Key Points
Jumping translocations of 1q12 (JT1q12) provide a mechanism for the deletion of 17p in cytogenetically defined high-risk myeloma.
Sequential JT1q12s introduce unexpected copy number gains and losses in receptor chromosomes during subclonal evolution.
Abstract
Multiple myeloma (MM) is a B-cell malignancy driven in part by increasing copy number alterations (CNAs) during disease progression. Prognostically significant CNAs accumulate during clonal evolution and include gains of 1q21 and deletions of 17p, among others. Unfortunately, the mechanisms underlying the accumulation of CNAs and resulting subclonal heterogeneity in high-risk MM are poorly understood. To investigate the impact of jumping translocations of 1q12 (JT1q12) on receptor chromosomes (RCs) and subsequent clonal evolution, we analyzed specimens from 86 patients selected for unbalanced 1q12 aberrations by G-banding. Utilizing spectral karyotyping and locus-specific fluorescence in situ hybridization, we identified 10 patients with unexpected focal amplifications of an RC that subsequently translocated as part of a sequential JT1q12 to one or more additional RCs. Four patients exhibited amplification and translocation of 8q24 (MYC), 3 showed amplification of 16q11, and 1 each displayed amplification of 18q21.3 (BCL2), 18q23, or 4p16 (FGFR3). Unexpectedly, in 6 of 14 patients with the combination of the t(4;14) and deletion of 17p, we identified the loss of 17p as resulting from a JT1q12. Here, we provide evidence that the JT1q12 is a mechanism for the simultaneous gain of 1q21 and deletion of 17p in cytogenetically defined high-risk disease.
Introduction
Multiple myeloma (MM) is a plasma cell disorder that is characterized by complex genetic alterations, including both primary and secondary cytogenetic aberrations.1 To help optimize clinical diagnostic testing and provide guidelines for cytogenetic risk categories, the International Myeloma Working Group (IMWG) recommends the use of 3 markers, including the t(4;14), deletion 17p, and gain of 1q21.2 Two of these markers, the t(4;14) and del17p, when identified together by interphase fluorescence in situ hybridization (iFISH), are considered cytogenetically defined high-risk disease.2 Interestingly, the deletion of 17p and gain of 1q21 are both secondary chromosomal copy number alterations (CNAs) known to occur during disease progression. In fact, molecular profiling studies have demonstrated that CNAs, including the deletion 17p and gain of 1q21, accumulate during tumor progression and contribute to clonal heterogeneity and poor response to therapy.3-10 These and other CNAs can involve whole-chromosome gains or losses, whole-arm gains or losses, or very short (focal) amplifications or deletions.11 In MM, the most common segmental losses include deletions of 1p, 6q, 8p, 13q, 16q, 17p, and 22q, whereas the most common gains are of 1q, 6p, 9q, 11q, 12q, 15q, 17q, and 19q.3-10 Of the focal copy number (CN) gains, the one most frequently found in proliferative, relapsed, and/or refractory myeloma is the gain of the 1q21 region, which occurs in about 40% of cases.12 Among the amplified and/or deregulated genes in this region are MCL1, MUC1, IL6Ra, BCL9, ANP32E,CKS1B, S100A4, PSMD4, and UBE2Q1, as well as many others.3-16 As a result of this amplification, interstitial copy number gains of 1q21 have been suggested as one of a number of potential “driver” lesions of myeloma progression due to the numerous potentially relevant oncogenes in this region.17
The primary mechanism for amplification of 1q21 in MM is the whole-arm CN aberration known as the jumping translocation 1q (JT1q12).18-21 A JT1q12 occurs when a duplication of the 1q12 pericentromeric region translocates (jumps) as a donor chromosome (DC) segment to one or more receptor chromosomes (RCs). In most cases, JT1q12 becomes a stable clonal aberration; however, the JT1q12 can also translocate to a second RC or, alternatively, “jump off” a RC and be lost from the cell in the form of a single or multiple micronuclei.19 Although any number of possible combinations of JT1q12 aberrations are possible, 3 primary types occur. The first type is a whole-arm 1q12 translocation to the telomere of an RC resulting in only a CN gain of 1q21 and no loss in the RC. A second type is a whole-arm 1q12 translocation resulting in a simultaneous CN gain of 1q21 and also a whole-arm CN loss in the RC.18 The third type of 1q21 CN gain involves the focal amplification of the 1q12∼23 region by direct or inverted duplications and/or a breakage-fusion-bridge cycle mechanism.22
The aim in this study was to clarify the impact of JT1q12 on CNAs involving RCs. We have identified a novel type of the JT1q12 aberration in which the same JT1q12 sequentially translocates from one RC to a second RC and, in some cases, focally amplifies a segment of the original RC that becomes part of the second translocation. The same JT1q12s can also be a source of deletions in RCs and, therefore, can introduce both CN gains and losses. Unexpectedly, in 6 of 14 patients with t(4;14), we identified the JT1q12 as the cause of the deletion of 17p, providing evidence for a novel mechanism for the introduction of cytogenetically defined high-risk disease.
Patients and methods
The institutional review board of the University of Arkansas for Medical Sciences approved the research studies, and all subjects provided written informed consent approving the use of their samples for research purposes. This study was conducted in accordance with the Declaration of Helsinki.
Metaphase chromosomes were prepared from bone marrow, fine-needle aspirate, and plural effusion specimens and processed for G-band analysis as previously described.23 Karyotype designations of cytogenetically related clones are described according to International System for Human Cytogenetic Nomenclature 2013 guidelines.24
Patient sample selection was based on 2 factors: (1) the presence of at least 1 additional copy of 1q21 identified by G-banding and suspected unresolved aberrations in the routine clinical studies, and (2) the availability of suitable quality archived methanol:acetic acid–fixed cytogenetic cell pellet for further FISH and/or spectral karyotyping (SKY) analysis. Initially, 86 patients with gains of 1q21 aberrations were screened by locus-specific metaphase FISH to identify adverse immunoglobulin heavy chain (IGH) translocations and TP53 deletions cryptic to G-banding. Specimens with unresolved aberrations by FISH were further scrutinized by SKY.
FISH and SKY
Probes used to demark the pericentromeric region of chromosome 1 included α sat (1p11.1-1q11.1) and satII/III (1q12) (Vysis, Downers Grove, IL). Two probe sets for detection of IGH translocations included IGH/FGFR3 dual-color, dual-fusion probe set and the IGH dual-color, break-apart rearrangement probe (Vysis). Locus-specific probes used included 4p16 (FGFR3), 8q24 (C-MYC), 11q23 (MLL), 16q11.2,18q24 (BCL2), and 18q23 (Vysis). Human bacterial artificial chromosome clones were allocated according to publication of the human genome resource of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/projects/genome/guide/human/).25 Both FISH probe preparation and SKY procedures were performed as previously described.26,27 Image acquisition for FISH and SKY was performed using a SD200 Spectracube (Applied Spectral Imaging, Carlsbad, CA) mounted on a Zeiss Axioplan II microscope (Gottingen, Germany). 4′,6 diamidino-2-phenylindole images were captured and then inverted and enhanced by SKY View software to produce G-band–like patterns on the chromosomes. Original magnification of all G-band and FISH images was ×1000. Original magnification for SKY images was ×630. In cases with segmental aberrations identified by SKY, locus-specific probes to these regions (eg, MYC) were used for calculation of CNAs.
