To the editor:
Recently, the Met408-Del420 haplotype in the OCT1 gene (the combination of methionine at codon 408 and a deletion of another methionine at codon 420) was associated with lack of transport activity for imatinib and tetraethylammonium (TEA+) and with poor clinical outcome in chronic myeloid leukemia patients.1
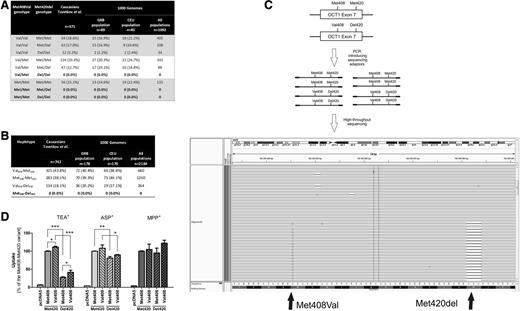
To explore the relevance of the Met408-Del420 haplotype in other clinical contexts, we genotyped 371 Caucasians who were previously studied for the effects of OCT1 polymorphisms on pharmacokinetics or efficacy of metformin, tropisetron, or tramadol.2-4 To our great surprise, none of the homozygous Met408 carriers carried the Met420-deletion allele (Figure 1A). In comparison, 52% of the homozygous Val408 carriers carried the Met420 deletion (12 homozygous and 63 heterozygous carriers; P < 10−6, χ2 test for comparison with homozygous Met408 carriers). This suggested a lack of the Met408-Del420 haplotype as confirmed with PHASE software to infer the individual haplotypes (Figure 1B). Furthermore, we performed haplotype-specific resequencing of all double-heterozygous carriers of the Met408 and the Met420-deletion alleles to exclude the possibility that they may carry the Met408-Del420 haplotype. We amplified the 2 polymorphic loci, which are 36-bp apart, in a single polymerase chain reaction (PCR) amplicon and resequenced the products using semiconductor-based sequencing. In all double-heterozygous individuals in our population (n = 47; Figure 1A), we found only the Val408-Del420 and not the Met408-Del420 haplotype (Figure 1C). To confirm that this finding was not restricted to our population, we also analyzed the Met408Val and Met420del genotypes in the samples of the 1000 Genomes Project.5 The haplotype Met408-Del420 was not present in other European populations or worldwide (Figure 1A-B).
Haplotype combinations of the Met408Val and Met420del polymorphisms in the OCT1 gene in humans and their functional activity. (A-B) Combined representation of the Met408Val and Met420-deletion genotypes (A) and haplotypes (B) of Caucasians from our previous clinical studies and from selected (CEU and GRB) or all populations from the 1000 Genomes Project (the individual genotype data that was used in the calculations was obtained from http://www.ncbi.nlm.nih.gov/variation/tools/1000genomes/).5 Individual haplotypes were inferred using PHASE version 2.1.6 The genotype/haplotype frequencies are given as number (and %) of all individuals in the corresponding population. CEU, population of Utah residents (CEPH) with Northern and Western European ancestry; GRB, population of British in England and Scotland. (C) Strategy (upper 2 panels) and an example of the results (lower panel) of haplotype sequencing analyses using semiconductor-based massively parallel sequencing. A 155-bp region from the chromosome 6 was amplified via PCR. The region contained both the Met408Val and Met420-deletion loci. This haplotype-specific single-molecule DNA analysis enabled us to experimentally verify the presence of different haplotype combinations in individuals heterozygous for the Met408Val and Met420-deletion polymorphisms. The PCR products were purified, pooled together, subjected to template preparation using Ion OneTouch, and sequenced using Ion PGM (both from Life Technologies, Darmstadt, Germany). We obtained on average 19 000 sequencing reads per chromosome that covered both loci. The results were visualized using Integrative Genomics Viewer version 2.3 (Broad Institute; http://www.broadinstitute.org/igv/). Each gray line represents a single sequencing read. The reads containing the Val408 allele are denoted with a G indicating a disagreement with the Met408 allele that is present in the human reference sequence (that is given in the bottom of the figure). The presence of the Met420 deletion is designated with a black dash indicating a disagreement with the presence of ATG in the human reference sequence. This is a representative example of part of the reads of a single sample demonstrating that individuals heterozygous for both Met408 and Met420 deletion did not carry the 2 alleles on the same