Abstract
Although the prognosis of chronic myeloid leukemia (CML) patients treated with imatinib is good, many fail to develop an optimal response or lose one. This heterogeneity could be attributed to the presence of human organic cation transporter-1 (hOCT1) single nucleotide polymorphisms (SNPs). In the present study, we analyzed the effect of 23 hOCT1 SNPs on imatinib treatment outcome in newly diagnosed CML patients using MassARRAY sequencing and pyrosequencing. The only SNP associated with outcome was M420del (rs35191146), with patients with the M420del demonstrating an increased probability of imatinib treatment failure. In CML cell lines transfected with M420del and/or M408V, M420del significantly decreased imatinib uptake, but this effect was countered if the M408V (rs628031) SNP was also present. A similar effect was seen for the uptake of the hOCT1 substrates TEA+ and ASP+. Finally, apparent hOCT1 mRNA levels were studied using both our earlier primers covering the M420del and another set that did not. Different mRNA expression was observed, explaining the disparity in published data on the prognostic importance of hOCT1 mRNA and highlighting the importance of avoiding common SNP sites in primer design. These data demonstrate that the common M420del SNP can modulate the outcome of imatinib treatment.
Key Points
M420del is the only hOCT1 SNP linked with outcome in imatinib-treated CML. Its assessment at diagnosis may be useful to tailor TKI therapy.
The relevance of M420del is confirmed in a functional assay, in which it decreased imatinib uptake, whereas M408V counters this effect.
Introduction
Membrane transporters are major determinants of drug pharmacokinetics and pharmacodynamics. The human organic cation transporter 1 (hOCT1, SLC22A1), an influx transporter, is responsible for the uptake of imatinib into chronic myeloid leukemia (CML) cells.1-3 Imatinib, a tyrosine kinase inhibitor (TKI), is now the standard of care for the treatment of chronic phase CML. The majority of patients achieve complete cytogenetic response (CCR) and approximately half achieve a major molecular response (MMR) at 3 years of treatment.4 However, there are still some patients who never achieve a CCR while others lose an initial response. The frequency of imatinib resistance and intolerance is estimated at 30%-40%.5-8 Several mechanisms of resistance and/or intolerance have been described, including mutations in the kinase domain and amplification of the BCR-ABL gene. In addition, the level of expression and/or activity of hOCT1 are powerful predictors of the clinical response to imatinib, so low expression or activity is a determinant of imatinib resistance. Studies of hOCT1 activity have focused on imatinib uptake with and without inhibitors.1-3 Although this may give useful information on hOCT1 function and the amount of hOCT1 present, there are very little data on single nucleotide polymorphisms (SNPs) affecting the function of hOCT1 in CML.
hOCT1 exhibits several nonsynonymous SNPs that affect the transport of the hOCT1 substrates ASP+ [4-(4-(dimethylamino)styryl)-N-methylpyridinium], MPP+ (3-methyl-4-phenylpyridinium), and TEA+ (tetraethylammonium).9-11 SNPs in hOCT1, in particular the deletion of methionine at codon 420 (M420del; rs35191146), can affect the action and pharmacokinetics of metformin, an antidiabetic drug and a well-known hOCT1 substrate.12,13 Several studies have investigated the association between the presence of hOCT1 SNPs and clinical response to imatinib with somewhat contradictory results.14-17 However, all of these studies focused only on a small (up to 8) and differing selection of SNPs, some of which are infrequent in the general population. None of these studies correlated clinically significant SNPs with studies on imatinib uptake in CML cells, raising the possibility that these may be chance observations.
In the present study, we investigated the genotype frequencies and effects of 23 hOCT1 SNPs on the clinical response to imatinib treatment in a large cohort of newly diagnosed CML patients. Of these, the most common exonic hOCT1 SNPs were M420del and M408V (rs628031), with an allelic frequency of approximately 18.5% and 59.8%, respectively, in European-Americans according to the National Center for Biotechnology Information (NCBI). We report herein that M420del is associated with imatinib treatment failure, which was confirmed in a functional assay of imatinib uptake showing that M408V may counteract this effect. Furthermore, we show that the disparity in the data concerning the predictive value of hOCT1 mRNA levels on outcome may be because differing PCR primer locations are inadvertently detecting M420del. This is compatible with the notion that patient-to-patient variation is predominantly because of hOCT1 SNPs (and their resultant effect on function/activity) rather than gene expression.
Methods
Patient clinical response and outcome
A total of 336 newly diagnosed chronic-phase CML patients treated with imatinib were examined in this study. Of these, 180 were from the Royal Liverpool University Hospital (Liverpool, United Kingdom), 11 were from the Royal Free Hospital (London, United Kingdom), 19 from the Hammersmith Hospital (London, United Kingdom), 70 were from the University of Glasgow (Glasgow, United Kingdom), and 56 were from the University of Heidelberg (Heidelberg, Germany). All patients received imatinib 400 mg daily from diagnosis, preceded only by up to 6 weeks of hydroxycarbamide. Complete clinical data were available for a total of 195 patients (median age, 50 years [range, 19-80]; 52% were male) and thus were assessable for analyses of association of SNP with clinical outcome. Each patient gave informed consent in accordance with the Declaration of Helsinki. The study, which was approved by the Liverpool Local Research Ethics Committee, commenced enrollment in June 2000 with follow-up until July 2011.
To investigate the association between hOCT1 SNPs and clinical response, tests of association with each of the following outcomes were undertaken: (1) progression-free survival (PFS): an event was defined as progression to accelerated phase (AP), blast crisis (BC), or death; (2) event-free survival (EFS): an event was defined as either loss of complete hematologic or cytogenetic response, progression to AP or BC, or death; and (3) time to treatment failure (TTF): an event was a change of treatment because of either unsatisfactory response or intolerance to imatinib.
hOCT1 SNP selection and genotyping
The hOCT1 SNPs included in this study fulfilled 1 or more of the following criteria: (1) were previously published SNPs with known function for hOCT1 substrates; (2) were recorded in the NCBI (www.ncbi.nlm.nih.gov) SNP database and on the HapMap SNP site (www.hapmap.org); and (3) were SNPs with a minor allele frequency (MAF) of > 5% according to NCBI. This yielded a total of 47 SNPs. However, 24 of those SNPs were found to be tagged to others (cutoff point: r2 = 0.81), so 23 individual SNPs were selected for study in MassARRAY sequencing (Sequenom). Multiplex SNP assays were designed using the MassARRAY Designer software (www.mysequenom.com) that can automatically design both PCR and MassEXTEND primers.18 The M408V SNP was found to be tagged to rs594709 (intronic SNP) with a D′ = 1 and r2 = 0.89. The forward, reverse, and extension primers used are provided in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). One 18-plex assay and 1 4-plex assay were designed. The M420del SNP failed the primer design software and was therefore individually screened by PCR pyrosequencing. The locations of all of the 23 SNPs analyzed, including M408V, are illustrated in Table 1.