Results
Complete G-band karyotype results for all 86 patients are provided in supplemental Table 1, available on the Blood Web site. Karyotype modal chromosome numbers were classified as either hyperdiploid with 47 to 75 chromosomes or nonhyperdiploid with <46 and/or >75 chromosomes. Recurring whole-arm translocations by G-banding were found in 32 patients. Twelve showed a der(1;16)(q12;q11), 7 a der(1;19)(q12;q10), 9 a der(1;17)(q12;q10), and 2 each a der(1;6)(q12;q10), der(1;8)(q12;q10), or der(1;9)(q12;q10). Recurring telomeric JT1q12s were found in 24 patients: 6 with der(2)t(1;2)(q12;q37), 2 with der(3)t(1;3)(q12;q29), 4 with der(8)t(1;8)(q12;q24), 4 with der(15)t(1;15)(q12;q10), 6 with der(18)t(1;18)(q12;q23), and 2 with der(22)t(1;22)(q12;q10). Fourteen patients showed direct duplications of 1q21∼22, 12 with isochromosome 1q, and 2 had inverted duplications 1q21∼23 (supplemental Table 1).
Metaphase FISH analysis for IGH translocations identified 27 patients with t(4;14), 11 with t(14;16), 4 each with t(11;14) or t(6;14), and 1 with t(14;20) (supplemental Table 2). Thirty-one patients showed no evidence of an IGH translocation (supplemental Table 2). CN for 1q21 included 28 patients with a CN of 3, 25 with a CN of 4, 14 with a CN of 5, 7 with a CN of 6, 7 with a CN of 7 to 8, 2 with a CN of 9 to 10, and 1 patient with a CN of 12. CNAs for 1q12 were similar to CNAs for 1q21 in all patients except patients 4, 7, 17, 19, 46, 48, 53, 60, 64, 68, 73, 76, 81, and 84, where 7 were associated with the loss of 1q12 and 7 with the loss of 1q21. Locus-specific CNAs for 17p (TP53) identified 31 patients with a CN of 1, 53 with a CN of 2, and 2 with a CN of 3 (supplemental Table 2).
JT1q12 introduced unexpected CNAs in 16 patients (Table 1). Ten of these patients showed segmental gains of a RC detected by SKY and/or FISH. Of these 10 patients, 4 (patients 1, 31, 52, and 62) showed gains of 8q24, 3 (patients 17, 59, and 70) of 16q11, and 1 each of 18q21 (patient 15), 18q23 (patient 34), and 4p16 (patient 32) involving amplifications of RC cosegregating with the 1q in stemlines or subclones. Seven patients (12, 18, 34, 36, 69, 77, and 86) showed deletions of 17p resulting from JT1q12, 6 of which occurred in clones with t(4;14) and 1 in a clone with t(11;14) (Table 1).
Results of patients studied in further detail with designations of stemlines and subclones, IGH translocations, and CNAs for 1q12, 1q21, 17p, and additional CNAs identified in RCs by FISH and SKY
| Patient # . | Clones . | IGH translocation . | CN 1q12 . | CN 1q21 . | CN 17p . | CNAs in RC . |
|---|---|---|---|---|---|---|
| Patients with der(1;8) | ||||||
| 1 | Stemline | Normal | 3 | 3 | 2 | MYC × 3,del(3q) |
| 31 | Stemline | Normal | 3 | 3 | 1 | MYC × 2 |
| Subclone | Normal | 4 | 4 | 1 | MYC × 3,del(7q) | |
| 52 | Stemline | t(4;14) | 3 | 3 | 2 | MYC × 3 |
| 62 | Stemline | t(14;16) | 8 | 8 | 1 | MYC × 8 |
| Patients with der(1;16) | ||||||
| 17 | Stemline | Normal | 5 | 5 | 1 | 16q11 × 4 |
| 59 | Stemline | t(4;14) | 8 | 8 | 1 | 16q11 × 8,del(11p) |
| Subclone | t(4;14) | 11 | 11 | 1 | 16q11 × 10,del(11p) | |
| 70 | Stemline | t(4;14) | 3 | 3 | 1 | 16q11 × 3 |
| Patients with complex sequential CNAs | ||||||
| 15 | Stemline | t(1;14) | 8 | 8 | 2 | BCL2 × 3, MLL × 4 |
| 32 | Stemline | t(4;14) | 8 | 8 | 1 | 1p36 × 5, FGFR3 × 5 |
| Patients with der(1;17) | ||||||
| 12 | Stemline | t(4;14) | 3 | 3 | 1 | TP53 × 1 |
| Subclone | t(4;14) | 2 | 2 | 1 | TP53 × 1 | |
| Subclone | t(4;14) | 3 | 3 | 1 | TP53 × 1 | |
| 18 | Stemline | t(11;14) | 3 | 3 | 1 | TP53 × 1 |
| 34 | Stemline | t(4;14) | 3 | 3 | 3 | TP53 × 3 |
| Subclone | t(4;14) | 4 | 4 | 2 | TP53 × 2,18q23 × 3 | |
| 36 | Stemline | t(4;14) | 4 | 4 | 1 | TP53 × 1 |
| Subclone | t(4;14) | 4 | 4 | 2 | TP53 × 2 | |
| 69 | Subclone | t(4;14) | 3 | 3 | 1 | TP53 × 1 |
| Subclone | t(4;14) | 2 | 2 | 1 | TP53 × 1 | |
| 77 | Stemline | t(4;14) | 3 | 3 | 1 | TP53 × 1 |
| 86 | Stemline | t(4;14) | 5 | 5 | 3 | TP53 × 3 |
| Subclone | t(4;14) | 5 | 5 | 2 | TP53 × 2 |
| Patient # . | Clones . | IGH translocation . | CN 1q12 . | CN 1q21 . | CN 17p . | CNAs in RC . |
|---|---|---|---|---|---|---|
| Patients with der(1;8) | ||||||
| 1 | Stemline | Normal | 3 | 3 | 2 | MYC × 3,del(3q) |
| 31 | Stemline | Normal | 3 | 3 | 1 | MYC × 2 |
| Subclone | Normal | 4 | 4 | 1 | MYC × 3,del(7q) | |
| 52 | Stemline | t(4;14) | 3 | 3 | 2 | MYC × 3 |
| 62 | Stemline | t(14;16) | 8 | 8 | 1 | MYC × 8 |
| Patients with der(1;16) | ||||||
| 17 | Stemline | Normal | 5 | 5 | 1 | 16q11 × 4 |
| 59 | Stemline | t(4;14) | 8 | 8 | 1 | 16q11 × 8,del(11p) |
| Subclone | t(4;14) | 11 | 11 | 1 | 16q11 × 10,del(11p) | |
| 70 | Stemline | t(4;14) | 3 | 3 | 1 | 16q11 × 3 |
| Patients with complex sequential CNAs | ||||||
| 15 | Stemline | t(1;14) | 8 | 8 | 2 | BCL2 × 3, MLL × 4 |
| 32 | Stemline | t(4;14) | 8 | 8 | 1 | 1p36 × 5, FGFR3 × 5 |
| Patients with der(1;17) | ||||||
| 12 | Stemline | t(4;14) | 3 | 3 | 1 | TP53 × 1 |
| Subclone | t(4;14) | 2 | 2 | 1 | TP53 × 1 | |
| Subclone | t(4;14) | 3 | 3 | 1 | TP53 × 1 | |
| 18 | Stemline | t(11;14) | 3 | 3 | 1 | TP53 × 1 |
| 34 | Stemline | t(4;14) | 3 | 3 | 3 | TP53 × 3 |
| Subclone | t(4;14) | 4 | 4 | 2 | TP53 × 2,18q23 × 3 | |
| 36 | Stemline | t(4;14) | 4 | 4 | 1 | TP53 × 1 |
| Subclone | t(4;14) | 4 | 4 | 2 | TP53 × 2 | |
| 69 | Subclone | t(4;14) | 3 | 3 | 1 | TP53 × 1 |
| Subclone | t(4;14) | 2 | 2 | 1 | TP53 × 1 | |
| 77 | Stemline | t(4;14) | 3 | 3 | 1 | TP53 × 1 |