chromosome. (D) Comparison of the uptake activity of hOCT1 carrying all theoretically possible combinations of the Met420del and Met408Val polymorphisms. The uptake was measured in human embryonic kidney 293 cells stably transfected with the hOCT1 variants or with an empty control plasmid pcDNA5. The stably transfected cells were generated using the FlpIn system for targeted chromosomal integration (Life Technologies) following a procedure described before.7 The uptake was measured as the total cellular accumulation after a 2-minute incubation at 37°C with 1 µM MPP+, 5 µM TEA+, or 5 µM ASP+. The concentrations used were below (MPP+ and TEA+) or close to (ASP+) the known KM values for the substrates. The concentrations of TEA+ and MPP+ used in this study were identical to the one used by Giannoudis et al1 and Shu et al,8 respectively. Two minutes of incubation were within the linear range for time dependence of the OCT1-mediated uptake. Shown are means and standard error of the means of at least 3 independent experiments, each performed in duplicate. Met420 deletion caused a significant 70% decrease in TEA+ uptake. The decrease was less strong (18.6%) when ASP+ was used and missing when MPP+ was used as a substrate. In comparison, the exchange of Met408 to Val408 caused only a 12% increase in TEA+ uptake. The effects of Met408Val substitution were similar in size both with the Met420 and Del420 background and vice versa. This suggests independent additive effects and a lack of epistatic interaction between the 2 polymorphisms.
Haplotype combinations of the Met408Val and Met420del polymorphisms in the OCT1 gene in humans and their functional activity. (A-B) Combined representation of the Met408Val and Met420-deletion genotypes (A) and haplotypes (B) of Caucasians from our previous clinical studies and from selected (CEU and GRB) or all populations from the 1000 Genomes Project (the individual genotype data that was used in the calculations was obtained from http://www.ncbi.nlm.nih.gov/variation/tools/1000genomes/).5 Individual haplotypes were inferred using PHASE version 2.1.6 The genotype/haplotype frequencies are given as number (and %) of all individuals in the corresponding population. CEU, population of Utah residents (CEPH) with Northern and Western European ancestry; GRB, population of British in England and Scotland. (C) Strategy (upper 2 panels) and an example of the results (lower panel) of haplotype sequencing analyses using semiconductor-based massively parallel sequencing. A 155-bp region from the chromosome 6 was amplified via PCR. The region contained both the Met408Val and Met420-deletion loci. This haplotype-specific single-molecule DNA analysis enabled us to experimentally verify the presence of different haplotype combinations in individuals heterozygous for the Met408Val and Met420-deletion polymorphisms. The PCR products were purified, pooled together, subjected to template preparation using Ion OneTouch, and sequenced using Ion PGM (both from Life Technologies, Darmstadt, Germany). We obtained on average 19 000 sequencing reads per chromosome that covered both loci. The results were visualized using Integrative Genomics Viewer version 2.3 (Broad Institute; http://www.broadinstitute.org/igv/). Each gray line represents a single sequencing read. The reads containing the Val408 allele are denoted with a G indicating a disagreement with the Met408 allele that is present in the human reference sequence (that is given in the bottom of the figure). The presence of the Met420 deletion is designated with a black dash indicating a disagreement with the presence of ATG in the human reference sequence. This is a representative example of part of the reads of a single sample demonstrating that individuals heterozygous for both Met408 and Met420 deletion did not carry the 2 alleles on the same chromosome. (D) Comparison of the uptake activity of hOCT1 carrying all theoretically possible combinations of the Met420del and Met408Val polymorphisms. The uptake was measured in human embryonic kidney 293 cells stably transfected with the hOCT1 variants or with an empty control plasmid pcDNA5. The stably transfected cells