Key Points
Mutations in genes of the cohesin complex are recurrent mutations in AML with a strong association with NPM1 mutations.
Cohesin gene mutations have no clear prognostic impact in AML patients.
Abstract
Mutations in the cohesin complex are novel, genetic lesions in acute myeloid leukemia (AML) that are not well characterized. In this study, we analyzed the frequency, clinical, and prognostic implications of mutations in STAG1, STAG2, SMC1A, SMC3, and RAD21, all members of the cohesin complex, in a cohort of 389 uniformly treated AML patients by next generation sequencing. We identified a total of 23 patients (5.9%) with somatic mutations in 1 of the cohesin genes. All gene mutations were mutually exclusive, and STAG1 (1.8%), STAG2 (1.3%), and SMC3 (1.3%) were most frequently mutated. Patients with any cohesin complex mutation had lower BAALC expression levels. We found a strong association between mutations affecting the cohesin complex and NPM1. Mutated allele frequencies were similar between NPM1 and cohesin gene mutations. Overall survival (OS), relapse-free survival (RFS), and complete remission rates (CR) were not influenced by the presence of cohesin mutations (OS: hazard ratio [HR] 0.98; 95% confidence interval [CI], 0.56-1.72 [P = .94]; RFS: HR 0.7; 95% CI, 0.36-1.38 [P = .3]; CR: mutated 83% vs wild-type 76% [P = .45]). The cohesin complex presents a novel pathway affected by recurrent mutations in AML. This study is registered at www.clinicaltrials.gov as #NCT00209833.
Introduction
Over the last several years, our knowledge of genes being mutated in acute myeloid leukemia (AML) patients has not only expanded due to the results of next generation and whole genome sequencing efforts, but we have also learned that mutations in AML occur in specific pathways. These pathways can be categorized according to their function.1,2 In the last year, genes in the cohesin complex have been described as novel mutations occurring in 13% of AML patients,1 suggesting that the cohesin-complex presents an important pathway in the pathogenesis of AML. Genes that belong to the cohesin complex in somatic vertebrate cells are SMC1A, SMC3, RAD21 (SCC1), STAG2 (SA-2), and STAG1 (SA-1)3 ; these genes form a ring structure that regulates chromosome segregation during meiosis and mitosis. Thus, the cohesin-complex is an essential structure during cell division.4 Undoubtedly, cell division is one of the key processes for every tumor cell including AML blasts due to the increased proliferation potential of malignant cells. Interestingly, more recent data suggests that cohesin genes have additional functions within the cell such as double-strand DNA repair and regulation of transcription.5 It is known that germline mutations lead to cohesinopathies that are characterized by growth and developmental disorders in regard to the role of cohesin genes in the pathogenesis of human disease.6 Mutations in the cohesin-complex have already been described in colorectal cancer, and there has been a link between these mutations and chromosomal instability.7 Further investigation is needed to determine which role the cohesin-complex fulfills in AML and whether cohesin mutations have clinical implications. The aim of this study was to investigate the frequency, clinical implications, and prognostic influence of mutations in the cohesin complex in the context of other prognostic markers in a cohort of 389 uniformly treated AML patients.
Patients, materials, and methods
Patients
Diagnostic bone marrow or peripheral blood samples were analyzed from 389 adult patients (aged 17-60 years) with de novo (n = 348) or secondary AML (n = 41, and of these, 35 patients had antecedent myelodysplastic syndrome and 6 patients had treatment-related secondary AML) with French-American-British classification M0-M2 or M4-M7. These patients were entered into the multicenter treatment trial AML SHG 0199 (#NCT00209833, June 1999 to September 2004, n = 276) or AML SHG 0295 (February 1995 to May 1999, n = 113) for whom pretreatment cell samples were available. Patients with PML-RARA or t(15;17)-positive AML were excluded from these trials. All patients received intensive, response-adapted double induction and consolidation therapy. Details of the treatment protocols have been previously reported.8,9 Peripheral blood mononuclear cells from 30 healthy volunteers were used to assess the frequency of germline single nucleotide variants (SNVs). Written informed consent was obtained according to the Declaration of Helsinki, and the studies were approved by the institutional review board of Hannover Medical School, Hannover, Germany.
Cytogenetic and molecular analysis
Pretreatment samples from all patients were studied centrally by G- and R-banding analysis. Chromosomal abnormalities were described according to the International System for Human Cytogenetic Nomenclature.10 Other relevant genes were assessed for frequently occurring mutations or expression levels as previously described (ie, FLT3-ITD,8 nucleophosmin1 [NPM1],8 DNMT3A,11 IDH1,12 IDH2,13 and MLL514 ). In the subgroup of cytogenetically normal AML (CN-AML), additional mutation analyses were performed for CEBPA,15 MLL-PTD,16 WT1, and WT1 SNP rs16754,17 NRAS,8 and expression levels of BAALC,18 ERG,19 EVI1,20,21 MN1,22 MLL5,14,17 and WT117 were quantified as previously described using complementary DNA from the KG1A cell line (BAALC, ERG, MLL5), plasmids (MN1,23 WT117 ), or from a patient sample (EVI1) to construct a relative standard curve using ABL as a housekeeping gene (Ipsogen, Marseille, France).