| 86 | Stemline | t(4;14) | 5 | 5 | 3 | TP53 × 3 |
| Subclone | t(4;14) | 5 | 5 | 2 | TP53 × 2 |
Focal amplification of MYC by JT1q12
Four patients (1, 31, 52, and 62) showed a JT1q12 translocated to the distal end of chromosome 8 to band 8q24 (Table 1). In all cases, locus-specific FISH for MYC (8q24) confirmed that the 1q12 pericentromeric heterochromatin was juxtaposed to 8q24 (Figure 1). In patients 1 and 31, SKY analysis identified a segment of 8q24 inserted between a second RC and a second copy of JT1q12. In these cases, the original JT1q12 remained on the 8q24 and additional copies of MYC and JT1q12 were translocated to a second RC, thus simultaneously increasing the CN for both MYC and 1q21. For example, in patient 1, a JT1q12 was detected on 2 different RCs in the same clone, demonstrating intraclonal heterogeneity introduced by the JT1q12. In this patient, 1 JT1q12 was juxtaposed to 8q24 and a second copy of the same JT1q12 translocated MYC to a second RC at 3q23 (Figure 1A). The second JT1q12 translocation resulted in a deletion of chromosome 3 distal to band q23. In a second patient (31), 2 specimens were analyzed. In the first specimen, a single JT1q12 was translocated to MYC (Figure 1B). In a subsequent specimen analyzed 3 months later, the original JT1q12 was still present on 8q24 and a second JT1q12 translocation including an extra copy of MYC was identified on a second RC at 7q23. The second JT1q12 resulted in a deletion of 7q distal to 7q23 in the new subclone (Figure 1C).
Partial karyotypes of 2 patients demonstrating initial JT1q12 to RC MYC and a second sequential JT1q12 to different RCs. FISH probes 1q12 (red) and 1q21 (green) are shown on the left, SKY image-classified colors for chromosomes 8 (orange) and 1q (yellow) are in the middle, and the FISH probe for MYC (red) is on the right. (A) Patient 1 shows a single copy of JT1q12 translocated to 8q24 (left bracket), and in the same clone, a second translocation of the JT1q12 has translocated an extra copy of MYC to 3q (right bracket). (B) Patient 31 demonstrates a JT1q12 on chromosome 8q24 as the sole 1q aberration in the stemline. (C) In a second specimen analyzed 3 months later, a second translocation of the JT1q12 translocated an additional copy of MYC to 7q23. The sequential translocations of JT1q12 in both cases resulted in a CN increase for both MYC and 1q21 and deletions in the second RCs. Note: In all figures, FISH hybridizations are shown in inverse 4′,6 diamidino-2-phenylindole to delineate chromosome similarities to the G-banding patterns used in routine studies.
Partial karyotypes of 2 patients demonstrating initial JT1q12 to RC MYC and a second sequential JT1q12 to different RCs. FISH probes 1q12 (red) and 1q21 (green) are shown on the left, SKY image-classified colors for chromosomes 8 (orange) and 1q (yellow) are in the middle, and the FISH probe for MYC (red) is on the right. (A) Patient 1 shows a single copy of JT1q12 translocated to 8q24 (left bracket), and in the same clone, a second translocation of the JT1q12 has translocated an extra copy of MYC to 3q (right bracket). (B) Patient 31 demonstrates a JT1q12 on chromosome 8q24 as the sole 1q aberration in the stemline. (C) In a second specimen analyzed 3 months later, a second translocation of the JT1q12 translocated an additional copy of MYC to 7q23. The sequential translocations of JT1q12 in both cases resulted in a CN increase for both MYC and 1q21 and deletions in the second RCs. Note: In all figures, FISH hybridizations are shown in inverse 4′,6 diamidino-2-phenylindole to delineate chromosome similarities to the G-banding patterns used in routine studies.
Whole-arm JT1q12 to 16q11
Whole-arm unbalanced translocations of JT1q12 to 16q11 (RC) were identified in 13 patients (6, 9, 17, 21, 24, 26, 33, 56, 59, 63, 70, 75, and 82) (supplemental Table 1). This translocation results in the simultaneous CN gain of 1q21 and deletion of 16q, which also results in the loss of heterozygosity for CYLD at 16q12 and WWOX at 16q23. In most cases, the RC der(1;16) chromosome is a stable clonal aberration; however, in 3 patients, we identified the clonal progression of 16q11 as part of subsequent JT1q12s (patients 17, 59, and 70) (Table 1). For example, in patient 17, we identified the focal amplification of 16q11 translocated to 18q as part of a second translocation of the same JT1q12 (Figure 2A). The der(20) in this cell line did not show a signal for 16q11 and, therefore, was not a sequential JT1q12 (Figure 2A). In a second more complex example, patient 59, 2 sequential specimens were analyzed. In the original specimen, the JT1q12 translocated to 16q11 (first RC) where a coamplification 16q11 and 1q12∼23 occurred (Figure 2B). The coamplified segments of 16q11 and 1q12∼23 then translocated to 11p (second RC), resulting in the deletion of the entire 11p (Figure 2B). In the original stemline, 8 copies of 1q21 and 7 copies of 16q11 were present. In the subsequent specimen analyzed 6 months later, a subclone had emerged showing JT1q12s with different segments of 16q11 and 1q12∼23 that had translocated back to both of the previously normal chromosomes 1q (Figure 2C). One JT1q12 segment translocated to band 1q23 (third RC) (Figure 2C, far left), and the second segment involving 16q11 and 1q12 had translocated to the chromosome 1 with the deletion in 1p (fourth RC) (Figure 2C). In the subclone, the CN for 1q21 increased from 8 to 11 and for 16q11 increased from 7 to 9.
