologic relapse after a 5-mo complete hematologic response |
| ALL-33-03 | 12/1/2005 | NIL | 3 | G250E (∼100), F317L (∼50), Y253H (∼30) | G250E (99.99), F317L(ttc>tta)(43.63), Y253H (26.36), V299L (4.84), L248R (1.89), F317L(ttc>ctc)(1.01) | G250E (35.54), G250E+F317L(ttc>tta) (31.90), G250E+Y253H (14.86), G250E+F317L(ttc>tta)+Y253H (9.84), G250E+V299L (2.72), G250E+L248R (1.13), G250E+Y253H+V299L (0.99), G250E+F317L(ttc>tta)+V299L (0.77), G250E+F317L(ttc>tta)+L248R (0.76), G250E+F317L(ttc>ctc) (0.70), G250E+F317L(ttc>tta)+V299L+Y253H (0.36), G250E+F317L(ttc>ctc)+Y253H (0.31) | Progressive disease |
For SS results, mutation-relative abundance was assessed on the basis of variant peak height. In the TKI column, the TKI being administered at the time of analysis is indicated. In the Line column, the number of different lines of TKI therapy that had been administered to the patient is indicated. Disease status and response at each time point are also detailed. Response definitions as in Baccarani et al14 and Vignetti et al.23
In patient ALL-29, “T315?” denotes that 2 overlapping peaks at adjacent positions (c/t at 1091 and t/g at 1092) of codon 315 were identified in the SS chromatogram and the resulting amino acid substitution(s) could not be resolved (see supplemental Figure 9). In patients MBC-11, ALL-23, ALL-32, and ALL-33, the same amino acid changes were found to result from different nucleotide substitutions at the same codons (specified in parentheses). N.A. indicates that clonal analysis could not be possible because of multiple mutations located >450 bp apart. N.D. indicates that amplification with fusion primers was unsuccessful and the sample could not be analyzed with UDS.
ALL, Ph+ acute lymphoblastic leukemia; AP, accelerated phase; CP, chronic phase; DAS, dasatinib; FISH, fluorescence in situ hybridization; IM, imatinib; LBC, lymphoid blast crisis; MBC, myeloid blast crisis; N.A., not assessable; N.D., not done; NIL, nilotinib; PON, ponatinib; SCT, stem cell transplantation.
Cutoff set at variants ≥1%, see “Materials and methods” for details.
Percentage calculated after manual visual inspection of nucleotide sequences at the specific positions where the variants ≥1% were identified.
Samples from 15 CML patients who had achieved stable optimal response to imatinib14 were also analyzed by UDS, for comparison. None were found to harbor point mutations at a cutoff level of ≥1%.
A complex clonal texture is uncovered by clonal analysis of samples harboring multiple mutations by UDS
When multiple mutations fell in KD regions ≤450 bp, it was possible to design “ad hoc” amplicons mapping to those regions (supplemental Figure 1) and take advantage of the clonal nature of the sequence reads generated by UDS to see whether multiple mutations belonged to the same (compound mutations) or to different (polyclonal mutations) BCR-ABL transcripts (hence, defined 1 or multiple Ph+ populations). Supplemental Figures 8 and 9 show 2 representative examples. The great majority of the samples turned out to be a complex mosaic of populations harboring the mutations alone as well as in combination (Table 3). Single mutants were 136 of 274 (49.6%); compound mutants were almost as frequent (Figure 1).
Relative frequency of single as opposed to compound mutants. Compound mutants harboring 2 paired mutations were almost as frequent (38.3%) as single mutants (49.6%) and were by far more frequent than triple and quadruple (they accounted for 105 of the 138 [76%] compound mutants overall identified). Mutated Ph+ populations harboring 3 or 4 mutations in the same BCR-ABL molecule were occasionally detected, but in 1 case only they were found to have achieved clonal dominance over those with 1 or 2 mutations.
Relative frequency of single as opposed to compound mutants. Compound mutants harboring 2 paired mutations were almost as frequent (38.3%) as single mutants (49.6%) and were by far more frequent than triple and quadruple (they accounted for 105 of the 138 [76%] compound mutants overall identified). Mutated Ph+ populations harboring 3 or 4 mutations in the same BCR-ABL molecule were occasionally detected, but in 1 case only they were found to have achieved clonal dominance over those with 1 or 2 mutations.