Analysis of cohesin mutations
Leukemic cells from peripheral blood or bone marrow were collected from patients at diagnosis, and genomic DNA was extracted, whole genome amplified (GenomePlex whole genome amplification kit, Sigma-Aldrich, Seelze, Germany) and polymerase chain reaction amplified for 119 amplicons with not more than 5 amplicons per well using standard conditions. The primers for all exons of the cohesin genes are listed in supplemental Table 1, available on the Blood Web site. All amplicons from 1 patient were pooled and randomly ligated (Quick Ligation Kit, New England Biolabs, Ipswich, MA). The long concatenated DNA was then sheared into 100 to 250 bp fragments using the Covaris System (Covaris, Woburn, MA) to obtain randomly fragmented sequences, and was size selected for 200 bp fragments using Agencourt AMPure XP reagent (Agencourt Bioscience Corp., Beverly, MA). Patient-specific barcodes and sequencing primers P1 and P2 were ligated to these fragments. The fragments were loaded and amplified with the SOLiD sequencing control beads during emulsion polymerase chain reaction. The beads were then added to the Flow Chip for sequencing in the SOLiD system (Life Technologies, Darmstadt, Germany), and sequenced according to the manufacturer’s protocol (Life Technologies). Individual reads were 75 bp long. Reads were assigned to their patient-specific barcode, and sequences were analyzed twice separately using the 2010 DNAnexus software and the following pipeline of bioinformatics software. The color-space reads were aligned with NovoalignCS24 and genotyped with GATK’s Unified Genotyper.25 SNV and indel discovery was performed across all samples using standard parameters and a maximum coverage of 10 000. The mean coverage of all amplicons was 2484 reads per amplicon. The resulting list of candidate SNVs was filtered with R.23 First, mutations outside the coding region were excluded. Second, known single nucleotide polymorphisms (SNPs) were removed (dbSNP, version 137). Third, mutations with a quality score of <8000 or an allele frequency of <15% were excluded. The remaining mutations were validated by Sanger sequencing.
To determine the coverage of genomic intervals, the data were processed with BEDTools26 and analyzed in R. Mutations were validated by Sanger sequencing and are only reported if they were detected by Sanger sequencing either in genomic DNA or in an independently whole genome-amplified DNA sample. The somatic or germline status of mutations in genes of the cohesin complex was established by evaluating remission samples or T cells (CD3+CD11b−CD14−CD33−) purified from diagnostic samples by flow cytometry.
Statistical analysis
The definition of complete remission (CR), overall survival (OS), and relapse-free survival (RFS) followed recommended criteria.27 Primary analysis was performed on OS. Sensitivity analyses were performed on CR and RFS, and results are displayed for exploratory purposes. Median follow-up time for survival was calculated according to the method of Korn.28 OS endpoints measured from the date of entry into one of the prospective studies were death (failure) and alive at last follow-up (censored). RFS endpoints measured from the date of documented CR were relapse (failure), death in CR (failure), and alive in CR at last follow-up (censored).
Pairwise comparisons of variables were performed for exploratory purposes using the Kolmogorov-Smirnov test and Student t test for continuous variables and the χ-squared test for categorical variables. The Kaplan-Meier method and log-rank test were used to estimate the distribution of OS and RFS, and to compare differences between survival curves, respectively. Mutations in the analyzed genes were used as categorical variables. To determine the expression levels of EVI1, relative quantification was calculated using the equation 2−ΔΔCt, as previously described.20 To provide quantitative information on the relevance of results, 95% confidence intervals (CIs) of odds ratios (OR) and hazard ratios (HR) were computed. Two-sided P values <.05 were considered significant in the primary analysis and indicators for a trend in all additional analyses. Statistical analyses were performed with the statistical software package SPSS 20.0 (IBM Corp., Armonk, NY). Associations between gene mutations are represented by a Circos diagram (Figure 1).29
Results
Mutations in the cohesin gene complex
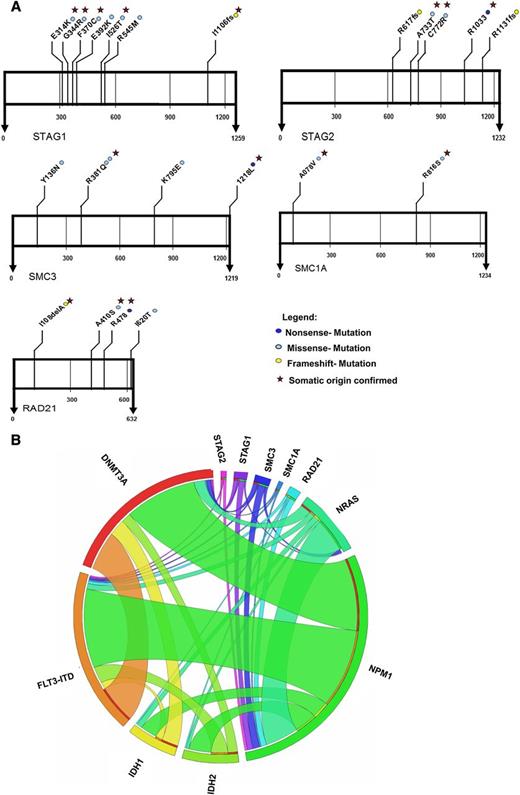