Partial karyotypes of 2 patients demonstrating additional CNAs originating from JT1q12 to16q11. Note: Normal chromosome 16 is not shown for these 2 patients. FISH probes 1q12 (red), 1q21 (green), and 16q11 (aqua) and SKY classification colors for chromosome 16 (light orange) are 1q (yellow), 18 (auburn) and 20 (blue) are shown. (A) The karyotype of patient 17 showed a total of 5 copies of JT1q12, including extra copies on a der(1;16), a der(18), and a der(20). The JT1q12 on the der(18) also demonstrated an extra copy of 16q11(aqua signal) was amplified and translocated from the der(16). (B) The stemline of patient 59 showed 1 normal chromosome 1 (left), a chromosome 1 with a 1p deletion (second from left), a chromosome 16 with the coamplification of 1q12∼23 and 16q11 (third from left), and a coamplified segment of 16q11 and 1q12∼23 translocated to 11p (right). The CN was 8 for both 1q21 and for 16q11 in this clone. (C) In a specimen analyzed 6 months later, a subclone emerged showing a coamplified segment of 16q11 and 1q12∼23 had translocated to the normal chromosome 1q at 1q25 (left) and a second smaller segment of 16q11 and JT1q12 to the other chromosome 1 with deletion of 1p. Note: In this subclone, the CN for 1q21 increased from 8 to11 and the CN of 16q11 increased from 8 to 10.
Partial karyotypes of 2 patients demonstrating additional CNAs originating from JT1q12 to16q11. Note: Normal chromosome 16 is not shown for these 2 patients. FISH probes 1q12 (red), 1q21 (green), and 16q11 (aqua) and SKY classification colors for chromosome 16 (light orange) are 1q (yellow), 18 (auburn) and 20 (blue) are shown. (A) The karyotype of patient 17 showed a total of 5 copies of JT1q12, including extra copies on a der(1;16), a der(18), and a der(20). The JT1q12 on the der(18) also demonstrated an extra copy of 16q11(aqua signal) was amplified and translocated from the der(16). (B) The stemline of patient 59 showed 1 normal chromosome 1 (left), a chromosome 1 with a 1p deletion (second from left), a chromosome 16 with the coamplification of 1q12∼23 and 16q11 (third from left), and a coamplified segment of 16q11 and 1q12∼23 translocated to 11p (right). The CN was 8 for both 1q21 and for 16q11 in this clone. (C) In a specimen analyzed 6 months later, a subclone emerged showing a coamplified segment of 16q11 and 1q12∼23 had translocated to the normal chromosome 1q at 1q25 (left) and a second smaller segment of 16q11 and JT1q12 to the other chromosome 1 with deletion of 1p. Note: In this subclone, the CN for 1q21 increased from 8 to11 and the CN of 16q11 increased from 8 to 10.
Complex sequential l JT1q12s
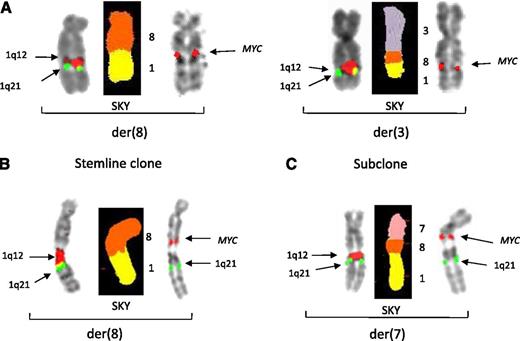
Two patients (15 and 32) showed complex sequential JT1q12s in which 3 independent chromosome segments were involved in JT1q12s. In these cases, different chromosome segments were identified both proximally and distally on the JT1q12. For example, in patient 15, an isochromosome 1q was identified with 11q23 (MLL) translocated to both ends (Figure 3A, left bracket). In this patient, one-half of the iso1q (1q12 breakpoint) first translocated to 18q23 (Figure 3A, middle bracket). In a translocation to a second RC, the JT1q12 carried both an amplified copy of BCL2 (proximal end) and of MLL (distal end) to 21p (Figure 3A, right bracket). In the second patient (32), the first translocation of JT1q12 was to 1p36 (not shown). A focal amplification of 1p36 and an inverted duplication of 1q21 occurred and this 2-chromosome segment of 1p36 and 1q23∼23 translocated to a der(12) (Figure 3B, left bracket). On the der(12), SKY detected (and FISH confirmed) an additional cryptic translocation of the band 4p16 (FGFR3) on the distal end of this JT1q12 (Figure 3B, left bracket). This 3-chromosome segment subsequently translocated to 2 additional RCs including chromosome 8p (Figure 3B, middle bracket) and 15p (Figure 3B, right bracket).
Progression of sequential JT1q12s in patients 15 and 32 demonstrating “passenger” segments on the JT1q12. (A) In patient 15, an isochromosome 1q (left bracket) is shown with FISH probes 1q12 (red) and 1q21 (green) on the left, SKY in the middle, and MLL (11q23) (yellow) on the right. The middle bracket shows a JT1q12 has translocated to the distal end of the long arm of 18, with the 11q23 (MLL) as a passenger. Note: der(18) on the right is probed for BCL2 (red). The bracket on the right shows the second jump of the same JT1q12 to chromosome 21p. As a result of this translocation, SKY identified an extra copy of both BCL2 and MLL duplicated and translocated to 21p. The resulting CNAs in this cell line were 3 copies of 1q21, 3 copies of BCL2, and 5 copies of MLL. (B) Patient 32 demonstrated JT1q12 translocations to 3 different RCs. A 3-chromosome segment involving 1p36, 1q12∼23, and 4p16 (FGFR3) translocated to the der(12) (left bracket). Two additional copies of JT1q12 with this 3-chromosome segment (1p36 proximally, 1q12∼23, and 4p [FGFR3] distally) translocated to both 8p (middle bracket) and 15p (right bracket).
