Abstract
Multiple myeloma is a plasma cell malignancy in which significant advances have been observed during the last 15 years. Our understanding of the disease has been advanced through its molecular characterization. We have also seen improvements in patient care with the development of 2 new classes of active agents, proteasome inhibitors and immunomodulatory drugs (IMiDs), resulting in a significant improvement in overall survival of myeloma patients such that it can now be debated as to whether some subsets of myeloma patients can be cured. However, the advances in our understanding of myeloma biology occurred in parallel with advances in treatment as opposed to being directly informed by the research. Moreover, the molecular characterization of malignant plasma cells would not have predicted the effectiveness of these novel therapies. We hypothesize that proteasome inhibitors and IMiDs are highly active because malignant plasma cells are constrained by many of the characteristics of their normal counterparts and these novel therapies target both normal plasma cell biology and the cancer biology of myeloma. Thus, a better understanding of normal plasma cell biology will likely yield as many actionable targets as mapping the genomic landscape of this disease.
Introduction
Multiple myeloma is the second most common hematologic malignancy. In 2014, the expectations are that >24 000 new cases will be diagnosed and >11 000 deaths will occur in the United States.1 Until the 1990s, few advances in treatment of the disease occurred; thus, myeloma was incurable with a median survival of 2 to 3 years. However, beginning in the mid-1990s, with the introduction of high-dose melphalan and autologous bone marrow transplantation, patient survival began to improve.2 Further gains were made in the 2000s with the introduction of highly active agents with mechanisms of action independent of DNA damage.3 Thalidomide and bortezomib were the first examples of active agents, and subsequently, second in class agents for both immunodulatory drugs (IMiDs) and proteasome inhibitors have been US Food and Drug Administration approved.4 Combinations of these 2 classes of novel agents result in responses in nearly all patients and are now routinely used for the treatment of newly diagnosed myeloma followed by stem cell transplant.5 Together, this has significantly improved the overall survival of myeloma patients, and with a sizable percentage (20-30%) of patients surviving for >10 years,6,7 it is no longer appropriate to generically call myeloma an incurable disease.
Improved understanding of the biology of myeloma ushered in an era of therapies designed to target signaling pathways associated with the growth and survival of malignant plasma cells.8 However, these approaches have not been nearly as successful as the use of proteasome inhibitors and IMiDs and have yet to result in the approval of any agent for the treatment of myeloma. Thus, rational approaches, based on high-quality basic/translational research, have not been nearly as effective as using 2 classes of agents that either targeted the proteasome, a molecular machine present in every cell, or functioned through an until recently unknown mechanism that was initially believed to be related to angiogenesis.9 Even more surprising, the cataloging of genomic changes associated with myelomagenesis has provided little insight as to why proteasome inhibition or IMiDs would be effective in the setting of malignant vs normal plasma cells.10-12 Because changes in the expression, copy number, or “activating” mutations have not been identified in the 2 molecular targets of proteasome inhibitors (PSMB5) or IMiDs (CRBN), neither could be considered targets based oncogene addiction. Thus, it is unlikely that we would be using either of these classes of agents if treatment decisions were solely based on molecular profiling of the disease.
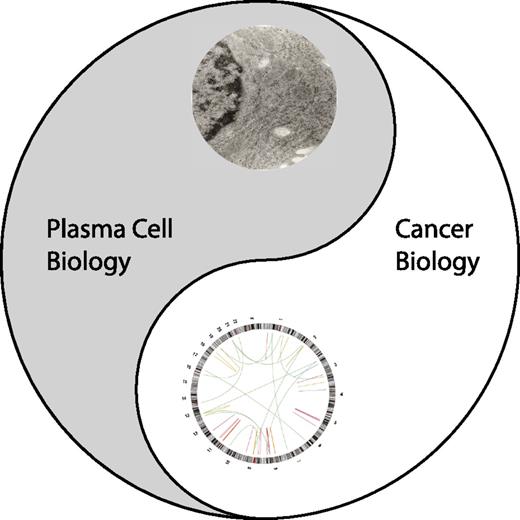
If the genomic landscape of the disease does not predict why these classes of agents are the most active in myeloma, are there other characteristics of the disease that are being effectively targeted? We believe the answer to this question is yes and review the evidence that myeloma cells are the product of 2 distinct but complementary biologies that can both be targeted therapeutically. Thus, like Yin and Yang of Taoist philosophy, the genomic changes associated with cancer exist in balance with the biology of normal plasma cells, and both can be effectively targeted in the treatment of this disease (Figure 1).