In our cohort, we identified mutations in genes of the cohesin complex in 23 AML patients (5.9%). The most commonly mutated gene in this complex was STAG1 with 7 patients harboring a mutation in this gene. The second most frequently mutated genes in the cohesin complex were SMC3 and STAG2, and we found 5 patients with these mutations, respectively. Additionally, we identified 4 patients with mutations in RAD21 and 2 patients with mutations in SMC1A (Figure 1). Interestingly, all mutations were mutually exclusive among each other. No mutation hotspot was identified in any of the genes (Figure 1 and supplemental Tables 2-6). STAG2 and SMC1A are X-linked, whereas the other 3 genes are autosomal. Of the 5 patients with STAG2 mutations, 2 patients were female and 3 were male. Both patients with SMC1A mutations were male. The male patients had a functionally homozygous mutation for STAG2 and SMC1A.
Location and type of mutations in genes of the cohesin complex in 389 patients with AML, and associations of gene mutations in the AML patient cohort outlined by a Circos diagram.
Location and type of mutations in genes of the cohesin complex in 389 patients with AML, and associations of gene mutations in the AML patient cohort outlined by a Circos diagram.
Six of 7 mutations in STAG1 were missense mutations, while one patient showed a frameshift mutation (Figure 1 and supplemental Table 2). Two patients with mutations in STAG2 had missense mutations, 2 patients had frameshift mutations, and 1 patient had a nonsense mutation in this gene (Figure 1 and supplemental Table 3). Four patients were identified with missense mutations in SMC3 and 1 patient with the stop codon being changed to leucine (Figure 1 and supplemental Table 4). In RAD21, we found 2 patients with missense mutations, 1 patient with a frameshift mutation, and 1 with a nonsense mutation (Figure 1 and supplemental Table 5). The 2 mutations in SMC1A were both missense mutations (Figure 1 and supplemental Table 6). Only the mutation in R381Q of SMC3 was recurrent in 2 patients, whereas all other mutations were only identified once (Figure 1 and supplemental Table 4).
From a total of 23 putative mutations in cohesin genes, the somatic status could be confirmed in 16 by analyzing remission samples or T-cells (CD3+CD11b−CD14−CD33), which were purified from diagnostic samples by flow cytometry (supplemental Tables 2-6). For the remaining patients with mutations, the somatic origin could not be confirmed due to lack of suitable material. Besides these somatic mutations in the cohesin complex, in our analysis, we also identified 3 unannotated SNPs (supplemental Table 7). These SNPs have not been reported in dbSNP (version 137) and were not identified in peripheral blood mononuclear cells from 30 healthy controls.
Association of cohesin gene mutations with clinical characteristics
Most clinical and disease characteristics of patients with mutations in the cohesin complex were similarly distributed, as in patients with wild-type cohesin genes (Table 1).30 Only 1 patient with favorable cytogenetics had a mutation in a cohesin gene (SMC1A), whereas the majority of patients with cohesin gene mutations had intermediate risk cytogenetics, most showing a normal karyotype (supplemental Tables 1 and 2-6). Interestingly, we found a strong correlation between the cohesin gene and NPM1 mutations (57% of cohesin gene mutated patients had an NPM1 mutation; 9.5% of NPM1 mutated patients had a cohesin gene mutation compared with 4% of NPM1 wild-type patients; P = .029) (Table 1). Of 7 patients with STAG1 mutations, 3 patients also showed an NPM1 mutation. Of 5 STAG2 and SMC3 mutated patients, 3 carried a mutation in NPM1, respectively. Of the 4 RAD21 mutated patients, 3 harbored a concomitant NPM1 mutation, and 1 of the 2 SMC1 mutated patients was also NPM1 mutated (supplemental Tables 2-6). No correlation was observed between genes in the cohesin complex and other mutations such as FLT3-ITD, IDH1, IDH2, or NRAS mutations. BAALC expression was lower in patients with a mutation compared with patients without a mutation in a gene of the cohesin complex (P = .033) (Table 1). There was no significant difference between patients with or without a mutation in the cohesin complex with regard to gene expression of MN1, ERG, EVI1, MLL5, and WT1. To get a better understanding of when mutations in the cohesin complex occur during clonal evolution, we evaluated the allelic burden of mutations in the cohesin complex. Because of the strong association between NPM1 and cohesin mutations, the allelic ratio of mutated and wild-type NPM1 was compared with the allelic ratio of mutated and wild-type cohesin genes. Interestingly, we found a similar mutation burden between NPM1 and genes of the cohesin complex in most patients (supplemental Figure 1), suggesting that cohesin gene mutations occurred in the same clone as NPM1 mutations.