The landscape of mutated populations is highly dynamic
Longitudinal quantitative follow-up of mutated populations painted an elaborate picture of how the relative frequency of competing populations can ebb and flow over time and with therapeutic intervention (Table 3). Some representative examples are illustrated in Figure 2. The switch from a TKI to another determined the fall of previously dominant population(s) and the rise of new dominant one(s), not necessarily preexisting at the time of switchover (or, at least, not always detectable at the time of switchover with the level of sensitivity allowed by our experimental approach). A new dominant population could be unrelated to the former, rather arising from an unmutated population (as exemplified in Figure 2A-B), or could result from the acquisition of new mutations by the former, which generated compound mutants (Figure 2B-C). In several cases, the evolution of the pattern of mutated populations suggested that the same mutation could have been acquired in parallel by independent populations (ie, 1 unmutated and 1 already harboring a mutation; Figure 2C-D). Selection/deselection of mutated populations could be strikingly rapid. Some compound mutants (M351T+F317L, Y253H+T315I, Y253H+F317L, F359V+T315I) were observed to have higher selective advantage over the respective single mutants. Other compound mutants (H396R+F317L, H396R+T315I, T315I+F317L, E355G+T315I, G250E+F317L, E255V+Y253H, E255V+T315I) were identified that did not overcome the respective single mutants. The E255K+T315I compound mutant was detected in 6 cases: in 3, it became dominant over the E255K and T315I mutants, whereas in another 3 it did not. The triple and quadruple compound mutants detected fluctuated at low levels and were never able to gain dominance, except in patient LBC-17, suggesting that accumulation of >2 mutations, when tolerated, has almost always limited selective advantage.
Mutated populations rise and fall in dominance over time in relation to therapeutic intervention. Graphical illustration of the kinetics of mutated population abundances in 4 representative cases. Arrows indicate the time points at which UDS was performed. Patient IDs are as in Table 3. (A) At the time of first relapse, 7 distinct imatinib-resistant mutated populations were detected. Dasatinib treatment cleared these mutants as quickly as in 2 months, but just as quickly a pan-resistant T315I mutant was found to have emerged. The patient achieved a transient hematologic response after 1 month but lost it shortly after detection of T315I. The question mark indicates that no T315I had been detected by UDS at the time of switchover (at a coverage of 4527 reads, suggesting that either it was present in <1/5000 transcripts, or that it was acquired some time later). (B) At the time of first relapse, a single imatinib-resistant Y253H mutant that accounted for almost 90% of BCR-ABL–positive cells was detected by SS and UDS. After 3 months of dasatinib therapy, the patient had achieved a complete cytogenetic response (no Ph+ metaphases detectable in the bone marrow by standard chromosome banding analysis) although residual disease remained detectable at the molecular level (as assessed by real-time quantitative [RT-Q]-PCR for BCR-ABL transcript). Neither the Y253H (known to be substantially sensitive to dasatinib) nor other mutants were detectable any longer by UDS. After 9 months on dasatinib, the patient was found to have lost the cytogenetic response. UDS showed the coexistence of 3 distinct compound mutants where Y253H was coupled with 3 well-known dasatinib-resistant mutations (a T315I and an F317L resulting from 2 different nucleotide substitutions). It might be hypothesized that the original Y253H-positive cells were never completely eliminated by dasatinib and persisted at very low levels (undetectable by UDS) until they happened to gain a selective advantage again, although de novo acquisition of mutations by previously unmutated cells cannot be ruled out. Interestingly, Y253H and T315I were already detectable by UDS 1 month before. (C) At the time of second relapse, after 6 months of dasatinib therapy, a dasatinib-resistant F317L mutation was detected. UDS portrayed a complex scenario with 3 distinct populations where the former imatinib-resistant Y253H and the newly acquired dasatinib-resistant F317L were present alone and in combination, although the Y253H+F317L compound mutant quantitatively dominated over the F317L- and Y253H-positive ones. It might be hypothesized that the same mutation was acquired in parallel by independent populations (ie, one unmutated and one already positive for the Y253H). (D) After 6 months of second-line dasatinib treatment, during which the patient achieved a complete cytogenetic response but no molecular response, the former imatinib-resistant H396R mutant plus 2 additional populations, 1 harboring a dasatinib-resistant F317L and 1 harboring H396R+F317L, were detected. Three months later, the F317L had become the dominant one, while H396R+F317L and H396R had declined. During dasatinib therapy, a pan-resistant T315I was also acquired because at the time of switchover to nilotinib, a T315I together with H396R+T315I and F317L+T315I compound mutants were already detectable by UDS (4.44%, 0.76%, and 0.22%, respectively). As quickly as in 2 months, nilotinib treatment selected the T315I mutant that expanded, achieving almost full dominance. IM, imatinib; DAS, dasatinib; NIL, nilotinib.