The Tao of myeloma. Yin represents the plasma cell biology. The eye of the Yin is an electron micrograph of the human myeloma cell line, MM.1s. Yang represents the cancer biology of myeloma. The eye of the Yang is a cirrcos plot of genomic structural variations observed in MM.1s.
The Tao of myeloma. Yin represents the plasma cell biology. The eye of the Yin is an electron micrograph of the human myeloma cell line, MM.1s. Yang represents the cancer biology of myeloma. The eye of the Yang is a cirrcos plot of genomic structural variations observed in MM.1s.
The Yin of myeloma: plasma cell biology
Yin represents the dark, soft, and more passive portion of the whole, and in the case of myeloma, this best exemplifies the normal plasma cell biology that is retained during myelomagenesis. Myeloma has long been characterized by its plasma cell biology, specifically by a change in plasma cells in the bone marrow and an increase in monoclonal antibody or immunoglobulin light chain (IgL) in the serum and/or urine. With the development of free IgL analysis, we can now say this is true for nearly all diagnosed myelomas.13 Thus, the pathognomonic marker of myeloma is not based on a genetic change associated with tumorigenesis, it is due to a clonal population of tumor cells that retain the normal function of a long-lived bone marrow plasma cell.
Plasma cells are the effector cells of the B-lymphocyte lineage. They result from the differentiation of B cells following activation through the antigen receptor or through Toll-like receptors in concert with costimulatory and cytokine signals.14,15 Through a differentiation program controlled by downregulation of Pax5 and up-regulation of 3 transcription factors, B lymphocyte-induced maturation protein-1, interferon regulatory factor 4 (IRF4), and x-box binding protein-1 spliced (XBP1s), the B cell with antigen recognition function and proliferative potential differentiates to one that no longer expresses surface antigen receptors but is able to constitutively secrete copious amounts of antibodies.16 Plasma cells that have undergone affinity maturation and class switch recombination in a germinal center reaction, home to the bone marrow where they can be maintained and provide protective immunity.17 This is the basis for the long-term efficacy of vaccines.18
Myeloma plasma cells are typically found in the bone marrow microenvironment, like normal plasma cells, for most of the disease course. Moreover, they are dependent on similar survival cues from that microenvironment. These interactions include support in the form of soluble cytokines produced by cells within the microenvironment and direct cell interactions (see Table 1 for a list of common survival factors in the bone marrow microenvironment for normal and myeloma plasma cells).19,20 Thus, developing therapies that interfere with many of the interactions or signaling mechanisms between myeloma cells and the microenvironment are in effect targeting the normal plasma cell biology of the myeloma cell. Intrinsically, the survival of long-lived plasma cells is maintained by Mcl-1. The Bcl-2/Bcl-xL inhibitor, ABT-737, is capable of blocking the differentiation of B cells but does not result in the loss of established plasma cells in the bone marrow, suggesting Mcl-1 dependence.21 Recently, a more direct proof of bone marrow plasma cell dependence on Mcl-1 was provided through a conditional knockout.22 Additionally, all human myeloma cell lines tested to date are either dependent on Mcl-1 or codependent on Mcl-1 and Bcl-2/xL.23-25 However, myeloma cells must be able to survive and grow in this microenvironment beyond the finite space that has been proposed for the long-lived plasma cell, as plasma cells typically constitute 1% of the bone marrow space, whereas intramedullary myeloma can often consume the majority of available space in the compartment. The most plausible explanation for this difference is that myeloma plasma cells are altering the microenvironment, resulting in an increase in its size or function. Alternatively, through the production of factors such as interleukin (IL)-6, upregulation of adhesion molecules (eg, ITGB7), and the expression of surface receptor/ligand pairs including CD28/CD86, the myeloma cells are contributing to their own microenvironment. Although these are also important for normal plasma cell survival,26,27 an increase in both CD28 and CD86 expression is observed in myeloma and with disease progression.28-30 Evidence for both possibilities exists, and importantly, these are not mutually exclusive events. However, in each case, the myeloma cells are using the same types of signals that normal plasma cells use to assure survival within the microenvironment.