Comparison of pretreatment characteristics between patients with and without mutations in the cohesin complex genes
| Characteristic . | Cohesin genes mutated (n = 23) no. (%) . | Cohesin genes wild-type (n = 366) no. (%) . | P . |
|---|---|---|---|
| Age, years | .13 | ||
| Median | 46 | 44.5 | |
| Range | 28-60 | 17-60 | |
| Sex | .58 | ||
| Male | 13 (57%) | 185 (51%) | |
| Female | 10 (43%) | 181 (49%) | |
| ECOG performance status | .37 | ||
| 0 | 5 (22%) | 90 (25%) | |
| 1 | 16 (70%) | 221 (60%) | |
| 2 | 1 (4%) | 53 (14%) | |
| Missing data | 1 (4%) | 2 (1%) | |
| FAB-subtype | .53 | ||
| M0 | 0 (0%) | 7 (2%) | |
| M1 | 8 (35%) | 59 (16%) | |
| M2 | 5 (22%) | 90 (25%) | |
| M4 | 7 (30%) | 111 (30%) | |
| M5 | 2 (9%) | 56 (15%) | |
| M6 | 0 (0%) | 6 (2%) | |
| M7 | 0 (0%) | 3 (1%) | |
| Missing data | 1 (4%) | 17 (5%) | |
| Type of AML | .32 | ||
| De novo | 22 (96%) | 326 (89%) | |
| Secondary | 1 (4%) | 40 (11%) | |
| Cytogenetic risk group† | .26 | ||
| Favorable | 1 (4%) | 61 (17%) | |
| Intermediate | 17 (74%) | 257 (70%) | |
| Adverse | 4 (17%) | 44 (12%) | |
| Missing data | 1 (4%) | 4 (1%) | |
| Peripheral blood blasts | .1 | ||
| Mean | 58.1 | 46.7 | |
| Missing data | 1 (4%) | 15 (4%) | |
| Bone marrow blasts | .6 | ||
| Mean | 74.1 | 71.4 | |
| Missing data | 0 (0%) | 24 (7%) | |
| WBC count | .66 | ||
| Median (x109/L) | 26.6 | 21.6 | |
| Range (x109/L) | 1.3-229 | 0.5-328 | |
| Missing data | 0 (0%) | 1 (0%) | |
| Hemoglobin | .09 | ||
| Median (g/L) | 8.4 | 9 | |
| Range (g/L) | 5.5-11.8 | 3-15.4 | |
| Missing data | 0 (0%) | 9 (2%) | |
| Platelet count | .26 | ||
| Median – (x109/L) | 39 | 48.5 | |
| Range – (x109/L) | 7-215 | 2.6-483 | |
| Missing data | 0 (0%) | 10 (3%) | |
| FLT3-ITD | .93 | ||
| Mutated | 6 (26%) | 98 (27%) | |
| Wild-type | 17 (74%) | 266 (73%) | |
| Missing | 0 (0%) | 2 (1%) | |
| NPM1 | .029 | ||
| Mutated | 13 (57%) | 124 (34%) | |
| Wild-type | 10 (43%) | 240 (66%) | |
| Missing | 0 (0%) | 2 (1%) | |
| NPM1/FLT3 mutation risk group | .024 | ||
| Low risk* | 9 (39%) | 71 (19%) | |
| High risk* | 14 (61%) | 293 (80%) | |
| Missing | 0 (0%) | 2 (1%) | |
| IDH1 | .15 | ||
| R132 mutated | 0 (0%) | 26 (7%) | |
| Wild-type | 22 (96%) | 276 (75%) | |
| Missing | 1 (4%) | 64 (17%) | |
| IDH2 | .43 | ||
| Mutated | 1 (4%) | 30 (8%) | |
| Wild-type | 20 (87%) | 271 (74%) | |
| Missing | 2 (9%) | 65 (18%) | |
| IDH1/2 | .11 | ||
| Mutated | 1 (4%) | 56 (15%) | |
| Wild-type | 20 (87%) | 244 (67%) | |
| Missing | 2 (9%) | 66 (18%) | |
| DNMT3A | .4 | ||
| Mutated | 6 (26%) | 68 (19%) | |
| Wild-type | 17 (74%) | 292 (80%) | |
| Missing | 0 (0%) | 6 (2%) | |
| NRAS | .48 | ||
| Mutated | 2 (9%) | 50 (14%) | |
| Wild-type | 18 (78%) | 263 (72%) | |
| Missing | 3 (13%) | 53 (14%) | |
| MN1 | .071 | ||
| Median copy number (relative MN1/ABL) | 9.03 | 15.15 | |
| Range (relative MN1/ABL) | 0.31-31.7 | 0.62-5134.5 | |
| BAALC | .033 | ||
| Median copy number (relative BAALC/ABL) | 1.70 | 4.70 | |