Mutated populations rise and fall in dominance over time in relation to therapeutic intervention. Graphical illustration of the kinetics of mutated population abundances in 4 representative cases. Arrows indicate the time points at which UDS was performed. Patient IDs are as in Table 3. (A) At the time of first relapse, 7 distinct imatinib-resistant mutated populations were detected. Dasatinib treatment cleared these mutants as quickly as in 2 months, but just as quickly a pan-resistant T315I mutant was found to have emerged. The patient achieved a transient hematologic response after 1 month but lost it shortly after detection of T315I. The question mark indicates that no T315I had been detected by UDS at the time of switchover (at a coverage of 4527 reads, suggesting that either it was present in <1/5000 transcripts, or that it was acquired some time later). (B) At the time of first relapse, a single imatinib-resistant Y253H mutant that accounted for almost 90% of BCR-ABL–positive cells was detected by SS and UDS. After 3 months of dasatinib therapy, the patient had achieved a complete cytogenetic response (no Ph+ metaphases detectable in the bone marrow by standard chromosome banding analysis) although residual disease remained detectable at the molecular level (as assessed by real-time quantitative [RT-Q]-PCR for BCR-ABL transcript). Neither the Y253H (known to be substantially sensitive to dasatinib) nor other mutants were detectable any longer by UDS. After 9 months on dasatinib, the patient was found to have lost the cytogenetic response. UDS showed the coexistence of 3 distinct compound mutants where Y253H was coupled with 3 well-known dasatinib-resistant mutations (a T315I and an F317L resulting from 2 different nucleotide substitutions). It might be hypothesized that the original Y253H-positive cells were never completely eliminated by dasatinib and persisted at very low levels (undetectable by UDS) until they happened to gain a selective advantage again, although de novo acquisition of mutations by previously unmutated cells cannot be ruled out. Interestingly, Y253H and T315I were already detectable by UDS 1 month before. (C) At the time of second relapse, after 6 months of dasatinib therapy, a dasatinib-resistant F317L mutation was detected. UDS portrayed a complex scenario with 3 distinct populations where the former imatinib-resistant Y253H and the newly acquired dasatinib-resistant F317L were present alone and in combination, although the Y253H+F317L compound mutant quantitatively dominated over the F317L- and Y253H-positive ones. It might be hypothesized that the same mutation was acquired in parallel by independent populations (ie, one unmutated and one already positive for the Y253H). (D) After 6 months of second-line dasatinib treatment, during which the patient achieved a complete cytogenetic response but no molecular response, the former imatinib-resistant H396R mutant plus 2 additional populations, 1 harboring a dasatinib-resistant F317L and 1 harboring H396R+F317L, were detected. Three months later, the F317L had become the dominant one, while H396R+F317L and H396R had declined. During dasatinib therapy, a pan-resistant T315I was also acquired because at the time of switchover to nilotinib, a T315I together with H396R+T315I and F317L+T315I compound mutants were already detectable by UDS (4.44%, 0.76%, and 0.22%, respectively). As quickly as in 2 months, nilotinib treatment selected the T315I mutant that expanded, achieving almost full dominance. IM, imatinib; DAS, dasatinib; NIL, nilotinib.