Progression of sequential JT1q12s in patients 15 and 32 demonstrating “passenger” segments on the JT1q12. (A) In patient 15, an isochromosome 1q (left bracket) is shown with FISH probes 1q12 (red) and 1q21 (green) on the left, SKY in the middle, and MLL (11q23) (yellow) on the right. The middle bracket shows a JT1q12 has translocated to the distal end of the long arm of 18, with the 11q23 (MLL) as a passenger. Note: der(18) on the right is probed for BCL2 (red). The bracket on the right shows the second jump of the same JT1q12 to chromosome 21p. As a result of this translocation, SKY identified an extra copy of both BCL2 and MLL duplicated and translocated to 21p. The resulting CNAs in this cell line were 3 copies of 1q21, 3 copies of BCL2, and 5 copies of MLL. (B) Patient 32 demonstrated JT1q12 translocations to 3 different RCs. A 3-chromosome segment involving 1p36, 1q12∼23, and 4p16 (FGFR3) translocated to the der(12) (left bracket). Two additional copies of JT1q12 with this 3-chromosome segment (1p36 proximally, 1q12∼23, and 4p [FGFR3] distally) translocated to both 8p (middle bracket) and 15p (right bracket).
JT1q12-associated deletions of 17p13
Fourteen patients (12, 13, 25, 32, 34, 36, 38, 39, 59, 69, 70, 77, 84, and 86) were identified with high-risk disease with the combination of cytogenetic markers of t(4;14) and del17p (supplemental Table 2). A total of 7 patients showed the deletion of 17p resulting from a JT1q12 (patients 12, 18, 34, 36, 69, 77, and 86), with 6 of the 7 (patients 12, 34, 36, 69, 77, and 86) also demonstrating a t(4;14) (Table 1). The progression of structural rearrangements resulting in the deletion of 17p by JT1q12 is depicted in supplemental Figure 1. Among the 8 patients without a t(4;14) but with deletions of 17p, 2 had different unbalanced translocations to 17p (patients 38 and 39), 2 had whole-chromosome 17 loss (patients 59 and 70), 2 had terminal deletions at 17p11 (patients 32 and 84), and 2 had interstitial deletions of TP53 cryptic to G-banding (patients 13 and 25). Interestingly, 5 of the 6 patients (patients 12, 34, 36, 69, and 86) with t(4;14) and del 17p showed subclonal heterogeneity relating to CNAs of 1q21. In 2 patients (12 and 69), the stemlines demonstrated a der(1;17) while subclones showed only the deleted 17p. In these patients, the JT1q12 had translocated off (jumped off) the 17 leaving only a terminally deleted 17p in the subclones, which masked the presence of a der(1;17) in the stemline. For example, in patient 12, the JT1q12 was identified as a der(17) in the stemline (Figure 4A), whereas in different subclones, different CNs for 1q21 are found (Figure 4B-C). In one subclone, a direct duplication of 1q21 was identified that maintained the CN of 1q21 at 3 (Figure 4B), whereas in a second subclone (Figure 4C), only normal 1qs were found, resulting in a CN of 2 for 1q21.
Partial karyotypes of patient 12 showing variable CNAs in different subclones for 1q21 in clones with deletion 17p. The direct loss of 17p occurred in the stemline by a whole-arm translocation of JT1q12. FISH probes on chromosome 1 are 1q12 (red) and 1q21 (green) and on chromosome 17 are TP53 (green) and ERB2 (red). SKY classified colors for 1q (yellow) and 17 (blue) are shown. (A) The stemline clone with chromosomes 1 (left) and 17 (right) showing JT1q12 replacing 17p (left bracket). This same pair of 17s was rehybridized 2 additional times with both SKY (middle dashed bracket) and FISH probes for TP53 and ERB2 (right dashed bracket). In this patient, 2 additional related subclones with variable CNAs for 1q21 were also identified. In both subclones, the JT1q12 on 17p has been lost (translocated off), leaving only the deleted 17p. In one subclone (panel B), the 1q21 CN remained at 3 due to the increase in CN of 1q21 in the normal chromosome 1, whereas in a second subclone (panel C), a CN of 2 was found for 1q21.
Partial karyotypes of patient 12 showing variable CNAs in different subclones for 1q21 in clones with deletion 17p. The direct loss of 17p occurred in the stemline by a whole-arm translocation of JT1q12. FISH probes on chromosome 1 are 1q12 (red) and 1q21 (green) and on chromosome 17 are TP53 (green) and ERB2 (red). SKY classified colors for 1q (yellow) and 17 (blue) are shown. (A) The stemline clone with chromosomes 1 (left) and 17 (right) showing JT1q12 replacing 17p (left bracket). This same pair of 17s was rehybridized 2 additional times with both SKY (middle dashed bracket) and FISH probes for TP53 and ERB2 (right dashed bracket). In this patient, 2 additional related subclones with variable CNAs for 1q21 were also identified. In both subclones, the JT1q12 on 17p has been lost (translocated off), leaving only the deleted 17p. In one subclone (panel B), the 1q21 CN remained at 3 due to the increase in CN of 1q21 in the normal chromosome 1, whereas in a second subclone (panel C), a CN of 2 was found for 1q21.
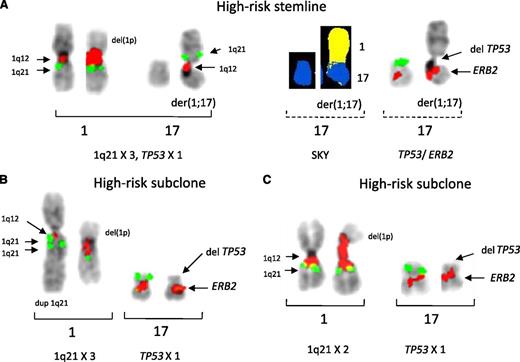
Three additional patients with the t(4;14) showed subclonal heterogeneity for CNAs for both 1q21 and 17p deletions (patients 34, 36, and 86). For example, in patient 36, we were able to analyze 2 specimens. In the first specimen, a single clone with 4 copies of 1q21 and 17p deletion due to a JT1q12 was identified (Figure 5A). Surprisingly, in a second specimen analyzed 5 months later (Figure 5B), a different single clone with normal 17s (2 copies TP53 and 4 copies of 1q21) was identified indicating the presence of an independent clone. This clone showed a direct tandem duplication of the entire 1q (2 copies of 1q) on 18p, indicating by virtue of the less-evolved cytogenetic aberrations that it is the progenitor clone. Therefore, we surmise the JT1q12, which resulted in the deletion of the 17p in the first specimen, was originally on the 18q. This finding indicates the 2 clones have coexisted in this patient and are a cytogenetic example of the alternating dominance of clones.28-30 Finally, in patient 34, a JT1q12 was translocated to 18q23 in the first specimen (Figure 6). In a subsequent specimen analyzed 20 months later, CNAs involving 3 different chromosomes resulted from a second translocation of the JT1q12 on 18q23 to 17p. The stemline of the original specimen showed 3 17s (TP53 × 3) and a JT1q12 on chromosome 18q23 (1q21 × 3) (Figure 6A). In the subclone, a second JT1q12 was detected on 17p (second RC) resulting in the deletion of 17p, including TP53 (Figure 6B). Strikingly, in the second jump of JT1q12, a cryptic insertion of 18q23 was detected by SKY between the 17q10 and JT1q12 (Figure 6B). This second translocation is another example of the simultaneous amplification and translocation of 2 chromosome segments (1q21 and 18q23) by a JT1q12 that causes a deletion in a third chromosome (17p) (Figure 6B). Taken together, these 6 patients (12, 34, 36, 69, 77, and 86) demonstrate the cosegregation of t(4;14), deletion of 17p, and gain of 1q21 and identify a mechanism for the simultaneous gain of 1q21 and the deletion of 17p.