Survival factors common to normal and myeloma plasma cells
| Survival factor . | Examples . |
|---|---|
| Membrane-bound factors (receptors) | CD80/CD86 (CD28) |
| Soluble factors (receptors) | Interleukin-6 (IL6R), APRIL (BCMA), tumor necrosis factor α (TNFR) |
| Intrinsic factors | MCL1 |
| Survival factor . | Examples . |
|---|---|
| Membrane-bound factors (receptors) | CD80/CD86 (CD28) |
| Soluble factors (receptors) | Interleukin-6 (IL6R), APRIL (BCMA), tumor necrosis factor α (TNFR) |
| Intrinsic factors | MCL1 |
Myeloma cells maintain the secretory function of normal plasma cells, yet it is not clear that they are as efficient in the secretion of antibodies as normal plasma cells. Although serum/urine concentrations of M-protein or IgL are markers of disease burden and changes can accurately reflect changes in tumor burden, this does not necessarily equate to similar levels of antibody production or secretion at the single cell level. Indeed if one compares the transcript levels of the antibody genes (IGH and IGL) in freshly isolated plasma cells, normal cells typically express higher mRNA levels than myeloma plasma cells (L.H.B., unpublished observation, December 2012). Moreover, the quality control of secretion from myeloma plasma cells appears to be compromised, as increased levels of free IgL are associated with myeloma even when myeloma cells are capable of secreting antibodies.31 Regardless, myeloma plasma cells appear to be unable to shed themselves completely of their secretory function in most cases, where cessation of antibody production would be predicted to allow the myeloma plasma cell to more efficiently use its resources for growth.
Finally, myeloma cells appear to be dependent on the same transcriptional program as normal plasma cells. Myeloma cells have been shown to express all 3 transcription factors that normal plasma cells are dependent on, and inhibition of any of these transcription factors or of inositol-requiring protein-1α (IREα), which is required for the production of XBP1s, results in myeloma cell death32-36 or loss of the mature phenotype.37 Furthermore, recent studies also demonstrated the dependency of myeloma cells on Aiolos and Ikaros, 2 transcription factors required for the generation of high-affinity long-lived plasma cells in the bone marrow.38
Taken together, the 3 most identifiable characteristics of a differentiated normal bone marrow plasma cells, (1) secretory function, (2) long-lived nature, and (3) dependence on microenvironment, as well as the transcriptional program that is required for plasma cell differentiation and maintenance, are all maintained to a significant extent in the myeloma plasma cell. Most importantly, these characteristics are maintained regardless of the genetic and genomic changes that result in myelomagenesis. There are 2 implications of these findings that have a significant impact on therapeutic approaches for the treatment of myeloma. First, the maintenance of normal plasma cell characteristics suggests that myeloma cells are as dependent on plasma cell biology as they are on the oncogenic changes that resulted in tumorigenesis. Although myeloma cells may not secrete antibodies or IgL with the same efficiency as normal plasma cells, it is not effective use of biosynthetic resources for a growing cell to dedicate a significant portion of their amino acid pool to the production of proteins that will be secreted. Thus, although many if not all growing cells undergo metabolic reprogramming to generate building blocks for continued growth,39 myeloma cells waste these resources. This only makes sense if the secretion of proteins is intimately tied to a genetic program that is necessary for the existence of the cell. Thus, targeting the existing plasma cell biology of the myeloma cell may be equally effective to targeting the cancer biology of the cell. Second, the genomic era of myeloma research has shown us that myeloma is not only a heterogeneic disease between patients, but it is also a multiclonal disease within a given patient.11,12,40-43 Although this gives us opportunities to target specific lesions within patients, it also suggests that, regardless of the alteration, the myeloma cell must maintain aspects of the plasma cell phenotype, as significant differences in these characteristics have not been associated with specific genomic changes in myeloma. Thus, although mapping the genomic landscape of the disease results in a greater fractionation of myeloma into smaller and smaller subsets, the characteristics of plasma cell biology lump myeloma back toward a single disease. Therefore, therapies targeted at myeloma plasma cell biology are more likely to be active in a larger proportion of myeloma patients.