| Range (relative BAALC/ABL) | 0.04-26.2 | 0.026-806.4 | |
| ERG | .64 | ||
| Median copy number (relative ERG/ABL) | 28.1 | 33.4 | |
| Range (relative ERG/ABL) | 0.67-125.3 | 0.07-1605.7 | |
| EVI1 | .47 | ||
| Expressers | 0 (0%) | 6 (2%) | |
| Nonexpressers | 14 (61%) | 157 (43%) | |
| Missing | 9 (39%) | 203 (55%) | |
| MLL5 | .85 | ||
| Median copy number (relative MLL5/ABL) | 34.3 | 38.2 | |
| Range (relative MLL5/ABL) | 14.04-174.5 | 4.45-498 | |
| WT1 | .29 | ||
| Median copy number (relative WT1/ABL) | 0.87 | 1.2 | |
| Range (relative WT1/ABL) | 0.035-5.88 | 0.001-231.3 |
| Characteristic . | Cohesin genes mutated (n = 23) no. (%) . | Cohesin genes wild-type (n = 366) no. (%) . | P . |
|---|---|---|---|
| Age, years | .13 | ||
| Median | 46 | 44.5 | |
| Range | 28-60 | 17-60 | |
| Sex | .58 | ||
| Male | 13 (57%) | 185 (51%) | |
| Female | 10 (43%) | 181 (49%) | |
| ECOG performance status | .37 | ||
| 0 | 5 (22%) | 90 (25%) | |
| 1 | 16 (70%) | 221 (60%) | |
| 2 | 1 (4%) | 53 (14%) | |
| Missing data | 1 (4%) | 2 (1%) | |
| FAB-subtype | .53 | ||
| M0 | 0 (0%) | 7 (2%) | |
| M1 | 8 (35%) | 59 (16%) | |
| M2 | 5 (22%) | 90 (25%) | |
| M4 | 7 (30%) | 111 (30%) | |
| M5 | 2 (9%) | 56 (15%) | |
| M6 | 0 (0%) | 6 (2%) | |
| M7 | 0 (0%) | 3 (1%) | |
| Missing data | 1 (4%) | 17 (5%) | |
| Type of AML | .32 | ||
| De novo | 22 (96%) | 326 (89%) | |
| Secondary | 1 (4%) | 40 (11%) | |
| Cytogenetic risk group† | .26 | ||
| Favorable | 1 (4%) | 61 (17%) | |
| Intermediate | 17 (74%) | 257 (70%) | |
| Adverse | 4 (17%) | 44 (12%) | |
| Missing data | 1 (4%) | 4 (1%) | |
| Peripheral blood blasts | .1 | ||
| Mean | 58.1 | 46.7 | |
| Missing data | 1 (4%) | 15 (4%) | |
| Bone marrow blasts | .6 | ||
| Mean | 74.1 | 71.4 | |
| Missing data | 0 (0%) | 24 (7%) | |
| WBC count | .66 | ||
| Median (x109/L) | 26.6 | 21.6 | |
| Range (x109/L) | 1.3-229 | 0.5-328 | |
| Missing data | 0 (0%) | 1 (0%) | |
| Hemoglobin | .09 | ||
| Median (g/L) | 8.4 | 9 | |
| Range (g/L) | 5.5-11.8 | 3-15.4 | |
| Missing data | 0 (0%) | 9 (2%) | |
| Platelet count | .26 | ||
| Median – (x109/L) | 39 | 48.5 | |
| Range – (x109/L) | 7-215 | 2.6-483 | |
| Missing data | 0 (0%) | 10 (3%) | |
| FLT3-ITD | .93 | ||
| Mutated | 6 (26%) | 98 (27%) | |
| Wild-type | 17 (74%) | 266 (73%) | |
| Missing | 0 (0%) | 2 (1%) | |
| NPM1 | .029 | ||
| Mutated | 13 (57%) | 124 (34%) | |
| Wild-type | 10 (43%) | 240 (66%) | |
| Missing | 0 (0%) | 2 (1%) | |
| NPM1/FLT3 mutation risk group | .024 | ||
| Low risk* | 9 (39%) | 71 (19%) | |
| High risk* | 14 (61%) | 293 (80%) | |
| Missing | 0 (0%) | 2 (1%) | |
| IDH1 | .15 | ||
| R132 mutated | 0 (0%) | 26 (7%) | |
| Wild-type | 22 (96%) | 276 (75%) | |
| Missing | 1 (4%) | 64 (17%) | |
| IDH2 | .43 | ||
| Mutated | 1 (4%) | 30 (8%) | |
| Wild-type | 20 (87%) | 271 (74%) | |
| Missing | 2 (9%) | 65 (18%) | |
| IDH1/2 | .11 | ||
| Mutated | 1 (4%) | 56 (15%) | |
| Wild-type | 20 (87%) | 244 (67%) | |
| Missing | 2 (9%) | 66 (18%) | |
| DNMT3A | .4 | ||