hOCT1 SNPs examined, defined by rs number and (where exonic) their amino acid (AA) translation
| Analyzed dbSNP ID . | AA translation . | Region . | Alleles . | HWpval . | Observed MAF . | MAF in NCBI . | Function . | Reference(s) . |
|---|---|---|---|---|---|---|---|---|
| rs6935207 | 5′near gene | G:A | 0.105 | A: 0.257 | A: 0.345 | |||
| rs9457840 | 5′near gene | T:C | 1.000 | C: 0.020 | C: 0.065 | |||
| rs6899549 | 5′near gene | T:C | 0.003 | C: 0.003 | C: 0.013 | |||
| rs34447885* | C14F | Exon 1 | C:T | 1.000 | 0 | T: 0.005 | Reduced MPP, increased Metformin transport | 12, 26 |
| rs12208357 | R61C | Exon 1 | C:T | 1.000 | T: 0.067 | T: 0.027 | Reduced MPP and Metformin transport | 9, 26 |
| rs45584532 | Intron | C:T | 1.000 | T: 0.177 | T: 0.101 | |||
| rs683369 | L160F | Exon 2 | C:G | 0.434 | G: 0.217 | G: 0.129 | Loss of response, imatinib failure | 14 |
| rs594709† | Intron | A:G | 0.623 | G: 0.368 | G: 0.313 | |||
| rs36103319 | G220V | Exon 3 | G:T | 1.000 | T: 0.002 | T: 0.001 | Reduced MPP transport | 9, 26 |
| rs3777392 | Intron | G:A | 1.000 | A: 0.092 | A: 0.090 | |||
| rs2282143 | P341L | Exon 6 | C:T | 1.000 | T: 0.02 | T: 0.058 | Reduced MPP and Metformin transport | 9, 26 |
| rs6937722* | Intron | G:A | 1.000 | A: 0.05 | A: 0.022 | |||
| rs35191146‡ | M420del | Exon 7 | G/GAT/- | 0.412 | (-): 0.154 | (-): 0.155 | Reduced MPP and Metformin transport | 12 |
| rs1867350 | Intron | G:A | 0.088 | A: 0.124 | A: 0.144 | |||
| rs9295122* | Intron | C:T | 0.757 | T: 0.42 | T: 0.411 | |||
| rs622342 | Intron | A:C | 0.593 | C: 0.359 | C: 0.272 | |||
| rs11753995* | Intron | G:A | 1.000 | A: 0.149 | A: 0.099 | |||
| rs34059508 | G465R | Exon 9 | G:A | 0.048 | A: 0.031 | A: 0.008 | Reduced MPP and Metformin transport | 9, 26 |
| rs35270274 | R488T | Exon 9 | G:T | 1.000 | T: 0.002 | T: 0.011 | ||
| rs622591 | Intron | G:A | 0.384 | A: 0.219 | A: 0.337 | |||
| rs9457846 | 3′UTR | G:A | 0.335 | A: 0.09 | A: 0.002 | |||
| rs651164 | 3′UTR | G:A | 0.122 | A: 0.353 | A: 0.367 | |||
| rs2083867 | 3′UTR | A:G | 0.267 | G: 0.389 | G: 0.381 | |||
| rs628031 tagged to rs594709† | M408V | Exon 7 | G:A | 0.623 | A: 0.368 | A: 0.302 |
| Analyzed dbSNP ID . | AA translation . | Region . | Alleles . | HWpval . | Observed MAF . | MAF in NCBI . | Function . | Reference(s) . |
|---|---|---|---|---|---|---|---|---|
| rs6935207 | 5′near gene | G:A | 0.105 | A: 0.257 | A: 0.345 | |||
| rs9457840 | 5′near gene | T:C | 1.000 | C: 0.020 | C: 0.065 | |||
| rs6899549 | 5′near gene | T:C | 0.003 | C: 0.003 | C: 0.013 | |||
| rs34447885* | C14F | Exon 1 | C:T | 1.000 | 0 | T: 0.005 | Reduced MPP, increased Metformin transport | 12, 26 |
| rs12208357 | R61C | Exon 1 | C:T | 1.000 | T: 0.067 | T: 0.027 | Reduced MPP and Metformin transport | 9, 26 |
| rs45584532 | Intron | C:T | 1.000 | T: 0.177 | T: 0.101 | |||
| rs683369 | L160F | Exon 2 | C:G | 0.434 | G: 0.217 | G: 0.129 | Loss of response, imatinib failure | 14 |
| rs594709† | Intron | A:G | 0.623 | G: 0.368 | G: 0.313 | |||
| rs36103319 | G220V | Exon 3 | G:T | 1.000 | T: 0.002 | T: 0.001 | Reduced MPP transport | 9, 26 |
| rs3777392 | Intron | G:A | 1.000 | A: 0.092 | A: 0.090 | |||
| rs2282143 | P341L | Exon 6 | C:T | 1.000 | T: 0.02 | T: 0.058 | Reduced MPP and Metformin transport | 9, 26 |
| rs6937722* | Intron | G:A | 1.000 | A: 0.05 | A: 0.022 | |||
| rs35191146‡ | M420del | Exon 7 | G/GAT/- | 0.412 | (-): 0.154 | (-): 0.155 | Reduced MPP and Metformin transport | 12 |
| rs1867350 | Intron | G:A | 0.088 | A: 0.124 | A: 0.144 | |||
| rs9295122* | Intron | C:T | 0.757 | T: 0.42 | T: 0.411 | |||
| rs622342 | Intron | A:C | 0.593 | C: 0.359 | C: 0.272 | |||
| rs11753995* | Intron | G:A | 1.000 | A: 0.149 | A: 0.099 | |||
| rs34059508 | G465R | Exon 9 | G:A | 0.048 | A: 0.031 | A: 0.008 | Reduced MPP and Metformin transport | 9, 26 |
| rs35270274 | R488T | Exon 9 | G:T | 1.000 | T: 0.002 | T: 0.011 | ||
| rs622591 | Intron | G:A | 0.384 | A: 0.219 | A: 0.337 | |||
| rs9457846 | 3′UTR | G:A | 0.335 | A: 0.09 | A: 0.002 | |||
| rs651164 | 3′UTR | G:A | 0.122 | A: 0.353 | A: 0.367 | |||
| rs2083867 | 3′UTR | A:G | 0.267 | G: 0.389 | G: 0.381 | |||
| rs628031 tagged to rs594709† | M408V | Exon 7 | G:A | 0.623 | A: 0.368 | A: 0.302 |
Our observed MAF and the reported MAF from NCBI are given.
HWpval indicates the HWE values.
SNPs run in a 4-plex reaction and
SNPs run by pyrosequencing (the rest of the SNPs were run on an 18-plex reaction).
M408V was tagged to rs594709 with a D′ = 1 and r2 = 0.89.
Sequenom MassARRAY
Genotyping was performed using the Sequenom iPLEX platform and iPLEX Gold reaction according to the manufacturer's instructions. Twenty nanograms of DNA in each well of a 384-well plate were used. The first step was a multiplex PCR followed by shrimp alkaline phosphatase treatment to neutralize any unincorporated dNTPs. The iPLEX Gold reaction cocktail consisting of extension primer, enzyme, buffer, and mass-modified nucleotides was then added to the amplification product. During the reaction, allele-specific extension products of different masses were produced, depending on the analyzed sequence. Before mass spectrometry, resin (Sequenom) was added to the mixture to remove any extraneous salts that could interfere with the downstream MALDI-TOF analysis. The cleaned products were then transferred onto a SpectroCHIP by the MassARRAY nanodispenser and analyzed by the MALDI-TOF mass spectrometer. Duplicate samples and 4 negative controls (water) were included in each plate to check the quality of genotyping and genotypes for each sample were also manually checked.
PCR pyrosequencing
Genotyping for M420del was performed by PCR pyrosequencing. The primers used for the PCR reaction (SLC22A1-F, SLC22A1-R, and SLC22A1-S; Table 2) were created by Pyromark Assay Design 2.0 (QIAGEN). These generated a 92-bp product with the SNP of interest in the middle and the pyrosequencing primer just before the M420 SNP. Fifty nanograms of genomic DNA were amplified with a 10μM concentration of each primer and MasterMix (Promega) using the following conditions: 95°C for 2 minutes followed by 40 cycles of 94°C for 45 seconds, 51°C for 25 seconds, 72°C for 20 seconds, and a final step of 72°C for 10 minutes. The PCR products were then mixed initially with bead-binding buffer and then with annealing buffer, the sequencing primer, dNTPs, and the substrate/enzyme mixture. The 96-well plate was then transferred to the Pyrosequencer (QIAGEN).