Further insights into BCR-ABL KD sequence evolution: a phylogenetic approach
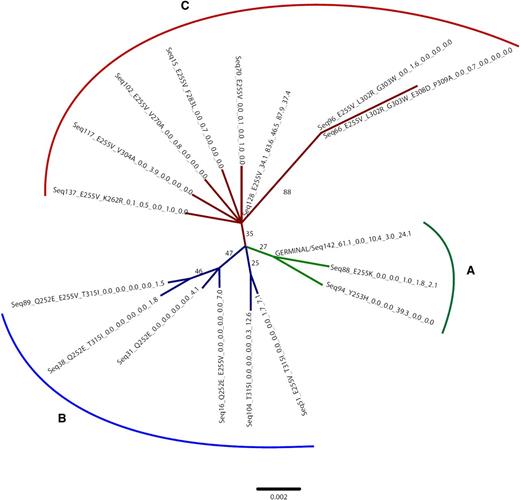
Figure 3 shows the neighbor joining bootstrap consensus tree for sequences recovered at 5 time points for patient ALL-30. The tree can be considered as consisting of 3 clusters. Reassuringly, within each cluster, both the timing of first observation of each sequence and the types of amino acid substitutions present are fairly consistent, suggesting that, despite the low bootstrap support for key branches, relevant hypotheses can be generated from the phylogenetic analysis. The tree is consistent with E255V occurring once, along the branch dividing cluster A from the rest of the tree. This substitution, which affords resistance to imatinib and nilotinib, is observed in the majority of the sequences in clusters B and C, and was already frequent in ALL-30-01 after 6 weeks of nilotinib treatment (seq128). Indeed, in ALL-30-02 (after 6 weeks on imatinib), seq128 is the dominant haplotype, although other amino acid substitutions are observed in cluster C combined with E255V. The tree and the timing of emergence of these haplotypes are both consistent with the hypothesis that they evolved from clones represented by seq128. In ALL-30-03, 2 other imatinib- and nilotinib-resistant mutations (E255K and Y253H) are observed. However, the tree indicates that these derive from an unmutated haplotype rather than on the predominant E255V background. The final clinically relevant substitution, the pan-resistant T315I, is observed in cluster B, often in conjunction with E255V (and Q252E whose possible clinical relevance is unknown). T315I thus appears to have arisen at the base of this cluster on an E255V background, rather than from the seq128 haplotype observed in cluster C. This observation suggests that, as in the cases of E255K and Y253H, new TKI-resistant mutations can arise from low frequency, possibly slow-proliferating clones that do not represent the majority of the circulating haplotypes at earlier time points. Interestingly, sequences 31, 38, and 104 do not exhibit E255V, suggesting that after withdrawal of imatinib/nilotinib treatment, reversion of this mutation can occur. Indeed, the relatively high frequency of seq104 might be consistent with the hypothesis that such a reversion is positively selected in the context of T315I under dasatinib treatment. An alternative interpretation of the distribution of T315I would be that it arose on an unmutated background, and that E255V subsequently arose independently in some lines within cluster B (as well as on the branch linking cluster C with clusters A and B).
Neighbor joining bootstrap consensus tree (K2P distances) for nucleotide sequences recovered at 5 time points for patient ALL-30 (samples ALL-30-01 through ALL-30-05 in Table 3). The patient was initially enrolled in a clinical study testing the use of nilotinib and imatinib administered in rotation in 6-week cycles. Sample ALL-30-01 (time point 1) was collected after the first (nilotinib) cycle. Sample ALL-30-02 (time point 2) was collected after the second (imatinib) cycle. Sample ALL-30-03 (time point 3) was collected after the fourth cycle. Sample ALL-30-04 was collected after the patient had discontinued the nilotinib-imatinib rotation schedule for hematologic relapse and had received 2 weeks of dasatinib treatment. Sample ALL-30-05 was collected after 6 weeks of dasatinib treatment and a few days before the exitus. Branches with <25% support are collapsed to polytomies. Amino acid substitutions are mapped onto the tree and shown for each sequence along with the percentage of reads that each sequence represented at each time point. “Germline” indicates the unmutated sequence. The scale bar shows substitutions per site. The colors identify the 3 sequence clusters that can be recognized (A-C; see “Further insights into BCR-ABL KD sequence evolution: a phylogenetic approach” for details).