Finally, there is evidence for the existence of less mature cells in multiple myeloma, although it remains somewhat unclear if this pool represents a true cancer stem cell population. Further understanding of this population and its role in myeloma pathogenesis will provide additional means to target the cancer biology of myeloma.44
The Yang of myeloma: cancer biology
Yang represents the more aggressive portion of the whole and in myeloma would best be represented by the changes associated with transformation. These changes have been extensively discussed in several recent reviews on the genomic landscape of myeloma45,46 and will not be covered in significant detail. A summary of these changes is provided in Table 2. However, the identification of gains and losses of chromatin, translocations, gene expression patterns within subsets of myeloma, epigenetic changes, or activating and inactivating mutations within specific genes did not provide the rationale for the use of IMiDs or proteasome inhibitors nor has this information led to the development of a single therapeutic agent that has contributed to the improvement in patient survival. Indeed, trials with the inhibitors of FGFR3, which is overexpressed in the majority of patients harboring the t(4;14), were disappointing (these studies were not designed to only target patients with mutant FGFR3).47 However, promising preclinical data with bromodomain inhibitors that target the expression of c-myc may translate into effective myeloma treatment.48 Moreover, 4% of myeloma patients have actionable B-Raf mutations, and this is translating rapidly to clinical testing.10 There has already been a report of a patient successfully treated with vermurafenib.49 Thus far, genetic and genomic alterations in myeloma have been far more important in defining the prognosis for a patient. It has also provided information about why certain therapies appear to be more effective in subsets of myeloma populations. For example, bortezomib is more effective in patients that have mutations or changes in gene copy number that result in activation of the noncanonical nuclear factor (NF)-κB pathway.50 Additionally, understanding the biology of the t(4;14) translocation has resulted in a better appreciation of why these patients do poorly with melphalan treatment. One of the genes that is activated due to this translocation, WHSC1 (MMSET), a histone methyltransferase whose activation results in global changes in histone methylation patterns, has been shown to regulate the DNA damage response through the regulation of histone H4 lys20 dimethylation (H4K20me2) and the recruitment of 53BP1 to sites of double-strand breaks.51
Genomic alterations observed in myeloma
| Event . | Examples . |
|---|---|
| Immunoglobulin translocations (cyclin D family) | 11q13, 6p21, 12p13 |
| Immunoglobulin translocations (MAF family) | 16q23, 20q12, 8q24 |
| Immunoglobulin translocations (MMSET/FGFR3) | 4p16 |
| MYC translocations (juxtaposition to plasma cell superenhancers) | IGH, IGL, IGJ, IGK, PRDM1, XBP1, FAM46C, NSMCE2, TXNDC5, KRAS |
| Hyperdiploid (trisomy) | 3, 5, 7, 9, 11, 15, 19, 21 |
| Chromosome deletions | 13p, 17p, 1p, 6q |
| Chromosome gains | 1q |
| Activating mutations | BRAF, NRAS, KRAS, FGFR3* |
| Inactivating mutations | TP53, FAM46C, DIS3, RB1, EGR1, SP140 |
| Genes inactivated by homozygous deletion | CDKN2C, KDM6A |
| Mutations that activate the nuclear factor-κB pathway | TRAF2, TRAF3, CYLD, BIRC2, BIRC3 |
| Mutations in genes involved in plasma cell biology | XBP1, PRDM1, IRF4 |
| Event . | Examples . |
|---|---|
| Immunoglobulin translocations (cyclin D family) | 11q13, 6p21, 12p13 |
| Immunoglobulin translocations (MAF family) | 16q23, 20q12, 8q24 |