List of primers (5′-3′) used
| Mutagenesis |
| M420delF: GCAGCCTGCCTCGTCATTTTTATCTCACCTGACC |
| M420delR: GGTCAGGTGAGATAAAAATGACGAGGCAGGCTGC |
| M408F: GCATCTACCCCATGGCCATGTCAAATTTGTTGGCG |
| M408R: CGCCAACAAATTTGACATGGCCATGGGGTAGATGC |
| PCR pyrosequencing |
| SLC22A1-F: GGCCATGTCAAATTTGTT |
| SLC22A1-R (biotin labelled): GTTGTCTGCTTTCATCATTTC |
| SLC22A1-S (Pyrosequencing): CGGGGGCAGCCTGCCTC |
| PCR direct sequencing |
| OCT634F(PCR & Sequencing): CATCACCATTGACCGCGTGG |
| OCT762R: CCCACACTTCGATTGCCTGG |
| mRNA quantification and sequencing |
| OCT3F: GGGCAGCCTGCCTCGTCATG |
| OCT3FN: GGGCAGCCTGCCTCGTCAT |
| OCT1503R: ACCTCCCTCAGCCTGAAGAC |
| OCT1181F (sequencing): TCATCACCATTGACCGCGTG |
| OCT1413R (sequencing): 5TCCCACCTATGTCACACAGG |
| Mutagenesis |
| M420delF: GCAGCCTGCCTCGTCATTTTTATCTCACCTGACC |
| M420delR: GGTCAGGTGAGATAAAAATGACGAGGCAGGCTGC |
| M408F: GCATCTACCCCATGGCCATGTCAAATTTGTTGGCG |
| M408R: CGCCAACAAATTTGACATGGCCATGGGGTAGATGC |
| PCR pyrosequencing |
| SLC22A1-F: GGCCATGTCAAATTTGTT |
| SLC22A1-R (biotin labelled): GTTGTCTGCTTTCATCATTTC |
| SLC22A1-S (Pyrosequencing): CGGGGGCAGCCTGCCTC |
| PCR direct sequencing |
| OCT634F(PCR & Sequencing): CATCACCATTGACCGCGTGG |
| OCT762R: CCCACACTTCGATTGCCTGG |
| mRNA quantification and sequencing |
| OCT3F: GGGCAGCCTGCCTCGTCATG |
| OCT3FN: GGGCAGCCTGCCTCGTCAT |
| OCT1503R: ACCTCCCTCAGCCTGAAGAC |
| OCT1181F (sequencing): TCATCACCATTGACCGCGTG |
| OCT1413R (sequencing): 5TCCCACCTATGTCACACAGG |
F indicates forward primers; and R, reverse primers. Sequencing primers are also indicated.
Results for the M420del status were obtained from 177 samples with adequate clinical information; 18 samples failed to amplify because of poor quality or lack of DNA. Results from 90 samples were also confirmed by direct sequencing using the ABI 3130 genetic analyzer (Applied Biosystems by Life Technologies). The PCR-pyrosequencing product could not be used for the direct sequencing reaction because the primers were located very close to the SNP of interest. Another set of primers was designed (Table 2) and used with 50 ng of DNA at 95°C for 2 minutes, followed by 18 cycles of 95°C for 20 seconds, 60°C for 10 seconds, 68°C for 3 minutes and 37 seconds, and a final step of 68°C for 5 minutes. The sequencing mixture consisted of 2 μL of PCR product, 3.2μM forward primer, the BigDye Terminator Version 1.1 Cycle Sequencing kit (Applied Biosystems), and 5× sequencing buffer. The reaction was cycled for 30 cycles at 96°C for 15 seconds, 55°C for 15 seconds, and 60°C for 4 minutes. The sequencing products were cleaned and concentrated using the DNAclean & Concentrate-25 Kit (Zymo Research, Cambridge Bio Science).
Cell lines and site-directed mutagenesis
Our previously established KCL22-hOCT1 cell line3,19 carries the M420 and V408 (valine, V, at position 408 instead of methionine, M) alleles, verified by direct sequencing (Lark Technologies). The M420del and M408 (methionine, M, at position 408 instead of valine, V) alleles were introduced to the pcDNA-hOCT1 plasmid by site-directed mutagenesis using the Stratagene QuikChange Lightning kit (Agilent Technologies) following the manufacturer's instructions. The primers bearing the specific SNPs were created by the QuikChange Primer Design Program (Agilent Technologies) and synthesized by Invitrogen (Life Technologies). Their sequences are illustrated in Table 2. The new plasmids were transfected by AMAXA nucleofection (Lonza) into KCL22 cells grown in RPMI 1640 medium (Biosera) medium supplemented with 1% l-glutamine, penicillin/streptomycin, and 10% FCS (Biosera). Stable cell lines were generated that expressed similar levels of hOCT1 mRNA by real-time PCR, as described previously, whereas a stable cell line carrying the empty vector pcDNA3.1 (mock-transfected cells) was used as a control for the uptake experiments.3,19
Studies of radiolabeled imatinib uptake
14C-imatinib (kind gift of Novartis, Basel, Switzerland) was used in all experiments at 1 μg/mL, and nonradiolabeled imatinib was added to a final concentration of 5μM, similar to the expected peak plasma levels achievable in CML patients. Briefly, 1 × 106 cells were added to the 14C-imatinib containing transport medium with or without the hOCT1 inhibitor amantadine (500μM) and incubated for 30 minutes.3,19 Radioactivity was then measured both in the cell pellets and supernatant using a scintillation counter as described previously.1,3,19 Experiments were performed on 6 different occasions. hOCT1 activity was determined by subtracting the uptake in the presence of amantadine from that in unmanipulated cells in each cell line, as described previously.2,20 To investigate whether the hOCT1 variants were functionally active, the hOCT1 substrates 14C-TEA+ (Moravek Biochemicals) at 5μM and ASP+ (Sigma-Aldrich) at 500μM were used as described previously.9,10,21
hOCT1 transcript levels
For hOCT1 mRNA level analysis, 13 “test” and 53 “validation” samples were taken from newly diagnosed, treatment-naive chronic-phase patients who subsequently received imatinib. hOCT1 transcript levels were quantified by real-time quantitative RT-PCR on a LightCycler (Roche Diagnostics). In our original primer set, OCT3F/OCT1503R,3 the forward primer OCT3F covers the M420del SNP site. The new primer set, OCT3FN/OCT1503R, was designed with similar PCR efficiency, but shifted away from M420del SNP by 1 base. Furthermore, the primer set OCT1181F/OCT1413R was designed for confirmation sequencing. All primer sequences are given in Table 2. The expression of hOCT1 was measured using LightCycler FastStart DNA MasterPlus SYBR Green I (Roche Diagnostics) as described previously.1 PCR products were purified by QIAquick PCR purification kit (QIAGEN) and sequenced directly using PCR primers (Beckman Coulter Genomics). The hOCT1 mRNA level is presented as a ratio of hOCT1 to GAPDH transcripts and normalized to a standard VBL100 hOCT1 mRNA level as described previously, which was considered as an arbitrary unit of 1.0.3
hOCT1 protein determination
hOCT1 protein levels were analyzed by flow cytometry on intact fixed cells and by Western blotting. A total of 1 × 106 cells was resuspended in 500 μL of 2% paraformaldehyde (VWR) and fixed for 10 minutes at 37°C, followed by 90% methanol (Fisher Scientific) on ice for 30 minutes. Cells were then washed with PBS containing 0.5% BSA and incubated with hOCT1 2C5 Ab (1:200) at room temperature for 1 hour. After washing twice, cells were resuspended in 100 μL of PBS-BSA buffer with the fluorescein-labeled anti–mouse secondary Ab Alexa Fluor 488 (Invitrogen) for 30 minutes in the dark. Cells were then washed and analyzed using flow cytometry (FACSCalibur; BD Biosciences) and CellQuest Pro 3.3 software for data analysis.