Neighbor joining bootstrap consensus tree (K2P distances) for nucleotide sequences recovered at 5 time points for patient ALL-30 (samples ALL-30-01 through ALL-30-05 in Table 3). The patient was initially enrolled in a clinical study testing the use of nilotinib and imatinib administered in rotation in 6-week cycles. Sample ALL-30-01 (time point 1) was collected after the first (nilotinib) cycle. Sample ALL-30-02 (time point 2) was collected after the second (imatinib) cycle. Sample ALL-30-03 (time point 3) was collected after the fourth cycle. Sample ALL-30-04 was collected after the patient had discontinued the nilotinib-imatinib rotation schedule for hematologic relapse and had received 2 weeks of dasatinib treatment. Sample ALL-30-05 was collected after 6 weeks of dasatinib treatment and a few days before the exitus. Branches with <25% support are collapsed to polytomies. Amino acid substitutions are mapped onto the tree and shown for each sequence along with the percentage of reads that each sequence represented at each time point. “Germline” indicates the unmutated sequence. The scale bar shows substitutions per site. The colors identify the 3 sequence clusters that can be recognized (A-C; see “Further insights into BCR-ABL KD sequence evolution: a phylogenetic approach” for details).
Discussion
UDS has revolutionized the way we can approach the study of drug-resistant cellular populations. Virology is one of the fields that has benefited most from the possibility of highlighting heretofore-undetectable minor mutated variants and performing haplotype analysis, thus allowing for characterization and monitoring of population diversity in HIV and hepatitis viruses.24 There seem to be striking similarities between viral populations and Ph+ leukemia populations, in that they both tend to accumulate mutations to escape antiviral or TKI therapy, respectively. In both scenarios, testing for resistance-associated mutations is important to guide selection of the most appropriate treatment regimen. We have now found that there may be a high degree of heterogeneity, recalling that of viral quasispecies, in BCR-ABL KD sequences from patients failing multiple sequential TKIs. In 55% of the samples, “major” mutations (detectable by SS) were found to be only “the tip of the iceberg”: UDS revealed that additional “minor” (<10%-15%) mutations might be present, even by using a quite “conservative” lower abundance cutoff of 1%. In 51% of the cases, minor mutations could be recognized as poorly sensitive either to the TKI being administered or to the previous TKI received. They most likely corresponded either to outgrowing mutations anticipating an imminent relapse (as for the pan-resistant T315I in samples CP-01-02, AP-10-01, ALL-26-01, ALL-28-01, ALL-29-04) or, more rarely, to “withdrawing” mutants not (yet) entirely deselected by the change in TKI (as in CP-03-06, CP-04-03, CP-06-02, and CP-06-03). In other cases, they could rather be seen as the result of “secondary route(s)” toward resistance followed by some Ph+ cells as an alternative to the one(s) leading to the dominant population(s) (as in AP-10-02, LBC-14-01, ALL-27-03, ALL-32-04, ALL-33-03). In a not negligible proportion of cases, however, minor mutations were either silent or never reported in association with TKI resistance. One would expect such mutations to be always colocalized (“passengers”) on BCR-ABL molecules already harboring a TKI-resistant mutation (“driver”; as in sample CP-03-05); however, this was not always the case, suggesting that, depending on the specific context, a mutant that per se would not be so markedly insensitive to treatment may somehow, at least temporarily, survive and expand to a certain extent, although it will never be able to achieve dominance.