| Immunoglobulin translocations (MMSET/FGFR3) | 4p16 |
| MYC translocations (juxtaposition to plasma cell superenhancers) | IGH, IGL, IGJ, IGK, PRDM1, XBP1, FAM46C, NSMCE2, TXNDC5, KRAS |
| Hyperdiploid (trisomy) | 3, 5, 7, 9, 11, 15, 19, 21 |
| Chromosome deletions | 13p, 17p, 1p, 6q |
| Chromosome gains | 1q |
| Activating mutations | BRAF, NRAS, KRAS, FGFR3* |
| Inactivating mutations | TP53, FAM46C, DIS3, RB1, EGR1, SP140 |
| Genes inactivated by homozygous deletion | CDKN2C, KDM6A |
| Mutations that activate the nuclear factor-κB pathway | TRAF2, TRAF3, CYLD, BIRC2, BIRC3 |
| Mutations in genes involved in plasma cell biology | XBP1, PRDM1, IRF4 |
FGFR3 mutations are only found in 5% of newly diagnosed patients with t(4;14).96
The Tao of myeloma therapy
Two biologies coexist within the myeloma plasma cell, and both appear to be required for myeloma cell survival. This provides 2 potential opportunities to target the disease. Thus far, rational attempts at targeting the cancer biology of myeloma have been unsuccessful. However, targeting the cancer biology is the likely explanation for why alkylating agents have been an important part of the antimyeloma armament. Rational targeting of plasma cell biology has not been tried beyond the use of steroids, and although these are active agents, they only work for a short duration. This begs the question of what do the most active therapies in myeloma target? Consistent with a balance between normal and cancer biology, it is tempting to speculate that proteasome inhibitors and IMiDs must be able to effectively target both the Yin and the Yang of myeloma. This explains why they both have activity in a larger proportion of myeloma patients than can be explained by a single genomic feature and also have activity in tumors that do not share the normal plasma cell biology of myeloma.
The rationale for inhibition of the proteasome as a therapeutic approach was based primarily on the blocking signaling that resulted in the activation of NF-κB.52 However, it was not predicted to be most effective in myeloma because of NF-κB activation within this disease. Indeed NF-κB is important in other tumor types as well, yet proteasome inhibitors are not as active in these cancers. We and others reasoned that it was the normal plasma cell biology, specifically quality control for the large amounts of immunoglobulin proteins produced, that was the primary reason why myeloma cells were sensitive to this class of drugs.53 These studies showed dysregulation of the unfolded protein response, as well as the role of protein load due to Ig production in proteasome inhibitor sensitivity.54-56 Similar findings were also shown in Waldenström macroglobulinemia,57 confirming a role for Ig metabolism in the response to proteasome inhibition. Importantly, recent studies have demonstrated that resistance to proteasome inhibitors is associated with accumulation of a cell population that is less differentiated and lacks plasma cell features.37,58,59 Additional studies have demonstrated that aggresome formation and autophagy provided a salvage pathway in the face of proteasome inhibitor-induced Ig accumulation.60,61 These studies provided the rationale for combining histone deacetylase (HDAC) inhibitors and autophagy inhibitors with bortezomib, which was highly active preclinically, and several clinical trials testing HDAC inhibitors and proteasome inhibitors are underway.62,63 These studies also resulted in the development of an HDAC6-selective antagonist (ACY-1215) to more specifically target the aggresome pathway in concert with proteasome inhibition.64 ACY-1215 also has preclinical activity in myeloma65 and is currently being evaluated in clinical trials.