Western blotting of biotinylated cell surface proteins was carried out as described previously.11,22 Briefly, proteins were isolated with the Pierce Cell Surface Protein Isolation kit (Thermo Scientific) following the manufacturer's protocol. Primary Abs were hOCT1 mouse mAb (2C5) at 1:1000 dilution and α-1 sodium/potassium ATPase (464.6) at 1:2500 (both from Abcam).
Statistical analysis
Association between hOCT1 SNPs and clinical response
Before the analyses of association, deviation from Hardy-Weinberg equilibrium (HWE) was tested for each SNP using the χ2 test, with P < .001 indicating deviation from HWE. To test for the association between each SNP in turn and the outcomes of PFS and EFS, a log-rank test was undertaken using the “survdiff” function in the R Survival statistical package.23 This tests for difference in survival between the 3 genotype groups wild-type homozygotes, heterozygotes, and mutant homozygotes. Where few (< 10) mutant homozygotes were present, these were analyzed with the heterozygotes as a single group. In testing for association between each SNP in turn and TTF, because patients may fail on imatinib for either unsatisfactory response or intolerance to imatinib, the Gray class of k-sample tests were used for comparing the cumulative incidence of the competing event cause between the 2 or 3 genotype groups24 using the “cuminc” function of the R Cmprsk 2.2-2 statistical package.25 Due to the high number of statistical tests undertaken, the false discovery rate (FDR) was calculated for each test. FDR < 0.05 was taken to indicate statistical significance.
For analyzing EFS and PFS, patients who stopped imatinib or switched treatment during follow-up were censored at the time of stopping or switching. However, these patients could be informative, as their reason for stopping/switching is related to the events investigated. Therefore, 2 sensitivity analyses were undertaken to determine whether censoring was informative. The first assumed that these patients were at high risk of an event and that all censored observations were therefore EFS/PFS events occurring immediately after censoring. The second assumed that these patients were at low risk of an event and assumed that all EFS/PFS events happened after the latest follow-up; their censoring time was changed to the time of last follow-up.
For the TTF outcome, some patients experienced an event before coming off of imatinib and were censored at this time point. A sensitivity analysis was undertaken assuming that the patient had come off of imatinib at the event time point. Unless stated otherwise in the Results section, the main analyses of association were robust to all sensitivity analyses undertaken.
Functional analysis of hOCT1 M420del and M408V SNPs
The Student t test (2-tail distribution) was used for the KCL22 cell line model studying the functional effect of SNPs on imatinib uptake.
Results
Association between hOCT1 SNPs and clinical response
Table 3 presents the prevalence of the 23 analyzed SNPs together with that expected within a normal population (www.ncbi.nlm.nih.gov).9,26 Three SNPs (S14F, G220V, and A488M/T) were monomorphic (ie, had only 1 allele) and could not therefore be analyzed for clinical association. The observed MAFs were in HWE and were comparable to those observed previously.
Genotyping data for hOCT1 SNPs: prevalence of hOCT1 SNPs
| SNP . | AA translation . | n . | WT Homo . | Het . | MT Homo . |
|---|---|---|---|---|---|
| rs594709 tagged to rs628031 | Intronic tagged to M408V | 292 | AA:114 | AG:141 | GG:37 |
| rs622342 | 291 | AA:122 | AC:129 | CC:40 | |
| rs622591 | 295 | AA:11 | AG:107 | GG:177 | |
| rs651164 | 293 | GG:129 | AG:121 | AA:4327 | |
| rs683369 | L160F | 296 | CC:178 | CG:106 | GG:12 |
| rs1867350 | 290 | GG:219 | AG:70 | AA:1 | |
| rs2083867 | 292 | AA:104 | AG:149 | GG:39 | |
| rs2282143 | P341L | 293 | CC:281 | CT:12 | TT:0 |
| rs3777392 | 292 | GG:240 | AG:50 | AA:2 | |
| rs35270274* | R488T | 290 | GG:289 | GT:1 | TT:0 |
| rs36103319* | G220V | 294 | GG:293 | GT:1 | TT:0 |
| rs6899549 | 296 | CC:294 | CT:0 | TT:2 | |
| rs6935207 | 292 | GG:167 | AG:100 | AA:25 | |
| rs9457840 | 295 | TT:283 | TC:12 | CC:0 | |
| rs9457846 | 205 | GG:168 | AG:37 | AA:0 | |
| rs12208357 | R61C | 293 | CC:255 | CT:37 | TT:1 |
| rs34059508 | G465R | 294 | GG:278 | AG:14 | AA:2 |
| rs45584532 | 294 | CC:199 | CT:86 | TT:9 | |
| rs6937722 | 309 | GG:279 | AG:29 | AA:1 | |
| rs9295122 | 307 | CC:103 | CT:150 | TT:54 | |
| rs34447885* | C14F | 308 | CC:308 | CT:0 | TT:0 |
| rs11753995 | 309 | GG:220 | AG:83 | AA:6 | |
| rs35191146† | M420del | 313 | N:224 | N/−:79 | −/−:10 |
| SNP . | AA translation . | n . | WT Homo . | Het . | MT Homo . |
|---|---|---|---|---|---|
| rs594709 tagged to rs628031 | Intronic tagged to M408V | 292 | AA:114 | AG:141 | GG:37 |
| rs622342 | 291 | AA:122 | AC:129 | CC:40 | |
| rs622591 | 295 | AA:11 | AG:107 | GG:177 | |
| rs651164 | 293 | GG:129 | AG:121 | AA:4327 | |
| rs683369 | L160F | 296 | CC:178 | CG:106 | GG:12 |
| rs1867350 | 290 | GG:219 | AG:70 | AA:1 | |
| rs2083867 | 292 | AA:104 | AG:149 | GG:39 | |
| rs2282143 | P341L | 293 | CC:281 | CT:12 | TT:0 |
| rs3777392 | 292 | GG:240 | AG:50 | AA:2 | |
| rs35270274* | R488T | 290 | GG:289 | GT:1 | TT:0 |
| rs36103319* | G220V | 294 | GG:293 | GT:1 | TT:0 |
| rs6899549 | 296 | CC:294 | CT:0 | TT:2 | |
| rs6935207 | 292 | GG:167 | AG:100 | AA:25 | |
| rs9457840 | 295 | TT:283 | TC:12 | CC:0 | |
| rs9457846 | 205 | GG:168 | AG:37 | AA:0 | |
| rs12208357 | R61C | 293 | CC:255 | CT:37 | TT:1 |
| rs34059508 | G465R | 294 | GG:278 | AG:14 | AA:2 |
| rs45584532 | 294 | CC:199 | CT:86 | TT:9 | |
| rs6937722 | 309 | GG:279 | AG:29 | AA:1 | |
| rs9295122 | 307 | CC:103 | CT:150 | TT:54 | |
| rs34447885* | C14F | 308 | CC:308 | CT:0 | TT:0 |
| rs11753995 | 309 | GG:220 | AG:83 | AA:6 | |
| rs35191146† | M420del | 313 | N:224 | N/−:79 | −/−:10 |
A total of 336 samples were analyzed for each SNP; the number of successful results is shown in column “n.”