were generated using the FlpIn system for targeted chromosomal integration (Life Technologies) following a procedure described before.7 The uptake was measured as the total cellular accumulation after a 2-minute incubation at 37°C with 1 µM MPP+, 5 µM TEA+, or 5 µM ASP+. The concentrations used were below (MPP+ and TEA+) or close to (ASP+) the known KM values for the substrates. The concentrations of TEA+ and MPP+ used in this study were identical to the one used by Giannoudis et al1 and Shu et al,8 respectively. Two minutes of incubation were within the linear range for time dependence of the OCT1-mediated uptake. Shown are means and standard error of the means of at least 3 independent experiments, each performed in duplicate. Met420 deletion caused a significant 70% decrease in TEA+ uptake. The decrease was less strong (18.6%) when ASP+ was used and missing when MPP+ was used as a substrate. In comparison, the exchange of Met408 to Val408 caused only a 12% increase in TEA+ uptake. The effects of Met408Val substitution were similar in size both with the Met420 and Del420 background and vice versa. This suggests independent additive effects and a lack of epistatic interaction between the 2 polymorphisms.
Secondly, we expressed all 4 possible Met408Val and Met420del combinations in human embryonic kidney 293 cells and measured transport of 1-methyl-4-phenylpyridinium+ (MPP+), TEA+, and 4-(4-dimethylamino) styryl-N-methylpyridinium+ (ASP+) (Figure 1D). We observed strong, substrate-specific effects of the Met420 deletion on OCT1 activity. However, the activity of the Met420 deletion was only marginally higher with valine408 than with methionine408. Our results are in line with the previously reported lack of relevant differences in MPP+ uptake8 and do not confirm the strong effects of the Met408Val polymorphisms on TEA+ uptake reported by Giannoudis et al.1 We did not use imatinib in this study because the validity of the Giannoudis et al data that suggest OCT1-mediated transport of imatinib has been questioned9 and recent attempts to demonstrate imatinib transport by OCT1 failed.10
Finally, careful inspection of the three-dimensional (3D) model published by Giannoudis et al1 used to explain the interactions between Met408Val and Met420del reveals that the model comprises only of 11 instead of 12 transmembrane helices (helix 1 is missing). Using the same approach and software, we obtained a 3D model with a helical cytoplasmic-loop domain between helices 6 and 7 that did not contain a β-sheet structure as described by Giannoudis et al. The long cytoplasmic loop is conserved in structurally homologous transporters like the glucose/H+ symporter (Protein Data Bank entry 4LDS) and XylE (Protein Data Bank entries 4GC0 and 4JA4). In all these structures, the intracellular loop forms a compact arrangement of 3 helices without β sheets. Consistently, secondary structure predictions using the software JPred also propose α-helical structures for the loop segment. Therefore, the conclusion of Giannoudis et al that the mutations alter the intracellular loop conformation seems very speculative given the fact that the applied modeling procedure seems to highly influence the predicted 3D structure.
In conclusion, our study demonstrates that the Met420 deletion exists exclusively together with the Val408 allele in humans and suggests that the Met420 deletion leads to substrate-specific loss of activity that is only marginally affected by the Met408Val polymorphism.
Authorship
Acknowledgments: The authors thank Ana Tzvetkova for her help in the bioinformatics analysis of the next-generation sequencing data. This study was financially supported by German Research Foundation grant GRK1034.
Contribution: M.V.T. designed the study, analyzed the data, produced the figures, and wrote the manuscript; T.S. performed experiments, analyzed the data and produced the figures; K.B. performed the experiments; T.M. analyzed data; and J.B. and H.K. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Mladen V. Tzvetkov, University of Göttingen, Institute of Clinical Pharmacology, Robert-Koch-Strasse 40, D-37075 Göttingen, Germany; e-mail: mtzvetk@gwdg.de.