| Mutated | 6 (26%) | 68 (19%) | |
| Wild-type | 17 (74%) | 292 (80%) | |
| Missing | 0 (0%) | 6 (2%) | |
| NRAS | .48 | ||
| Mutated | 2 (9%) | 50 (14%) | |
| Wild-type | 18 (78%) | 263 (72%) | |
| Missing | 3 (13%) | 53 (14%) | |
| MN1 | .071 | ||
| Median copy number (relative MN1/ABL) | 9.03 | 15.15 | |
| Range (relative MN1/ABL) | 0.31-31.7 | 0.62-5134.5 | |
| BAALC | .033 | ||
| Median copy number (relative BAALC/ABL) | 1.70 | 4.70 | |
| Range (relative BAALC/ABL) | 0.04-26.2 | 0.026-806.4 | |
| ERG | .64 | ||
| Median copy number (relative ERG/ABL) | 28.1 | 33.4 | |
| Range (relative ERG/ABL) | 0.67-125.3 | 0.07-1605.7 | |
| EVI1 | .47 | ||
| Expressers | 0 (0%) | 6 (2%) | |
| Nonexpressers | 14 (61%) | 157 (43%) | |
| Missing | 9 (39%) | 203 (55%) | |
| MLL5 | .85 | ||
| Median copy number (relative MLL5/ABL) | 34.3 | 38.2 | |
| Range (relative MLL5/ABL) | 14.04-174.5 | 4.45-498 | |
| WT1 | .29 | ||
| Median copy number (relative WT1/ABL) | 0.87 | 1.2 | |
| Range (relative WT1/ABL) | 0.035-5.88 | 0.001-231.3 |
ECOG, performance status of the Eastern Cooperative Oncology Group; FAB, French-American-British classification of AML; FLT3-ITD, internal tandem duplication of the FLT3 gene; no., number; P, P value from two-sided χ-squared tests for categorical variables and from 2-sided Student t or Kolmogorov-Smirnov tests for continuous variables.
The high-risk molecular group is defined as either NPM1wild-type/FLT3-ITDnegative, or NPM1wild-type/FLT3-ITDpositive, or NPM1mutated/FLT3-ITDpositive. The low-risk molecular group is defined by the presence of an NPM1 mutation and the absence of FLT3-ITD.
The cytogenetic risk group is defined according to Medical Research Council criteria.30
Clinical outcome in the total cohort of AML patients according to cohesin gene mutation status
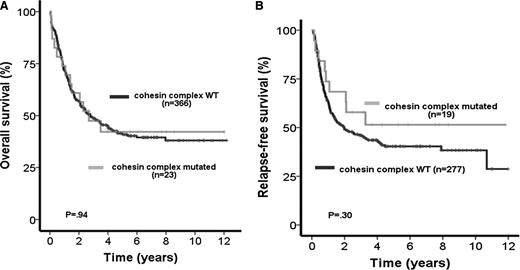
Median follow-up time for all patients was 5.1 years (range, 0.19 to 12.2 years). When considering all mutations in the cohesin complex as 1 group, OS and RFS were not influenced by the presence of cohesin mutations (OS: HR 0.98; 95% CI, 0.56-1.72; P = .94; Figure 2A) (RFS: HR 0.70; 95% CI, 0.36-1.38; P = .3, Figure 2B and Table 2). In addition, there was no difference between CR rates of mutated and wild-type patients (mutated 83% vs wild-type 76%; P = .45). Similar results were obtained when only considering patients with de novo AML (n = 348) with the exclusion of patients with secondary AML (OS: HR 0.96; 95% CI, 0.54-1.73; P = .89; supplemental Figure 2A), (RFS: HR 0.62; 95% CI, 0.3-1.25; P = .18; supplemental Figure 2B), and (CR: mutated 82% vs wild-type 78%; P = .65). For exploratory purposes, next we evaluated the prognostic influence of each gene in the cohesin complex separately in all patients, although this analysis is limited by the small number of mutated patients. STAG1, STAG2, SMC3, and RAD21 mutations had no influence on OS and RFS, whereas the analysis was not performed for SMC1A due to the low mutation frequency (Table 2).