N indicates M420 undeleted homozygotes; N/−, M420del heterozygotes; and −/−, M420del homozygotes.
Monomorphic SNPs that were excluded from the association analysis.
M420del SNP.
The overall median duration of follow-up was 61 months. During follow-up, 17 patients progressed to the advanced phase or died. Three additional patients lost a CCR, yielding 20 events for the EFS analysis. A further 30 patients stopped imatinib because of intolerance or rising disease burden without evidence of loss of CCR, thus constituting the events for TTF analysis. In the remaining 2 patients, the timing of their stopping the drug was unknown and these patients were therefore excluded from the analyses of association.
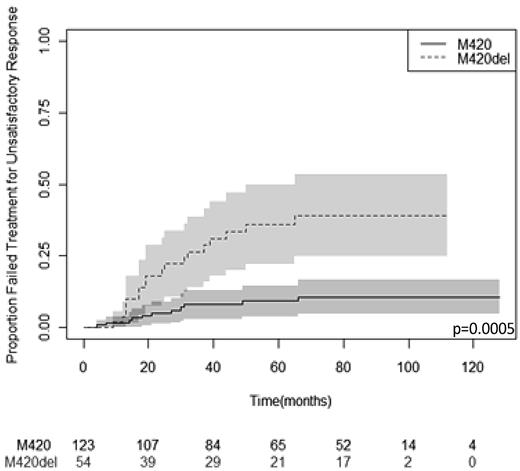
Using the Haploview 4.2 pharmacogenomic analysis software package, no nonspurious association was found between any SNP allele and the achievement of CCR or MMR at either 12 or 24 months. Results for the analysis of association between each SNP and PFS, EFS, and TTF are given in Table 4. Pyrosequencing data for M420del were obtained from 177 samples because 18 samples failed to amplify. The only statistically significant association was between M420del and TTF, in which M420del was associated with a higher risk of treatment failure (hazard ratio, 4.30; 95% confidence interval, 2.04-9.05); FDR = 0.004). This result was robust to all sensitivity analyses. Figure 1 illustrates this effect in a Kaplan-Meier plot.
Genotyping data for hOCT1 SNPs: correlation between hOCT1 SNPs and PFS, EFS, and TTF
| SNP . | AA translation . | n . | WT Homo . | Het . | MT Homo . | P . | |||
|---|---|---|---|---|---|---|---|---|---|
| PFS . | EFS . | TTF due to unsatisfactory response . | TTF due to intolerance . | ||||||
| rs594709 tagged to rs628031 | Intronic, tagged to M408V | 167 | 69 | 76 | 22 | .938 | 0.855 | .257 | .277 |
| rs622342 | 167 | 66 | 77 | 24 | .199 | 0.446 | .642 | .161 | |
| rs622591 | 169 | 100 | 65 | 4 | .464 | 0.777 | .707 | .399 | |
| rs651164 | 167 | 74 | 66 | 27 | .429 | 0.584 | .856 | .265 | |
| rs683369 | L160F | 170 | 99 | 62 | 9 | .649 | 0.942 | .502 | .873 |
| rs1867350 | 166 | 127 | 39 | 0 | .824 | 0.850 | .374 | .410 | |
| rs2083867 | 167 | 63 | 82 | 22 | .127 | 0.517 | .991 | .099 | |
| rs2282143 | P341L | 167 | 161 | 6 | 0 | .439 | 0.657 | .945 | .637 |
| rs3777392 | 167 | 141 | 25 | 1 | .869 | 0.673 | .736 | .816 | |
| rs6899549* | 170 | 168 | 2 | 0 | .012 | 0.031 | .119 | .832 | |
| rs6935207 | 168 | 91 | 60 | 17 | .548 | 0.746 | .132 | .098 | |
| rs9457840 | 169 | 163 | 6 | 0 | .426 | 0.697 | .927 | .639 | |
| rs9457846 | 112 | 91 | 21 | 0 | .074 | 0.122 | .212 | .118 | |
| rs12208357 | R61C | 168 | 145 | 23 | 0 | .722 | 0.540 | .486 | .328 |
| rs34059508 | G465R | 169 | 158 | 10 | 1 | .273 | 0.412 | .794 | .555 |
| rs45584532 | 168 | 109 | 51 | 8 | .784 | 0.848 | .929 | .089 | |
| rs6937722 | 181 | 158 | 22 | 0 | .537 | 0.755 | .783 | .391 | |
| rs9295122 | 182 | 64 | 84 | 33 | .115 | 0.119 | .584 | .406 | |
| rs11753995* | 182 | 127 | 51 | 3 | .488 | 0.478 | .045 | .128 | |
| rs35191146† | M420del | 177 | 123 | 48 | 6 | .856 | 0.699 | .00005 | .639 |
| SNP . | AA translation . | n . | WT Homo . | Het . | MT Homo . | P . | |||
|---|---|---|---|---|---|---|---|---|---|
| PFS . | EFS . | TTF due to unsatisfactory response . | TTF due to intolerance . | ||||||
| rs594709 tagged to rs628031 | Intronic, tagged to M408V | 167 | 69 | 76 | 22 | .938 | 0.855 | .257 | .277 |
| rs622342 | 167 | 66 | 77 | 24 | .199 | 0.446 | .642 | .161 | |
| rs622591 | 169 | 100 | 65 | 4 | .464 | 0.777 | .707 | .399 | |
| rs651164 | 167 | 74 | 66 | 27 | .429 | 0.584 | .856 | .265 | |
| rs683369 | L160F | 170 | 99 | 62 | 9 | .649 | 0.942 | .502 | .873 |
| rs1867350 | 166 | 127 | 39 | 0 | .824 | 0.850 | .374 | .410 | |
| rs2083867 | 167 | 63 | 82 | 22 | .127 | 0.517 | .991 | .099 | |
| rs2282143 | P341L | 167 | 161 | 6 | 0 | .439 | 0.657 | .945 | .637 |
| rs3777392 | 167 | 141 | 25 | 1 | .869 | 0.673 | .736 | .816 | |
| rs6899549* | 170 | 168 | 2 | 0 | .012 | 0.031 | .119 | .832 | |
| rs6935207 | 168 | 91 | 60 | 17 | .548 | 0.746 | .132 | .098 | |
| rs9457840 | 169 | 163 | 6 | 0 | .426 | 0.697 | .927 | .639 | |
| rs9457846 | 112 | 91 | 21 | 0 | .074 | 0.122 | .212 | .118 | |
| rs12208357 | R61C | 168 | 145 | 23 | 0 | .722 | 0.540 | .486 | .328 |
| rs34059508 | G465R | 169 | 158 | 10 | 1 | .273 | 0.412 | .794 | .555 |
| rs45584532 | 168 | 109 | 51 | 8 | .784 | 0.848 | .929 | .089 | |
| rs6937722 | 181 | 158 | 22 | 0 | .537 | 0.755 | .783 | .391 | |
| rs9295122 | 182 | 64 | 84 | 33 | .115 | 0.119 | .584 | .406 | |
| rs11753995* | 182 | 127 | 51 | 3 | .488 | 0.478 | .045 | .128 | |
| rs35191146† | M420del | 177 | 123 | 48 | 6 | .856 | 0.699 | .00005 | .639 |
A total of 195 samples had adequate clinical information. Distribution according to genotype is given (WT indicates wild-type; and MT, mutant). This is followed by the P value for the association analyses as explained in the statistical analysis section.
For the rs6899549 and rs11753995 SNPs, the apparently significant (P < .05) association with PFS and EFS and TTF, respectively, may be unreliable because of the small number of heterozygotes (2) and mutant homozygotes (3); the significance was lost after FDR analysis.