Partial karyotypes of patient 36 demonstrating similar but reconfigured CNs for 1q21 but a different CN for 17p in related clones. FISH probes 1q12 (red) and 1q21 (green), TP53 (green), and ERB2 (red) are shown. (A) In the first specimen, copies of JT1q12 were identified on both the 17p and 18p (top row, G-bands). FISH for 1q12 and 1q21 showed JT1q12 on both 17p and 18p with a 1q21 CN of 4 (middle row; normal chromosome 1 not shown). Probes for TP53 and ERB2 (bottom row) showed a CN of 1 for 17p. (B) In a second specimen analyzed 5 months later, normal chromosome 17 was identified by G-banding (top row), and a CN of 4 for 1q21 was found; however, both copies of JT1q12s were on the 18p (bottom row). This less-evolved clone indicates that the second copy of JT1q12 on the 18p in this clone is the JT1q12 that translocated to 17p, resulting in the deletion of TP53 in the previous specimen.
Partial karyotypes of patient 36 demonstrating similar but reconfigured CNs for 1q21 but a different CN for 17p in related clones. FISH probes 1q12 (red) and 1q21 (green), TP53 (green), and ERB2 (red) are shown. (A) In the first specimen, copies of JT1q12 were identified on both the 17p and 18p (top row, G-bands). FISH for 1q12 and 1q21 showed JT1q12 on both 17p and 18p with a 1q21 CN of 4 (middle row; normal chromosome 1 not shown). Probes for TP53 and ERB2 (bottom row) showed a CN of 1 for 17p. (B) In a second specimen analyzed 5 months later, normal chromosome 17 was identified by G-banding (top row), and a CN of 4 for 1q21 was found; however, both copies of JT1q12s were on the 18p (bottom row). This less-evolved clone indicates that the second copy of JT1q12 on the 18p in this clone is the JT1q12 that translocated to 17p, resulting in the deletion of TP53 in the previous specimen.
Partial karyotypes of patient 34 demonstrating the loss of 17p by 2 successive translocations of JT1q21. FISH probes 1q12 (red), 1q21 (green), 18 centromere (aqua), and 18q23 telomere (yellow) and on chromosome 17, TP53 (green), and ERB2 (red) are shown. SKY classification colors are 1q (yellow), 17 (blue), and 18 (auburn). (A) Two cell lines were detected in this stemline and a new subclone with additional aberrations of JT1q12 . The first abnormal specimen showed 3 normal copies of 17 (left bracket) and a JT1q12 translocated to the telomere of 18q [der(1;18), right bracket]. (B) In the second specimen analyzed 20 months later, a subclone was identified in which a second whole-arm JT1q12 resulted in the loss of 17p (left bracket). In this subclone, an unexpected focal amplification of 18q23 was detected by SKY (right bracket, middle) and confirmed by FISH (right bracket, far right) indicating the simultaneous segmental CN gains of 18qter, 1q21, and loss of 17p.
Partial karyotypes of patient 34 demonstrating the loss of 17p by 2 successive translocations of JT1q21. FISH probes 1q12 (red), 1q21 (green), 18 centromere (aqua), and 18q23 telomere (yellow) and on chromosome 17, TP53 (green), and ERB2 (red) are shown. SKY classification colors are 1q (yellow), 17 (blue), and 18 (auburn). (A) Two cell lines were detected in this stemline and a new subclone with additional aberrations of JT1q12 . The first abnormal specimen showed 3 normal copies of 17 (left bracket) and a JT1q12 translocated to the telomere of 18q [der(1;18), right bracket]. (B) In the second specimen analyzed 20 months later, a subclone was identified in which a second whole-arm JT1q12 resulted in the loss of 17p (left bracket). In this subclone, an unexpected focal amplification of 18q23 was detected by SKY (right bracket, middle) and confirmed by FISH (right bracket, far right) indicating the simultaneous segmental CN gains of 18qter, 1q21, and loss of 17p.
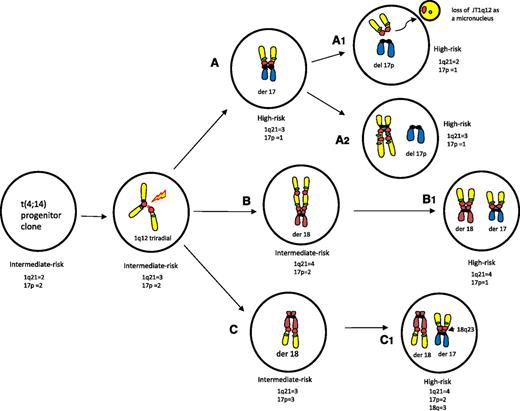
A model for the subclonal progression of JT1q12 and the evolution of cytogenetically defined high-risk disease
Based on the present and previous findings,18,22,27 we propose a model for the JT1q12 in the subclonal evolution of cytogenetically defined high-risk disease. Risk stratification of subclones is founded on the current IMWG2 iFISH probe recommendations. The key structural chromosome rearrangements of chromosomes 1 and 17 are depicted inside clones (Figure 7). The initial JT1q12 to a RC in a subclone has the potential to increase the CN of 1q21 and/or decrease CN of the first RC. Sequential JT1q12s, however, have the potential to induce both increased and decreased CNs for 1q21 and both gains and losses in the subsequent RCs. In this model, progenitor aneuploid clones with the t(4;14) (Figure 7, far left) undergo disruption of the 1q12 pericentromeric heterochromatin, resulting in the partial endoreduplication of 1q and the formation of 1q triradials (Figure 7, second from left). Extra copies of 1q can translocate to one or more RCs and become stable or, alternatively, undergo further sequential translocations. JT1q12s that progress through intermediate RCs represent a linear subclonal pathway to high-risk clones. Direct whole-arm translocations resulting in the der(1;17) are defined as high risk by IMWG criteria (Figure 7A). Further subclonal instability can also lower 1q21 CN if the JT1q12 translocates off the der(1;17) and is lost from the cell (patients 12 and 69), masking the previous CNAs of 1q21 (Figure 7A1). The JT1q12 can also reduplicate or translocate to different RCs (Figure 7A2), leaving the 17p deleted and the CN of 1q21 remaining stable. Clones B and C represent sequential JT1q12s when the JT1q12 first translocates to an intermediate RC, resulting only in the gain of 1q21 (intermediate-risk clones) (Figure 7B-C). The second JT1q12 can either reconfigure the CN for 1q21 (Figure 7 B1) or translocate a focal amplification of the first RC (Figure 7C1) to the second RC 17p and therefore introduce CNAs for 3 different chromosomes (del17p, +1q21, and +18q23; Figure 7C1).