Prognostic impact of cohesin mutations in all investigated AML patients. (A) OS in AML patients with wild-type (WT) or mutated genes of the cohesin complex. (B) RFS in AML patients with WT or mutated genes of the cohesin complex.
Prognostic impact of cohesin mutations in all investigated AML patients. (A) OS in AML patients with wild-type (WT) or mutated genes of the cohesin complex. (B) RFS in AML patients with WT or mutated genes of the cohesin complex.
Univariate analysis for OS and RFS in AML patients (n = 389) according to cohesin gene mutation status
| . | OS . | RFS . | ||||
|---|---|---|---|---|---|---|
| Endpoint . | HR . | 95% CI . | P . | HR . | 95% CI . | P . |
| Cohesin mutations combined | ||||||
| Cohesin mutated vs wild-type | 0.98 | 0.56-1.72 | .94 | 0.70 | 0.36-1.38 | .3 |
| Cohesin mutations separately | ||||||
| STAG1 mutated vs wild-type | 0.68 | 0.22-2.12 | .5 | 0.50 | 0.12-2 | .33 |
| STAG2 mutated vs wild-type | 1.53 | 0.57-4.11 | .4 | 1.26 | 0.4-3.93 | .7 |
| SMC3 mutated vs wild-type | 0.28 | 0.04-1.97 | .2 | 0.05 | 0-4.75 | .19 |
| RAD21 mutated vs wild-type | 1.47 | 0.47-4.60 | .51 | 1.02 | 0.25-4.1 | .98 |
| . | OS . | RFS . | ||||
|---|---|---|---|---|---|---|
| Endpoint . | HR . | 95% CI . | P . | HR . | 95% CI . | P . |
| Cohesin mutations combined | ||||||
| Cohesin mutated vs wild-type | 0.98 | 0.56-1.72 | .94 | 0.70 | 0.36-1.38 | .3 |
| Cohesin mutations separately | ||||||
| STAG1 mutated vs wild-type | 0.68 | 0.22-2.12 | .5 | 0.50 | 0.12-2 | .33 |
| STAG2 mutated vs wild-type | 1.53 | 0.57-4.11 | .4 | 1.26 | 0.4-3.93 | .7 |
| SMC3 mutated vs wild-type | 0.28 | 0.04-1.97 | .2 | 0.05 | 0-4.75 | .19 |
| RAD21 mutated vs wild-type | 1.47 | 0.47-4.60 | .51 | 1.02 | 0.25-4.1 | .98 |
HR >1 or <1 indicate an increased or decreased risk, respectively, of an event for the first category listed.
P, P value from univariate or multivariate Cox proportional hazards models.
In the subgroup of patients with CN-AML (n = 201), we identified 16 patients with mutations in the cohesin complex (8%). In this subgroup, we did not identify a difference in OS and RFS for patients with or without mutations in the cohesin complex (OS: HR 0.73; 95% CI, 0.34-1.57; P = .42) and (RFS: HR 0.47; 95% CI, 0.19-1.16; P = .1; supplemental Figure 3A-B). Again, no difference was found in CR rates (mutated 88% vs wild-type 78%; P = .39). Due to the strong association between NPM1 mutations and mutations in the cohesin complex, we studied the impact of cohesin mutations on OS and RFS in NPM1-mutated AML patients separately. In the group of NPM1-mutated AML patients (n = 137), OS and RFS were not influenced by the presence of mutations in the cohesin complex (OS: HR 1.17; 95% CI, 0.53-2.55; P = .7; supplemental Figure 3C) and (RFS: HR 0.85; 95% CI, 0.34-2.12; P = .72; supplemental Figure 3D). When considering the otherwise favorable prognostic group of patients with mutated NPM1, but wild-type FLT3, no significant difference for OS and RFS between patients with our without mutations in the cohesin complex was identified (OS: HR 0.65; 95% CI, 0.2-2.14; P = .48; supplemental Figure 3E) and (RFS: HR 0.32; 95% CI, 0.07-1.39; P = .13; supplemental Figure 3F).
Similar results were obtained when considering NPM1-mutated CN-AML patients or when looking at the different prognostic groups in the European LeukemiaNet classification (data not shown).31
We analyzed the effect of allogeneic transplantation on OS of patients with cohesin mutations. Our clinical trial protocol allowed an intent-to-treat analysis on the basis of donor availability in patients with CN-AML. The OS of cohesin-mutated patients with a related donor was similar compared with patients without a related donor (OS: HR 0.54; 95% CI, 0.06-4.8; P = .58; supplemental Figure 4A-B), suggesting that cohesin gene mutations did not influence the outcome of allogeneic transplantation.