Although the role of Ig metabolism suggests that myeloma cells are sensitive to proteasome inhibition as a result of their maintenance of normal plasma cell biology, the more compelling argument can be made by the effectiveness of proteasome inhibitors in the treatment of nonmalignant plasma cell disorders. There has been significant preclinical and clinical work demonstrating the effectiveness of bortezomib as an antiplasma cell agent in solid organ transplant and antibody-mediated autoimmunity. Antibody-mediated rejection (AMR) is the result of the presence or development of anti-HLA antibodies in organ transplant recipients.66 Currently, there are no US Food and Drug Administration-approved treatments for AMR,67 and based on the success of bortezomib in killing myeloma plasma cells, proteasome inhibition was tested in cases of AMR refractory to treatment with plasmapheresis and intravenous Ig in the presence or absence of rabbit anti-thymocyte globulin or rituximab following renal transplant.68 Bortezomib showed significant activity reducing anti-HLA antibodies by 50% within 2 weeks.69,70 Of the agents used for treatment of AMR in renal transplant, only bortezomib was shown to induce apoptosis of plasma cells in vitro and deplete bone marrow plasma cells in 2 patients. A larger trial using a bortezomib-based protocol has shown promising activity; however, not all studies have been positive, and there is no report of a randomized trial,68 although one is being initiated.71 Additionally results from the use of bortezomib to desensitize patients with anti-HLA antibodies prior to renal transplant have been equivocal.72
Use of bortezomib for plasma cell depletion in autoimmunity was initially shown in preclinical models of lupus.73 This has been extended to clinical use in other autoimmune disorders. A patient with hyper-IgG4 syndrome was successfully treated with the bortezomib-based myeloma treatment protocol CyBorD.74 More recently, a patient with thrombotic thrombocytopenic purpura who was refractory to plasma exchange and immunosuppressive agents responded to bortezomib.75
The activity of proteasome inhibitors in disrupting antibody catabolism in myeloma plasma cells, as well as their activity in normal plasma cells, could lead one to conclude that this class of drugs is primarily working through their ability to disrupt normal plasma cell biology. However, it is unlikely that this is the only reason that myeloma cells are so susceptible to this type of treatment. Although targeting the proteasome is pharmacologically how these agents function, the number of cell processes that are controlled by the ubiquitin proteasome system (UPS) would make it unlikely that a single consequence of proteasome inhibition would be responsible for its cell-killing activity. In addition to targeting dysregulated NF-κB in patients with mutations that result in increased NF-κB activity,50 proteasome inhibition may target another process where cancer cells, and specifically myeloma cells, are more dependent than their normal counterparts. Several lines of evidence suggest that myeloma, like other cancers, displays significant levels of genomic instability.76 This results in cells that have a higher level of dependence on DNA repair processes than normal cells and offers a likely explanation for why a slow-growing cell like the myeloma plasma cell is sensitive to alkylating agents. Several aspects of DNA repair processes require an active UPS; therefore, proteasome inhibitors can sensitize cells to DNA-damaging agents. The Fanconi anemia pathway is one pathway that requires ubiquitination of proteins for function/nuclear localization, and FANCD2 monoubiquitination is blocked by proteasome inhibition.77 Interestingly, FANCD2 expression is regulated in a proteasome/NF-κB–dependent mechanism in myeloma cells; therefore, inhibition of DNA repair may contribute to the activity of proteasome inhibitors.78 Consistent with this possibility, a small interfering RNA screen for modulators of the bortezomib response in a colon cancer line revealed several DNA damage response genes, particularly genes involved in homologous recombination (HR)-mediated DNA repair.79 In myeloma cells, bortezomib treatment results in a decrease in the nuclear pools of free ubiquitin resulting in decreased H2AX polyubitquitination by RNF8 and RNF168 at the sites of DNA double-stranded breaks. These ubiquitin tags on histones are critical for the recruitment and retention of RAD51 and BRCA1 at the DNA break site. Therefore, HR-dependent repair is blocked by proteasome inhibitors. This also renders cells susceptible to PARP1-2 inhibition and creates a contextual synthetic lethality between bortezomib and the PARP inhibitor ABT-888.80 The promising preclinical activity of this combination has translated to a clinical trial (clinicaltrials.gov #NCT01495351).
Together, these data suggest that bortezomib may have antitumoral activity in diseases like lymphoma because it can target DNA repair pathways and/or NF-κB activation, but it works best in myeloma because it can target both the cancer biology and the normal plasma cell biology. Moreover, if the targeting of the HR pathway is the result of decreased availability of ubiquitin, then one could argue that proteasome inhibitors may be the best way to simultaneously target these distinct biologies as the increased translational activity in the plasma cell results in a more susceptible ubiquitin pool when the proteasome is inhibited.