An even higher degree of complexity emerged when we tried to reconstruct the different haplotypes in the samples harboring multiple mutations (Table 3). In the early days of the second-generation TKI era, one study had suggested that compound mutations might be particularly insidious because they may be associated with enhanced oncogenic potential and TKI insensitivity than the separate mutants would exhibit.11 However, the clinical relevance of compound as against polyclonal mutations in patients failing multiple TKIs has long been underestimated. This is mainly because SS, the most widely used method for routine BCR-ABL KD mutation screening,3 precludes the determination of whether multiple mutations are colocated on the same BCR-ABL molecule: only SS of an appropriate number of bacterial colonies with cloned-in BCR-ABL KD would bypass this limitation, but the labor-intensiveness of this approach limits its use to exploratory studies of small patient cohorts.25 Our UDS approach revealed that compound and polyclonal mutations are not 2 mutually exclusive scenarios. This reminds the pattern observed for some patients with myeloproliferative disorders in which JAK2 and TET2 mutations were observed in the same clone and also in different clones.26 Thus, sequential changes in TKI-selective pressure result in heterogeneous mosaics of Ph+ populations harboring different mutations or mutation combinations. Longitudinal observation of the dynamics of these populations in vivo in relation to TKI treatment might suggest that some compound mutants (M351T+F317L, Y253H+T315I, Y253H+F317L) are selectively at an advantage over single mutants, whereas others (H396R+F317L, H396R+T315I, T315I+F317L) are not. On the other hand, the same E255K+T315I (the most frequent compound mutant identified in our samples) was found to achieve dominance in some cases but not in others, suggesting that the patient-specific context, including number and features of coexisting populations, may shape the fitness of a compound mutant and a general rule cannot easily be inferred.
Our results on the complexity of mutated populations and on their dynamics under sequential TKI treatment concur to depict a model in which evolution of BCR-ABL–positive cells is mainly shaped by TKI-selective pressure (whether microenvironment and/or the immune surveillance may also be playing a role is currently unknown) and the fitness of each mutated population is the net result of an “absolute” fitness (the ability to survive treatment depending on the intrinsic sensitivity to the specific TKI administered) and of a “relative” fitness (the ability to survive the competition with all other coexisting populations). For the majority of arising mutants, the fitness will luckily not be high enough to sustain clonal expansion (at least not over a certain threshold), and the population will rather face extinction. Other mutants will expand, although the switch to another TKI may later turn a “fit” into an “unfit” population and lead to an evolutionary “dead-end”. Rare cells within existing mutated populations will gain additional mutations (giving rise to compound mutants), and this will be either an advantage or a disadvantage depending on the type of mutation. Recent whole-genome sequencing studies have challenged the longstanding model of linear evolution of tumors through stepwise accumulation of genetic hits in a founding clone.27 In multiple myeloma, for example, tumor progression has been shown to proceed in a branching rather than in a linear manner, leading to substantial clonal diversity and coexistence of wide genetic heterogeneity.28,29 On a smaller scale, the same seems to apply to drug-resistant BCR-ABL mutants.
Two clinical caveats derive from our findings. We here show that information provided by SS may not always be sufficient to predict responsiveness to a TKI. SS is the currently recommended method for BCR-ABL KD mutation analysis, and its results do influence TKI selection3,30-33 : for example, if an imatinib-resistant patient was found to harbor a Y253H by SS, switching to dasatinib rather than to nilotinib should be considered, in light of the fact that this mutant has been shown both in vitro and in vivo to be fully sensitive to dasatinib but poorly sensitive to nilotinib.6,7 If the scenario turned out to be more complex with additional minor mutated populations, clinical efficacy of the selected TKI might be compromised, or only transient.34,35 Such a case should sound as a warning to the physician and a closer monitoring could be beneficial. We acknowledge that in very few instances the information added by UDS would alter TKI selection: in case one of these minor mutated populations were a T315I-positive one, for example, ponatinib might become preferable over dasatinib.4,36 However, if multiple low-level mutations were detected, it would become difficult to predict their role in relapse and progression and to place this information into a therapeutic decision algorithm. We also acknowledge that the clinical significance of low-level mutation detection remains, at present, unclear. Methods with sensitivities ranging between 1% and 10% have been set up and proposed by several authors.37-40 Nevertheless, the optimal lower detection limit allowing for identification of only those mutants that will outgrow and trigger relapse has not been established yet,41,42 although we believe that prospective application of UDS in a large series of patients might ultimately clarify this issue.