Discussion
In this analysis of 389 well-characterized patients with AML, we identified somatic mutations in the cohesin complex in 5.9% of patients. In our cohort, STAG1 followed by STAG2 and SMC3 mutations were the most frequently mutated genes in the cohesin complex, whereas SMC1A mutations were rare events. In the recent report of the Cancer Genome Atlas Research Network, no mutations in STAG1 were detected, whereas the mutation frequency in STAG2, SMC1A, SMC3, and RAD21 was slightly higher (2.5%-3.5%).1 The lower mutation frequency in our study may be explained by our approach that we only considered mutations that could be validated by Sanger sequencing and therefore are present in at least 10% to 20% of all cells. All mutations were heterozygous apart from STAG2 and SMC1A mutations in male patients, as both genes are X-linked. Mutations occurred throughout the genes without the presence of a mutational hotspot. Interestingly, mutations in this complex were mutually exclusive, similar to other mutations that belonged to 1 pathway similar to genes of the spliceosome complex or mutations in IDH1, IDH2, and TET2.13,32-34
To differentiate polymorphisms from somatic mutations, we studied T-cells purified from diagnostic samples by flow cytometry. In addition to the 23 somatic mutations, we identified 3 SNPs (2 in STAG1 and 1 in RAD21), which have not been described by dbSNP and were not found in mononuclear cells of 30 healthy donors. Although germline mutations in SMC1A, SMC3, and RAD21 have been associated with cohesinopathies, pathogenic STAG1 germline mutations have not been reported.35 All patients studied during remission lost the mutation at this time point. This data implements that cohesin mutations could be potentially used for minimal residual disease monitoring. However, further studies are needed to analyze the stability of these markers, as we also found 1 patient in which the SMC1A mutation from the time of diagnosis was neither present during remission nor at the time of relapse.
Most mutated patients had a normal karyotype, suggesting that cohesin gene mutations do not act through destabilization of chromosomal integrity in AML, but rather alternative mechanisms such as transcriptional control. Interestingly, cohesin genes have been found to bind to CCCTC-binding factor, a sequence-specific transcription factor36 that is known to interact with NPM1 in addition to regulating tumor suppressor loci.37 Importantly, cohesin genes can also affect transcription independently of CCCTC-binding factor.38
We compared the allelic burden of mutations in the cohesin complex with the allelic burden of NPM1 mutations, as the level of the allelic burden can indicate clonal hierarchy of different mutations.39 It has already been suggested that NPM1 is an early event in leukemogenesis.40 In our analysis, the allelelic burden of mutations in the cohesin genes were very similar to that observed for NPM1 mutations. This suggests that cohesin mutations may occur at a similarly early time point of leukemogenesis.
An interesting finding was the strong association between NPM1 mutations and the mutations in cohesin genes, as already indicated in the recent report of the Cancer Genome Atlas Research Network.1 In our analysis, the prognostic implications of mutations in the cohesin complex were not significant when considering all mutations in the complex together or individually.
Because of the strong association between NPM1 mutations and genes in the cohesin complex, we wanted to find out whether the cohesin mutations have an impact in the NPM1-mutated patient group. It could be possible that a negative prognostic impact of cohesin mutations might have been missed because of the favorable prognostic impact of NPM1 mutations and the strong association between these mutations. However, in CN-AML patients, in patients with mutated NPM1, and in the NPM1 mutated/FLT3 wild-type subgroup of patients, cohesin mutations had no impact on outcome. Taken together, our data suggest that cohesin mutations do not affect patient prognosis.
In summary, our results show that mutations in the cohesin complex are recurrent mutations in AML. We found a strong association between these mutations and mutations in NPM1. Cohesin complex mutations as a group had no prognostic impact in all and in cytogenetically normal AML patients.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgment
This study was supported by grants from Deutsche Krebshilfe (109003, 110284, and 110292), a grant from the Deutsche-José-Carreras Leukämie-Stiftung e.V. (DJCLS R 10/22), a grant from the German Federal Ministry of Education and Research (01EO0802) (IFB-Tx), and grants from the Deutsche Forschungsgemeinschaft (HE 5240/4-1, HE 5240/5-1, and TH 1779/1-1).
Authorship
Contribution: F.T. and M.H. designed the research; F.T., R.B., M.G., K.J.S., A.K., L.H., A.C., M.W., LW., R.Shahswar, F.D., and M.H. performed the research; F.T., R.Schlenk, K.D., H.D., J.K., A.G., and M.H. contributed patient samples and clinical data; G.G. and B.S. performed cytogenetic studies; F.T., C.W., M.D., A.G., and M.H. analyzed the data; F.T., A.G., and M.H. wrote the paper; and all authors read and agreed to the final version of the manuscript.
Conflict of interest disclosure: The authors declare no competing financial interests.
Correspondence: Felicitas Thol, Department of Hematology, Hemostasis, Oncology, and Stem Cell Transplantation, Hannover Medical School, Carl-Neuberg Strasse 1, 30625 Hannover, Germany; e-mail: thol.felicitas@mh-hannover.de.