M420del SNP (rs35191146), which is the only other SNP with a significant association (P = .0005; FDR = 0.004) with TTF. This result is robust to the sensitivity analyses.
Kaplan-Meier plot of TTF stratified by M420 deletion status. The median follow-up was 61 months. Patients carrying M420del had a significantly greater risk of treatment failure (P = .0005; FDR = 0.004). The 95% confidence limits are depicted by the shaded areas. Numbers of patients at risk of an event at successive 20-month time points are given below the x-axis.
Kaplan-Meier plot of TTF stratified by M420 deletion status. The median follow-up was 61 months. Patients carrying M420del had a significantly greater risk of treatment failure (P = .0005; FDR = 0.004). The 95% confidence limits are depicted by the shaded areas. Numbers of patients at risk of an event at successive 20-month time points are given below the x-axis.
Functional analysis of M420del and M408V
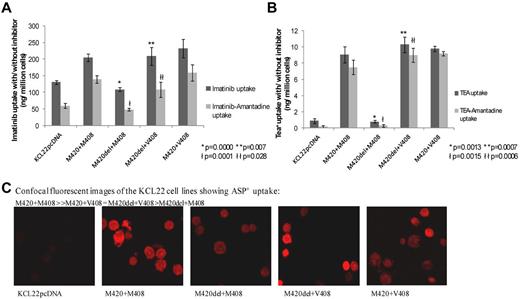
Figure 2 presents functional data on the uptake of imatinib (Figure 2A) and the hOCT1 substrates TEA+ (Figure 2B) and ASP+ (Figure 2C) in KCL22 cells transfected with various hOCT1 alleles of M420 and M408. In Figure 2A and B, the hOCT1 activity is shown in the light gray bars. Compared with the cell line with undeleted M420+M408, the line carrying the M420del+M408 alleles showed a significant decrease in imatinib uptake and hOCT1 activity (P = .0000 and .0001, respectively). In the line with both M420del+V408 alleles, uptake did not differ from the undeleted M420+M408 line, but was superior to that in cells carrying the M420del+M408 alleles (P = .007 for unmanipulated uptake and 0.028 for hOCT1 activity). TEA+ uptake showed a similar pattern to that of imatinib, with analogous P values (given in (Figure 2B). These data suggest that M408V can counteract the adverse effect of M420del on uptake and hOCT1 activity.
Functional assays of uptake into the KCL22 CML cell line transfected with the hOCT1 SNPs M420del and/or M408V. (A-B) Uptake of 14C imatinib (A; n = 6) and 14C TEA+ (B; n = 3). Each histogram bar represent the means ± SEM. In both panels, the dark gray bars indicate unmanipulated uptake only and the light gray bars show the uptake attributable to hOCT1 activity (obtained by subtracting the uptake with the hOCT1 amantadine from the unmanipulated uptake). In both panels, * and ł denote significantly decreased uptake and hOCT1 activity, respectively, in M420del+M408 cells compared with undeleted M420+M408 cells; ** and łł denote significantly increased uptake and hOCT1 activity, respectively, in M420del+V408 cells compared with M420del+M408 cells. Individual P values are given in the panels. (C) Uptake of the fluorescent hOCT1 substrate ASP+ by confocal microscopy. Uptake is still apparent in the M420del+M408 line compared with mock transfected control (KCL22pcDNA), confirming the presence and activity of functional hOCT1 in this line.
Functional assays of uptake into the KCL22 CML cell line transfected with the hOCT1 SNPs M420del and/or M408V. (A-B) Uptake of 14C imatinib (A; n = 6) and 14C TEA+ (B; n = 3). Each histogram bar represent the means ± SEM. In both panels, the dark gray bars indicate unmanipulated uptake only and the light gray bars show the uptake attributable to hOCT1 activity (obtained by subtracting the uptake with the hOCT1 amantadine from the unmanipulated uptake). In both panels, * and ł denote significantly decreased uptake and hOCT1 activity, respectively, in M420del+M408 cells compared with undeleted M420+M408 cells; ** and łł denote significantly increased uptake and hOCT1 activity, respectively, in M420del+V408 cells compared with M420del+M408 cells. Individual P values are given in the panels. (C) Uptake of the fluorescent hOCT1 substrate ASP+ by confocal microscopy. Uptake is still apparent in the M420del+M408 line compared with mock transfected control (KCL22pcDNA), confirming the presence and activity of functional hOCT1 in this line.
To exclude the possibility that hOCT1 was not expressed in the M420del+M408 line, uptake of the alternative hOCT1 substrate ASP+ was investigated by confocal microscopy (Figure 2C). Similar ASP+ uptake was seen in all 3 lines studied, including M420del+M408, confirming that hOCT1 is active in the M420del+M408 line and suggesting that SNPs may not alter the affinity of different hOCT1 substrates equally.
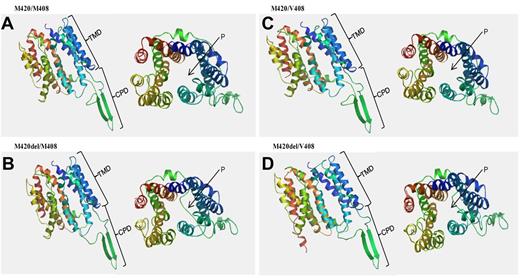
Conformational analysis using the Swiss Model software (http://swissmodel.expasy.org/) was used to produce 3-dimensional structures of hOCT1 (NM 003057) with various SNP alleles (Figure 3). If present, M420del confers altered protein folding in the cytoplasmic domain and a clear blockage of the transport pore. If V408 is present as well as M420del, this also alters the transmembrane domain, although pore blockage is less evident. These structural predictions within the cytoplasmic region and the pore are compatible with the findings shown in Figure 2 on the effect of M420del with or without V408 on hOCT1 substrate uptake and activity.
Conformational changes of the hOCT1 protein induced by the M420del and/or M408V SNPs. Model was created using the Swiss model software (http://swissmodel.expasy.org/). (A) Undeleted M420 with M408. (B) M420del with M408. (C) undeleted M420 with V408. (D) M420del with V408. In each panel, the left side depicts the lateral view of the folded protein and the right shows the dorsal view of the pore (P). Differences in protein folding are observed in the cytoplasmic domain (CPD) if M420del is present; the M420del+V408 combination also alters the transmembrane domain (TMD). A clear blockage of the pore (P) is also observed in the presence of M420del, although this is less marked when V408 is present.
Conformational changes of the hOCT1 protein induced by the M420del and/or M408V SNPs. Model was created using the Swiss model software (http://swissmodel.expasy.org/). (A) Undeleted M420 with M408. (B) M420del with M408. (C) undeleted M420 with V408. (D) M420del with V408. In each panel, the left side depicts the lateral view of the folded protein and the right shows the dorsal view of the pore (P). Differences in protein folding are observed in the cytoplasmic domain (CPD) if M420del is present; the M420del+V408 combination also alters the transmembrane domain (TMD). A clear blockage of the pore (P) is also observed in the presence of M420del, although this is less marked when V408 is present.