Characterization of alternative pathways and possible CNAs associated with the deletion 17p in progenitor clones with t(4;14). Key cytogenetic translocations resulting in CNAs for risk are indicated inside of clones. Note: FISH probes depicted in cartoon are 1q12 (red) and 1q21 (green) and chromosome arm colors are 1 (yellow), 17 (blue), 18 (auburn). Subclone A depicts clones of patient 12, B of patient 36, and C of patient 46. Arrows indicate probable derivation of subclones. The JT1q12 instability originates from a triradial of 1q (second from left), which results in an extra copy of the JT1q12 translocating to different RCs (subclones A-C). Subclone A shows that the JT1q12 can translocate directly to chromosome 17 resulting in a der(1;17) with a whole-arm loss of 17p and gain of 1q21 and a high-risk subclone. Subsequent subclones are also high risk, although JT1q12s can be lost as micronuclei (A1) and/or reconfigure on the same or different RC and 1q21 CN and remain stable (A2). In subclones B and C, different sequential translocations of JT1q12 result in high-risk subclones as well. Subclone B demonstrates the JT1q12 translocation first to18p in an intermediate risk clone and second to 17p, resulting in a high-risk subclone (B1). The detection of the high-risk clone (B1) 5 months before the detection of the intermediate-risk progenitor clone (B) in this patient suggests that alternating dominance or clonal tiding had occurred. In subclone C, the JT1q12 first translocates to the 18q23 telomere, resulting in a der(1;18) chromosome aberration and an intermediate-risk subclone. The second JT1q12 translocation from 18q23 to17 results in a high-risk clone with a focal amplification of 18q23, gain of 1q21, and deletion of 17p (C1).
Characterization of alternative pathways and possible CNAs associated with the deletion 17p in progenitor clones with t(4;14). Key cytogenetic translocations resulting in CNAs for risk are indicated inside of clones. Note: FISH probes depicted in cartoon are 1q12 (red) and 1q21 (green) and chromosome arm colors are 1 (yellow), 17 (blue), 18 (auburn). Subclone A depicts clones of patient 12, B of patient 36, and C of patient 46. Arrows indicate probable derivation of subclones. The JT1q12 instability originates from a triradial of 1q (second from left), which results in an extra copy of the JT1q12 translocating to different RCs (subclones A-C). Subclone A shows that the JT1q12 can translocate directly to chromosome 17 resulting in a der(1;17) with a whole-arm loss of 17p and gain of 1q21 and a high-risk subclone. Subsequent subclones are also high risk, although JT1q12s can be lost as micronuclei (A1) and/or reconfigure on the same or different RC and 1q21 CN and remain stable (A2). In subclones B and C, different sequential translocations of JT1q12 result in high-risk subclones as well. Subclone B demonstrates the JT1q12 translocation first to18p in an intermediate risk clone and second to 17p, resulting in a high-risk subclone (B1). The detection of the high-risk clone (B1) 5 months before the detection of the intermediate-risk progenitor clone (B) in this patient suggests that alternating dominance or clonal tiding had occurred. In subclone C, the JT1q12 first translocates to the 18q23 telomere, resulting in a der(1;18) chromosome aberration and an intermediate-risk subclone. The second JT1q12 translocation from 18q23 to17 results in a high-risk clone with a focal amplification of 18q23, gain of 1q21, and deletion of 17p (C1).
Discussion
MM is characterized by genetic heterogeneity that is contributed to by both primary and secondary cytogenetic aberrations. The accumulation of secondary CNAs is thought to occur in a Darwinian fashion and provide a selective pressure for the expansion of therapy-resistant clonal variants.3,10,17,28-31 Importantly, recent genomic studies have shown that cytogenetically defined high-risk patients show more changes involving both intraclonal and subclonal heterogeneity as well as alternating dominance of clones.28-30 Unfortunately, little is known about the origin or possible relationships between different types of chromosome aberrations and the accumulation of CNAs during clonal evolution of MM.
One possible explanation for the CNAs associated with the focal amplifications and deletions described here is the unique ability of 1q12 pericentromeric heterochromatin to duplicate itself and regions adjacent to it. Both direct and inverted duplications of 1q12∼23 region and unbalanced pericentromeric exchanges among chromosomes 1, 9, and 16 are well documented in both hematologic and solid tumors.32 We and others have previously speculated that the cause of these duplications and jumping translocations of 1q may be region-specific hypomethylation of the 1q12 pericentromeric heterochromatin.18-22 The 1q12 pericentromeric region is a distinctive region in the human genome in that it is the largest single block of late-replicating, highly repetitive satII/III DNA that is known to contain unstable segmental duplications (low copy repeats). It is thought that a 5-azacytidine fragile site at 1q12 may play a role in the incomplete replication (delayed condensation), breaks, and triradial configurations of 1q12.18,19 In addition, pediatric patients with immunodeficiency centromeric instability and facial anomalies syndrome have a defect in DNA methyltransferase 3B and develop triradial 1q12 aberrations similar to those seen in MM.33
The focal amplification and translocation of segments of RCs juxtaposed to 1q12 pericentromeric heterochromatin has not, to our knowledge, been previously reported. The amplification of the MYC locus is an example of a known secondary CNA aberration in MM. As a hallmark of proliferative myeloma, MYC amplification has been found to be a common difference between monoclonal gammopathy of undetermined significance and newly diagnosed MM patients.1,34 It is believed that abnormal CNAs of MYC and the associated dysregulation of MYC occur as progression events and are mediated in some cases by secondary MYC translocations, insertions of MYC, or gene duplications that do not involve B-cell–specific mechanisms.34 The finding of the cosegregating CN gains of the MYC locus and JT1q12 in the 4 patients identified here demonstrates that a subset of secondary translocations of MYC can result from JT1q12s. Additionally, focal amplifications and translocations involving BCL2 and FGFR3 identified in this study show this mechanism can potentially involve any RC segment and may help to partially explain the diversity of subclonal heterogeneity and genomic instability found in MM.
Whole-arm JT1q12s can result in the deletion of large chromosome regions that have been linked to the progression of MM. It has been reported by single-nucleotide polymorphism array that the most common segmental CN losses include the loss of 16q, 6q, and 8p and that the loss of 16q is associated with the loss of heterozygosity for CYLD and WWOX. We show here that these whole-arm deletions also cosegregate in subclones with the gain of 1q. Clinically, it has also been reported that the loss of 16q is associated with a worse overall survival.35 In this report, we identified the whole-arm loss of 16q by JT1q12 in 12 patients, 3 of which showed subclonal progression of CNAs in RCs for both 16q11 and 1q21 (Table 1).