Another “warning” from our observations is that sensitivity of a single or a compound mutant to a TKI in vivo might be dictated by more complex factors than the mere in vitro IC50 (half maximal inhibitory concentration) value. If the in vivo sensitivity of any single or compound mutant is influenced by competition with other coexisting mutants, physicians should be warned against using the in vitro IC50 values to estimate the degree of insensitivity of a specific Bcr-Abl mutant protein to a specific TKI.43,44
Many foresee that UDS will soon find routine diagnostic application, especially in the field of hematologic malignancies where the pivotal IRON study has recently provided first evidence of technical feasibility and high concordance of results across multiple laboratories.16 Although it is premature to predict whether UDS will replace SS as the gold standard for BCR-ABL KD mutation analysis, further evaluation of this technology is highly warranted. Currently, the ongoing IRON II study45 is setting-up and testing UDS strategies for the detection of a wide spectrum of mutations, including the BCR-ABL KD screening strategy herein presented.
The online version of this article contains a data supplement.
There is an Inside Blood commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the Interlaboratory Robustness of Next-generation sequencing (IRON) Phase II Study Group members for helpful discussions on the UDS assay.
This work was supported by European Leukemia Net, Fondazione Cassa di Risparmio in Bologna, Associazione Italiana contro le Leucemie, Programmi di Ricerca di Rilevante Interesse Nazionale 2009 (prot. 2009JSMKY), International Grant Agency Ministry of Health of the Czech Republic NT11555.
Authorship
Contribution: S.S. designed the research, performed experiments, analyzed and interpreted results, and wrote the paper; C.D.B. performed experiments and analyzed and interpreted results; K.M.P., A.B., I.I., C.V., M.T.B., and F. Cattina performed experiments; D.H. and M.I. performed phylogenetic analyses; F. Castagnetti, C.P., G.G., F.P., H.K., D.R., P.B., G.B., B.G., and G.R. provided patient samples and clinical data; A.K., T.H., and A.R. provided vital technical and bioinformatics support for the development of the UDS assay; M.C., M.B., and G.M. coordinated the clinical and research team activities and supervised the study; and all authors gave final approval for submission.
Conflict-of-interest disclosure: S.S. was a consultant for and received honoraria from Novartis, Bristol-Myers Squibb, and Ariad. F. Castagnetti, G.G., and G.R. were consultants for and received honoraria from Novartis and Bristol-Myers Squibb. A.K. received honoraria from Roche Diagnostics. T.H. and K.M.P. received research funding from Roche Diagnostics. M.B. and G.M. were consultants for and received honoraria from Novartis, Bristol-Myers Squibb, Ariad, and Pfizer. M.I. is a Roche employee. The remaining authors declare no competing financial interests.
Correspondence: Simona Soverini, Institute of Hematology “L. e A. Seràgnoli”, Via Massarenti 9, 40138 Bologna, Italy; e-mail: simona.soverini@unibo.it.

![Figure 1. Relative frequency of single as opposed to compound mutants. Compound mutants harboring 2 paired mutations were almost as frequent (38.3%) as single mutants (49.6%) and were by far more frequent than triple and quadruple (they accounted for 105 of the 138 [76%] compound mutants overall identified). Mutated Ph+ populations harboring 3 or 4 mutations in the same BCR-ABL molecule were occasionally detected, but in 1 case only they were found to have achieved clonal dominance over those with 1 or 2 mutations.