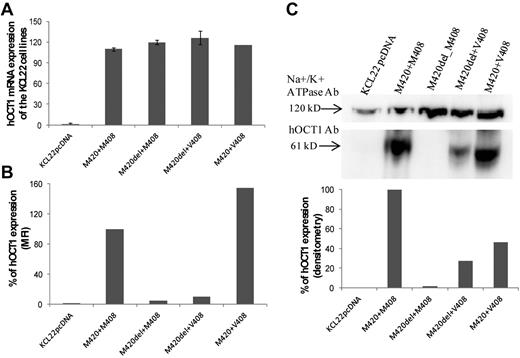
hOCT1 protein expression
The various hOCT1-transfected cell lines tested in the functional assays had similar levels of hOCT1 mRNA expression on real-time PCR (Figure 4A), suggesting that posttranscriptional changes account for the observed differences in imatinib uptake. hOCT1 protein expression for each line is shown in Figure 4B. The undeleted M420+M408 line was set as an arbitrary reference of 100% and the other transfected lines were analyzed relative to it. Low hOCT1 expression was seen in the 2 lines with M420del, whereas the cell line carrying undeleted M420+V408 had a higher apparent expression than the reference undeleted M420+M408. The 2C5 Ab bound to an epitope at 284-347 aa in the cytoplasmic region of hOCT1, which, as shown in Figure 3 may change conformation in the presence of M420del. Protein expression was therefore also studied by Western blot (Figure 4C), again using the undeleted M420+M408 line as a reference of 100%. The density of each band was normalized to the Na+/K+ ATPase-loading control. Similar to the flow cytometry data, analysis of cell-surface biotinylated hOCT1 revealed a marked reduction of hOCT1 in lines containing M420del. Interestingly, the line with undeleted M420+V408 also had apparent low hOCT1 expression, in contrast to the finding by flow cytometry. The findings shown in Figures 2, 3, and 4B and C are consistent with the view that M420 and M408 SNP alter hOCT1 protein folding and function.
hOCT1 mRNA and protein expression. (A) hOCT1 expression for the hOCT1 variants in relation to the mock-transfected cells (KCLpcDNA). (B-C) Protein expression carried out by flow cytometry (B) and Western blotting with densitometry (C, which also shows blots with their loading control, Na+/K+ ATPase). In both panels B and C, data are expressed relative to the undeleted M420+M408 line, which was arbitrarily defined as 100%.
hOCT1 mRNA and protein expression. (A) hOCT1 expression for the hOCT1 variants in relation to the mock-transfected cells (KCLpcDNA). (B-C) Protein expression carried out by flow cytometry (B) and Western blotting with densitometry (C, which also shows blots with their loading control, Na+/K+ ATPase). In both panels B and C, data are expressed relative to the undeleted M420+M408 line, which was arbitrarily defined as 100%.
Correlation of hOCT1 transcript levels with clinical response to imatinib
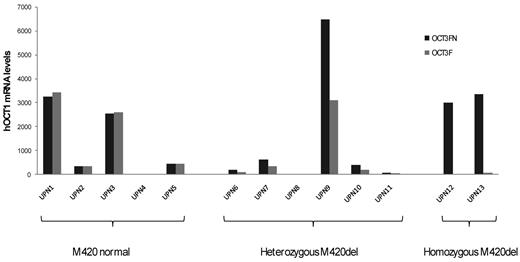
cDNA samples from 13 CML patients were tested with both the original (OCT3F/OCT1503R) and the new (OCT3FN/OCT1503R) primer sets. Because there is only 1 base different at the 3′ end of the forward primer, the PCR efficiency for these 2 pairs of primers was similar. In 5 cases without M420del, equal amounts of hOCT1 transcripts were detected by the 2 sets of primers. In 6 cases heterozygous for M420del, the apparent hOCT1 transcript level measured by the original primers was 50% lower than with the new primers. In 2 cases homozygous for M420del, hOCT1 transcripts were undetectable by the original primers, although these were detectable at low levels by the new primers (Figure 5). The original primers therefore detect only undeleted M420 transcripts, whereas the new primers detect both M420 and M420del transcripts.
Comparison of hOCT1 mRNA levels using original (OCT3F) and new (OCT3FN) primers. CML patients with undeleted M420 showed no differences in the mRNA expression using either primer, whereas the mRNA levels of patients heterozygous for the M420del almost doubled using the new primer. In M420del homozygotes, hOCT1 was detectable with the new primers but not with the original set. UPN indicates unique patient number.
Comparison of hOCT1 mRNA levels using original (OCT3F) and new (OCT3FN) primers. CML patients with undeleted M420 showed no differences in the mRNA expression using either primer, whereas the mRNA levels of patients heterozygous for the M420del almost doubled using the new primer. In M420del homozygotes, hOCT1 was detectable with the new primers but not with the original set. UPN indicates unique patient number.
To further assess the relationship between hOCT1 mRNA level and clinical outcome in the context of M420del, hOCT1 transcript levels were measured using both sets of primers in 53 “validation” newly diagnosed chronic-phase patients who then received first-line imatinib. All 53 samples were collected before imatinib treatment commenced. In cases heterozygous for M420del, transcript levels were significantly lower with the original than with the new primer sets (P = .03), whereas little difference was seen in cases without M420del. Using the original primers, at 120 months, patients with high hOCT1 mRNA expression had higher EFS than low expressors (81% vs 52%; P = .03) and also superior PFS (92% vs 60%; P = .01). However, using the new primers, these significant differences were lost, although a trend remained in favor of better outcome in high expressors (80 vs 60% for EFS, P = .16, and 92% vs 64% for PFS, P = .06). These data illustrate that the mRNA levels of M420 transcript (as measured by our original primers) were correlated with the outcome of treatment, whereas the actual hOCT1 transcript levels (as assessed here by our new primers) were not. This indicates that M420 function is more important than gene expression and highlights the significance of SNPs in the design of primers.
Exploratory analysis of the combined effect of M420del and M408V
Given our findings in the functional studies, exploratory analyses of the combined effect of genotype at M420del and M408V on the TTF were undertaken to determine whether the effect of the M420del SNP was altered by the genotype at the M408V SNP. First, a competing risks regression model was fitted including covariates to represent both SNPs (the baseline model).27 An interaction term between both SNPs was then added (the interaction model) and the 2 models compared using the likelihood ratio test. The P value obtained was nonsignificant (P = .20); therefore, there was no evidence in our cohort that the M408V SNP alters the effect of M420del on TTF. Next, patients were grouped according to genotype at both SNPs as follows: no M420 deletion and none or 1 copy of M408V (M420/M408); M420 deletion and none or 1 copy of M408V (M420del/M408); no M420 deletion and 2 copies of M408V (M420/M408V); or M420 deletion and 2 copies of M408V (M420del/M408V). The Gray class of k-sample tests were used for comparing the cumulative incidence of the competing event cause between the 4 genotype groups. The P value obtained for treatment failure because of unsatisfactory response was .0004. In Figure 6, the cumulative incident plot for the M420del/M408 group appears significantly different from that for the M420/M408 and M420/M408V groups, which are similar. This suggests that the significant association observed with combined genotype is driven purely by the association with the M420del. However, there are only 2 cases in the M420del/M408V group, so we cannot deduce whether they have similar outcome to the M420del/M408 group.
Interaction of M420del and M408V SNPs and their correlation with treatment failure. The median follow-up was 57 months. Patients were grouped according to their genotype at both SNPs as: no M420 deletion and none or 1 copy of M408V (M420/M408); M420 deletion and none or 1 copy of M408V (M420del/M408); no M420 deletion and 2 copies of M408V (M420/M408V); or M420 deletion and 2 copies of M408V (M420del/M408V). The number of patients at different time points is included under each 20-month follow-up point. There was insufficient information (n = 2) to prepare a cumulative incidence plot for the M420del/M408V genotype group. The P value is from global tests comparing survival across all genotype groups.
Interaction of M420del and M408V SNPs and their correlation with treatment failure. The median follow-up was 57 months. Patients were grouped according to their genotype at both SNPs as: no M420 deletion and none or 1 copy of M408V (M420/M408); M420 deletion and none or 1 copy of M408V (M420del/M408); no M420 deletion and 2 copies of M408V (M420/M408V); or M420 deletion and 2 copies of M408V (M420del/M408V). The number of patients at different time points is included under each 20-month follow-up point. There was insufficient information (n = 2) to prepare a cumulative incidence plot for the M420del/M408V genotype group. The P value is from global tests comparing survival across all genotype groups.