Although thalidomide (in combination with dexamethasone) was the first novel agent US Food and Drug Administration approved for the treatment of newly diagnosed myeloma, it is only in the last few years that we have come to appreciate how IMiDs can target both the myeloma plasma cell and function as immunodulatory agents. A landmark study demonstrated that thalidomide’s teratogenic activity was the result of its binding to CRBN, a component of the Cullin 4-containing complex.81 Zhu et al went on to show that CRBN was also the critical target for antimyeloma activity in preclinical models,82 and subsequent studies have shown a correlation between CRBN expression and IMiD clinical activity.83-85 Cullin ring complexes are part of the UPS; however, unlike targeting the proteasome, CRBN likely effects the fate of a smaller subset of proteins. Therefore, to target either cancer or plasma cell biology, it would need to be a negative regulator of a few key proteins that would be important for all myelomas. Recent findings are consistent with this possibility as 2 studies demonstrated that lenalidomide binding to CRBN modifies its degron binding and results in the targeting of 2 transcription factors that are important for plasma cell maintenance.38,86,87 Ikaros and Aiolos regulate the expression of IRF4, and studies have demonstrated that IMiDs downregulate IRF4 in myeloma cells.88,89 IRF4 can also regulate MYC expression, therefore providing a link to how IMiDs regulate the expression of MYC in myeloma. The role of MYC in myelomagenesis has become clearer through its ability to lead to a myeloma-like disease in a murine model,90,91 as well as the recent discovery of the prevalence of MYC translocations resulting in juxtaposition to superenhancers active in myeloma92,93 (Table 2). Thus, like proteasome inhibitors, IMiDs appear to be highly active in this disease because they target both of its biologies.
Conclusions
There are several implications to being able to target both the normal and cancer biologies of myeloma. First, it allows for a rational combination of agents that target each biology, as well as the development of additional drugs like proteasome inhibitors that appear to target both. A greater understanding of the myeloma genome will hopefully provide additional actionable targets for therapeutic intervention. Similarly, applying our current knowledge of the long-lived bone marrow plasma cell and identifying additional targets will also lead to new therapeutic opportunities in the myeloma. This has already begun with the preclinical development of inhibitors of the unfolded protein response sensor IRE1α,34,35 as well as targeting SLAMF7 and CD38 with antibody-based therapies94 and promising preclinical data demonstrating the potential of B cell maturation antigen for targeting by genetically modified T cells.95 Combining more selective plasma cell inhibitors with targeting proteins specifically mutated/activated in myeloma should allow one to achieve activity similar if not better than our current agents and with less toxicity than observed with proteasome inhibitors. A second implication of targeting 2 biologies, is that if you are selectively targeting 1 biology with therapy, then you are selecting against that biology for relapse. Relapse from proteasome inhibitors and IMiDs does appear to reflect a different disease than relapse from conventional chemotherapeutic agents as consistent with primarily targeting plasma cell biology. Extramedullary relapses appear to be more common than in the past. Additionally, one would predict that loss of immunoglobulin production is more likely to occur in patients treated with novel therapeutics. Hopefully, with more precise and balanced targeting of the Yin and Yang of myeloma, we will be able to build on the progress of the last decade and bring a larger proportion of our patients closer to a cure.
Acknowledgments
The authors were unable to cite all the original papers due to reference limits and apologize for not citing all relevant original papers. The authors thank Drs Esther Obeng and Scott Newman for the MM.1s EM and cirrcos plot used in the figure. The authors also thank members of the L.H.B and K.P.L. laboratories for critical review of the manuscript. L.H.B. is a Georgia Research Alliance Distiguished Cancer Scholar.
Authorship
Contribution: L.H.B., J.L.K., N.J.B., S.L., and K.P.L. participated in conceptualizing the review and helped in preparing the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Lawrence H. Boise, Department of Hematology and Medical Oncology, Winship Cancer Institute of Emory University, Emory University School of Medicine, 1365 Clifton Rd NE, Rm C4012, Atlanta, GA 30322; e-mail: lboise@emory.edu.