The most clinically significant finding in this report is the mechanistic association of gain of 1q21 with deletion of 17p. The IMWG currently recommends the use of 3 cytogenetic markers for risk stratification, including the t(4;14), deletion 17p, and gain of 1q21.2 Unfortunately, no consensus on a CN for 1q21 and risk stratification has been reached even in light of the evidence from genomic profiling and iFISH studies showing that the gain of 1q21 has prognostic significance3-9,36-41 As has been previously reported, the associated gain of 1q21 and CN losses in other chromosome segments may help explain the adverse reported outcome of patients with 1q gain.7 In fact, iFISH studies have shown the t(4;14) and deletions of 17p can cosegregate with gain of 1q and accumulate during disease progression.37 The clinical impact of these adverse lesions will presumably be modulated by the etiologic background of the myeloma clone, and, therefore, a +1q21 that is found in association with t(4;14) and del17p is thought to impart a worse prognosis than a +1q21 in association with hyperdiploidy.
We have shown here that sequential JT1q12 translocations can occur during the clonal evolution of MM and can introduce CNAs involving MYC, BCL2, MLL, and TP53, among others. The accumulation of these CNAs in RCs is an unexpected source for intraclonal and subclonal heterogeneity and may be one explanation for the clonal diversity found in this disease. To our knowledge, the JT1q12 is the only known cytogenetic lesion with the potential to act as a “subclonal driver” resulting in sequential CNAs of itself (1q21) and potentially in multiple RCs as well. Recent genome-wide studies provide evidence that clonal evolution of MM involves both linear and nonlinear patterns28-30 and that minor subclones can survive treatment, acquire new anomalies, and provide a reservoir for relapse.42 The diverse pathways for subclonal deletion of 17p identified in these patients supports these findings and demonstrates a mechanism for the accumulation of high-risk CNAs involving the combination of t(4;14), del17p, and +1q21.
The online version of this article contains a data supplement.
There is an Inside Blood commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the patients and staff of the Myeloma Institute for Research and Therapy. The expert editorial assistance of Diane Schrantz is greatly appreciated.
This work was supported in part by the National Institutes of Health, National Cancer Institute (PO1 grant CA 0055819).
Authorship
Contribution: J.R.S. conceived the hypothesis, analyzed and interpreted data, and wrote the manuscript; C.J.H., J.E., and D.J. analyzed the data; C.S., M.J., A.B., and G.S. performed chromosome analysis; E.T., J.L.L., and R.B. performed FISH and SKY studies; S.U., M.Z., S.W., and F.v.R. provided patient samples; and B.B. provided patient samples, interpreted data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jeffrey R. Sawyer, Cytogenetics Laboratory, Suite 200, Freeway Medical Tower, 5800 West 10th St, Little Rock, AR 72204; e-mail: sawyerjeffreyr@uams.edu.


![Figure 3. Progression of sequential JT1q12s in patients 15 and 32 demonstrating “passenger” segments on the JT1q12. (A) In patient 15, an isochromosome 1q (left bracket) is shown with FISH probes 1q12 (red) and 1q21 (green) on the left, SKY in the middle, and MLL (11q23) (yellow) on the right. The middle bracket shows a JT1q12 has translocated to the distal end of the long arm of 18, with the 11q23 (MLL) as a passenger. Note: der(18) on the right is probed for BCL2 (red). The bracket on the right shows the second jump of the same JT1q12 to chromosome 21p. As a result of this translocation, SKY identified an extra copy of both BCL2 and MLL duplicated and translocated to 21p. The resulting CNAs in this cell line were 3 copies of 1q21, 3 copies of BCL2, and 5 copies of MLL. (B) Patient 32 demonstrated JT1q12 translocations to 3 different RCs. A 3-chromosome segment involving 1p36, 1q12∼23, and 4p16 (FGFR3) translocated to the der(12) (left bracket). Two additional copies of JT1q12 with this 3-chromosome segment (1p36 proximally, 1q12∼23, and 4p [FGFR3] distally) translocated to both 8p (middle bracket) and 15p (right bracket).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/123/16/10.1182_blood-2013-12-546077/4/m_2504f3.jpeg?Expires=1763518944&Signature=VRw3TDG6Py5xHqjLgwQWTmIYbx6igiqKnLqu9-PqdQvj9VzR8X9ubb4PLXS8Z3ZJdDqNbBUYqNFloY2L2-kdr-ZXmCmQbi1AXvNJyftJ2IxHKHKMibzieRzoUAEfvKePzcGcYRWYGLy0jKdl126~ar4awRS9z3pzPD3fxVCno21Toxtlqu4LYZB10VfSF7308j4dH34S3Af-RYNuml-hfYdyML3d02gkbzxQqcFONJ~pvk-YMiE~1UvrAXLdeyxgMebsuWHDjvIVZoCQxDOMZj2phojoScWEzQ7MEKVSi9tBOudFUKQPwXK45FssA4v825EsK11uRfqPGlCau~cPzQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Figure 6. Partial karyotypes of patient 34 demonstrating the loss of 17p by 2 successive translocations of JT1q21. FISH probes 1q12 (red), 1q21 (green), 18 centromere (aqua), and 18q23 telomere (yellow) and on chromosome 17, TP53 (green), and ERB2 (red) are shown. SKY classification colors are 1q (yellow), 17 (blue), and 18 (auburn). (A) Two cell lines were detected in this stemline and a new subclone with additional aberrations of JT1q12 . The first abnormal specimen showed 3 normal copies of 17 (left bracket) and a JT1q12 translocated to the telomere of 18q [der(1;18), right bracket]. (B) In the second specimen analyzed 20 months later, a subclone was identified in which a second whole-arm JT1q12 resulted in the loss of 17p (left bracket). In this subclone, an unexpected focal amplification of 18q23 was detected by SKY (right bracket, middle) and confirmed by FISH (right bracket, far right) indicating the simultaneous segmental CN gains of 18qter, 1q21, and loss of 17p.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/123/16/10.1182_blood-2013-12-546077/4/m_2504f6.jpeg?Expires=1763518944&Signature=mMqtMIDuwhRwB1E-8ogg~3WH1ew~Tr9nqPUwNoBC7T0zbZi413jL0mUYFgez8u6ZFRYkttCqq~IJm-gX6EA-ykPhN~0muXNGa2oIN2mQarhZc-vQhLTicC0R265C19CFkKJ3i-mdRFWITtBj9IJXEvk-oeSLob~Dcl7q2tZ4tYoLZYvRZXFK5Xe7AvgFqgeIKSsDuBzjhAUX92kh-8xqqBh5CRnwSWfiFDrCxWH636FULAjkHC~CwnR8P3zoP5BYGsmCW1UoFMkveNGyNYZvzpaJgw7l9qCO76SLzeXb1HGoLq6z~nC3sf26ISl~L2su03jgNurM0~z-PGK9jPgtxg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)