Discussion
Several nonsynonymous SNPs in hOCT1 have been found to affect the transport of known hOCT1 substrates such as MPP+, TEA+, and ASP+. In addition, studies using Xenopus oocyte cell lines expressing hOCT1 variants, OCT1-deficient mice and mouse hepatocytes, and clinical samples have shown that hOCT1 SNPs affect the pharmacokinetics and therapeutic action of the antidiabetic drug metformin and may contribute to the observed variation in metformin response.12-14,28,29
Recently, the association between hOCT1 SNPs and clinical response to imatinib has been investigated. Kim et al examined 5 SNPs, 1 of which was synonymous, in 229 patients and found an association between the GG genotype of L160F (rs683369) and loss of response and imatinib failure.14 Bazeos et al showed in 132 CML patients that the infrequent (MAF = 0.7%) G401S SNP was correlated with a molecular response to imatinib. They also examined the SNPs R61C, P341L, P283L, R287G, C88R, and rs622342 (intronic SNP), but did not observe any clinical correlation.15 Zach et al analyzed 35 CML patients for the R61C SNPs and found no differences in response in CML patients heterozygous for this polymorphism.30 In 65 patients, Maffioli et al showed no correlation between R61C, M408V, and M420del and response to imatinib treatment, but the AA genotype of rs6935297, an intronic SNP that lies 5′ to the coding sequence, was not found in patients with inadequate imatinib response.17 White et al analyzed 136 newly diagnosed CML patients, but failed to show any correlation between hOCT1 SNPs (R61C, G401S, C88R, P341L, G220V, G465R, M408V, and M420del) and achievement of major molecular response, progression to advanced phase, or hOCT1 activity.16 However, all of these studies focused only on a small (1-8) and differing number of SNPs. The difference between our data and those of other investigators is likely because of the small number of cases with individual SNP alleles in all of the cohorts. Our data also differ from some in that all of our patients were in the chronic phase at initial presentation. A further factor may be the differing methodologies of SNP detection.
In the present study, we demonstrate 3 related findings. First, all 23 SNPs throughout the hOCT1 gene that either had an allele frequency of ≥ 0.05 or were previously studied were investigated in relation to treatment outcome in a large number of CML patients treated from diagnosis with imatinib. Patients carrying the M420del allele had a greater probability of imatinib failure because of unsatisfactory response than patients with undeleted M420. However, no SNP showed association with PFS or EFS, which is in agreement with previous studies.16,17 Therefore, our data endorse this emerging theme of correlation between SNPs and treatment failure, but not with disease outcome. This is consistent with the view that the M420del SNP may alter the pharmacodynamics without changing the systemic pharmacokinetic action of imatinib, reducing the efficacy of imatinib in the specific target leukemic cells. However, this may not result in altered survival because patients failing imatinib are typically switched to an alternative TKI that is handled independently of hOCT1 and its SNPs.19,31
Second, we investigated the function of the M420del and M408V SNP in relation to the uptake of imatinib in a transfected CML cell line model. Using a previously established functional assay, KCL22 cells stably expressing M420del/M408 alleles showed a significant decrease in imatinib uptake compared with cells with undeleted M420/M408, but in cells with both M420del/V408 alleles, uptake did not differ from the reference cells, suggesting that the V408 SNP can counteract the effect of the M420del. Similar data were seen for TEA+ and ASP+ uptake.
Differences in hOCT1 protein (but not mRNA) expression between the transfected lines were also seen. Interestingly, structural modeling predicts that the M420del and M408V SNPs alter the hOCT1 pore structure in a manner compatible with our functional data. Mutations in the intracellular domains of hOCT1 differentially influence the binding of various substrates, which is compatible with a binding pocket with overlapping interaction domains for different substrates and inhibitors.32 Similarly, the difference in apparent M420+V408 hOCT1 levels between Figure 4B and C highlight differential Ab binding in differing physicochemical conditions. These data highlight the importance of SNPs on protein folding and may explain the functional effects of both M420del and M408V in imatinib uptake; they also fit with functional studies on metformin uptake.12,13
To determine the clinical significance of the functional assays, we performed a post hoc analysis of the combined effect of M420del/M408V genotypes on treatment outcome. Our data reaffirmed the importance of M420del, but could not confirm whether M408V confers a protective effect when combined with the M420 deletion, partly because there were only 2 cases with the M420del/V408 alleles.
Finally, we demonstrate that the M420del SNP may be relevant to existing data on the value of hOCT1 mRNA assessment in predicting outcome of imatinib treatment. We have reported previously that patients with low apparent hOCT1 expression have inferior outcome on imatinib compared with those with higher levels.3 However, the terminal base of the forward PCR primer used in that work hybridized with codon 420; if present, M420del could therefore affect the apparent mRNA expression. When a forward primer is used with the same sequence but omitting this terminal base, the correlation between apparent expression and imatinib outcome is lost. This underlines our direct results demonstrating that M420del may affect both imatinib treatment outcome and imatinib uptake. Interestingly, White et al did not find that hOCT1 mRNA expression is correlated with outcome; their primers are located in exon 9 of hOCT1 (D.L. White, personal communication), whereas ours are in exon 7.
The present data on all 23 common hOCT1 SNPs extend earlier reports and underline their relevance to the clinical response to imatinib. The data broadly agree with the effects of hOCT1 SNPs on metformin in polycystic ovary syndrome,33 although not in diabetes,29 despite suggestive laboratory data.13 The results question whether a “one size fits all” strategy of imatinib at 400 mg daily is optimal in CML. Interestingly, there are conflicting clinical data on the advantage of higher imatinib doses (600-800 mg daily). It is plausible that this may reflect differing proportions of hOCT1 SNP between trials. If so, higher imatinib doses may overcome limitations on imatinib uptake, and there is some evidence for this from the Australian TIDEL I trial.2
In conclusion, we demonstrate in the present study the functional significance of both the M420del and M408V hOCT1 polymorphisms and show their importance in determining clinical outcome in imatinib-treated CML. These SNPs, particularly M420del, may have potential use in tailoring TKI therapy at diagnosis, because patients carrying M420del might be better treated by a TKI that does not rely on hOCT1 for transport. This requires testing in prospective trials.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Jack Clark and Abhinav Kishore for technical assistance; Dr Steven Lane for the initial statistics on M420del; Drs Kathleen Till and Mark Glenn for assistance with the confocal microscope; and the Kay Kendall Leukaemia Fund (grant 321).
T.L.H. is supported by a C11074/A11008 Cancer Research UK program grant.
Authorship
Contribution: A.G. designed the study, performed the experiments, analyzed the functional data, and wrote the manuscript; L.W. collected the clinical information, performed the experiments, and analyzed the mRNA data; A.L.J. performed the statistical analyses of association between SNPs and clinical outcomes and contributed significantly to the manuscript; G.X. and T.L. designed the pyrosequencing primers, demonstrated the use of pyrosequencing and direct sequencing, and helped with the analysis; A.D. analyzed the data; S.P. designed the Sequenom MassARRAY primers; J.-E.Z. demonstrated the use of the Sequenom MassARRAY primers and helped with the analysis; G.A. performed the experiments; T.L.H., L.F., P.D.K., M.C.M., and R.E.C. provided clinical material and associated clinical data; and M.P. and R.E.C. designed the study and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Prof Richard E. Clark, Department of Haematology, Royal Liverpool University Hospital, Prescot Street, Liverpool L7 8XP, United Kingdom; e-mail: clarkre@liverpool.ac.uk.