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/122/9/10.1182_blood-2013-03-487728/4/m_1634f1.jpeg?Expires=1765937612&Signature=I0EjbBeV5IRxHucrcZhPpzeVjdnzm-NaclAeWP7azi2j0EYwDfkgLZWNn0zobF-Ha1ZdziVhiZyXKp5vS3mbtYrKxp6K6A~oA2tfMDPO3ejSwH2C2D3GB3gr6z95ReeVr3Fw8hgMeujiKhAsk4h3NjQIbn6jX~MZwxS6~2a7I8fzioVkMRUHHuC3clzzwAituernu6oZpMVqbuC7-n6R0Gy-uMCudP9VJuhhy-wcN2uQXnlYHgswwI6KTi1L6ugJAbSV316u-xthY7VQ~tmuScMwg6OJHtPcgGkNyINF3~9fjB8wD6vPhYY4ZmLkkWAJaPybnqdg4S9~9oEXhfrjYQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 2. Mutated populations rise and fall in dominance over time in relation to therapeutic intervention. Graphical illustration of the kinetics of mutated population abundances in 4 representative cases. Arrows indicate the time points at which UDS was performed. Patient IDs are as in Table 3. (A) At the time of first relapse, 7 distinct imatinib-resistant mutated populations were detected. Dasatinib treatment cleared these mutants as quickly as in 2 months, but just as quickly a pan-resistant T315I mutant was found to have emerged. The patient achieved a transient hematologic response after 1 month but lost it shortly after detection of T315I. The question mark indicates that no T315I had been detected by UDS at the time of switchover (at a coverage of 4527 reads, suggesting that either it was present in <1/5000 transcripts, or that it was acquired some time later). (B) At the time of first relapse, a single imatinib-resistant Y253H mutant that accounted for almost 90% of BCR-ABL–positive cells was detected by SS and UDS. After 3 months of dasatinib therapy, the patient had achieved a complete cytogenetic response (no Ph+ metaphases detectable in the bone marrow by standard chromosome banding analysis) although residual disease remained detectable at the molecular level (as assessed by real-time quantitative [RT-Q]-PCR for BCR-ABL transcript). Neither the Y253H (known to be substantially sensitive to dasatinib) nor other mutants were detectable any longer by UDS. After 9 months on dasatinib, the patient was found to have lost the cytogenetic response. UDS showed the coexistence of 3 distinct compound mutants where Y253H was coupled with 3 well-known dasatinib-resistant mutations (a T315I and an F317L resulting from 2 different nucleotide substitutions). It might be hypothesized that the original Y253H-positive cells were never completely eliminated by dasatinib and persisted at very low levels (undetectable by UDS) until they happened to gain a selective advantage again, although de novo acquisition of mutations by previously unmutated cells cannot be ruled out. Interestingly, Y253H and T315I were already detectable by UDS 1 month before. (C) At the time of second relapse, after 6 months of dasatinib therapy, a dasatinib-resistant F317L mutation was detected. UDS portrayed a complex scenario with 3 distinct populations where the former imatinib-resistant Y253H and the newly acquired dasatinib-resistant F317L were present alone and in combination, although the Y253H+F317L compound mutant quantitatively dominated over the F317L- and Y253H-positive ones. It might be hypothesized that the same mutation was acquired in parallel by independent populations (ie, one unmutated and one already positive for the Y253H). (D) After 6 months of second-line dasatinib treatment, during which the patient achieved a complete cytogenetic response but no molecular response, the former imatinib-resistant H396R mutant plus 2 additional populations, 1 harboring a dasatinib-resistant F317L and 1 harboring H396R+F317L, were detected. Three months later, the F317L had become the dominant one, while H396R+F317L and H396R had declined. During dasatinib therapy, a pan-resistant T315I was also acquired because at the time of switchover to nilotinib, a T315I together with H396R+T315I and F317L+T315I compound mutants were already detectable by UDS (4.44%, 0.76%, and 0.22%, respectively). As quickly as in 2 months, nilotinib treatment selected the T315I mutant that expanded, achieving almost full dominance. IM, imatinib; DAS, dasatinib; NIL, nilotinib.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/122/9/10.1182_blood-2013-03-487728/4/m_1634f2.jpeg?Expires=1765937612&Signature=fti~QhZG6j8VBYx5cG9GAE2D0YitT8wQboTNrKnM46S-v3LBIytuTuzVjfCVxn2o7LyqG-pQA4L8DNNgQWQ1f5bl7esWSxm8qACuTpS1qKmp3Gjwx0GY~xZ6fM~P28l7fhHJWU-Xep1dNqZDicm1sI7SGYa2qhVc0zBes--y8x7Z4vc~qV-r1bqVu6BQ3SNCGwMgkZ0N7uQF2cb1nXX4BGg646rCitnPZ1qsvHFbOoL6SU~FdeyZMRo4ZAwLnJyzkM5ijGWVEahBo9NKTmPVO2kLuxx9PqeqtO9eGpr~tg1qE6J~hv~-mkcl0Se~1ue1245YhOzc4r4Kl28enCGqKw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)