Key Points
The Myc oncoprotein targets central regulators of the SUMOylation machinery, resulting in a hyper-SUMOylation state in Myc-induced lymphoma.
Targeting SUMOylation by genetic or pharmacologic means represents a novel therapeutic option for lymphomas with MYC involvement.
Abstract
Myc oncogenic transcription factors (c-Myc, N-Myc, and L-Myc) coordinate the control of cell growth, division, and metabolism. In cancer, Myc overexpression is often associated with aggressive disease, which is in part due to the destruction of select targets by the ubiquitin-proteasome system (eg, SCFSkp2-directed destruction of the Cdk inhibitor p27Kip1). We reasoned that Myc would also regulate SUMOylation, a related means of posttranslational modification of proteins, and that this circuit would play essential roles in Myc-dependent tumorigenesis. Here, we report marked increases in the expression of genes that encode regulators and components of the SUMOylation machinery in mouse and human Myc-driven lymphomas, resulting in hyper-SUMOylation in these tumors. Further, inhibition of SUMOylation by genetic means disables Myc-induced proliferation, triggering G2/M cell-cycle arrest, polyploidy, and apoptosis. Using genetically defined cell models and conditional expression systems, this response was shown to be Myc specific. Finally, in vivo loss-of-function and pharmacologic studies demonstrated that inhibition of SUMOylation provokes rapid regression of Myc-driven lymphoma. Thus, targeting SUMOylation represents an attractive therapeutic option for lymphomas with MYC involvement.
Introduction
Myc oncoproteins (c-Myc, N-Myc, and L-Myc) are overexpressed in over half of all tumor types by virtue of chromosomal amplifications or translocations or via mutations in pathways that normally control Myc expression.1,2 Myc oncoproteins function as basic/helix-loop-helix/leucine zipper transcription factors that, under physiological conditions, coordinate cell growth and metabolism with cell division. When overexpressed, Myc oncoproteins accelerate cell proliferation, augment growth (mass), and direct the cancer metabolic phenotype. In this scenario, Myc also blocks terminal differentiation and promotes tumor angiogenesis, which reflects the widespread selection for Myc activation in various malignancies.3,4
In addition to directly controlling transcription of a large cast of targets, Myc indirectly affects the translation and turnover of proteins.5 One prominent example is the activation of the ubiquitin-proteasome system (UPS), in particular SCFSkp2-mediated suppression of p27Kip1, which functions as a central inhibitor of cyclin-dependent kinase activity. Accordingly, low p27Kip1 protein levels are associated with aggressive cancer growth and poor prognosis in humans,6 and loss of p27Kip1 accelerates Myc-driven lymphomagenesis.7 Conversely, loss of Cks1 augments p27Kip1 levels and impairs Myc-induced proliferation and lymphomagenesis.8
Small ubiquitin-like modifier (SUMO) conjugation to cellular proteins is a second prominent posttranslational modification that controls protein function, subcellular localization, and/or expression. The SUMO proteases (SENP) deconjugate SUMOylated proteins and thus play essential roles in maintaining proper levels of SUMOylated and un-SUMOylated substrates.9-11 Notably, SUMO homeostasis goes awry in various carcinomas.12,13 Further, SUMOylation and the expression of SUMO-conjugating enzyme Ube2i and the SUMO ligase PIAS1 is markedly elevated in multiple myeloma, and this is associated with poor prognosis.14
Therapeutics that block Myc transcription functions are not available in the clinic.15 However, cells transformed by oncogenes like Myc rely on physiological pathways to perform essential cellular functions, a phenotype termed nononcogene addiction. By definition, these pathways are not mutated but operate at a critical level close to exhaustion in cancer cells. Thus, they can be targeted in a synthetic lethal manner to kill cancer cells yet spare normal cells that can resort to parallel pathways.16 This strategy has proven effective in several models of Myc-driven tumorigenesis.17-19
A major and immediate downstream effect of Myc activation is a dramatic increase in the protein synthetic capacity of the cell,5,20 and genetic strategies that restore rates of protein synthesis to normal levels suppress Myc-induced tumorigenesis.21 Thus, modulating protein synthesis control could be a promising therapeutic approach.22,23 However, the components of the translation machinery that can be therapeutically targeted to exploit this addiction of Myc-driven cancer cells are largely undefined. In contrast, loss-of-function studies indicate that inhibition of at least some of the components of the UPS or SUMO posttranslational modification systems is an attractive strategy for targeting Myc-driven malignancies.8,24
Here, we report that Myc dramatically augments SUMOylation in mouse and human Myc-driven B-cell lymphoma by transcriptionally inducing nearly all of the genes encoding the regulators and components of the SUMO pathway. Further, targeting SUMOylation in neoplastic Myc-expressing B cells provokes cell-cycle arrest, polyploidy, and apoptosis. Finally, the hyper-SUMOylation state is required for the development and maintenance of Myc-driven lymphoma. Collectively, these findings suggest that targeting the SUMOylation pathway is an attractive strategy for eradicating tumors with MYC involvement.
Materials and methods
Cells, cell culture, and colony-forming assay
P493-6 cells were provided by G. Bornkamm (Munich, Germany).25 Eµ-Myc lymphoma cells were established from single-cell tumor suspensions. BL2, BL70, Raji, and Daudi Burkitt lymphoma (BL) cell lines (DMSZ) were cultured in RPMI/10% fetal calf serum. Raji-Eco BL lines were transfected with MSCV-rtTA-IRES-EcoReceptor-PGK-puro and selected for expression of reverse-type tetracycline-controlled transactivator (rtTA) and the ecotropic receptor by growth in puromycin-containing medium.26 Colony-forming assays were performed using MethoCult M3434 (Stem Cell Technologies) and counted on day 10.
Mice and tumor analysis
Animal experiments were performed in accordance with the local/regional animal ethics committee approvals. Eµ-Myc transgenic mice (C57Bl/6)27 and C57Bl/6 syngeneic recipients (Harlan Laboratories) were observed daily for signs of morbidity and palpable lymphomas. To obtain wild-type (wt) and precancerous Eµ-Myc B cells, mice were humanely euthanized at 4 to 6 weeks of age and organs harvested. Cells were resuspended, incubated with B220-MicroBeads, and enriched by magnetic cell sorting for B cells (Miltenyi Biotech).
For lymphoma transplant studies, syngeneic C57Bl/6 mice were IV injected with 2 × 104 Eµ-Myc lymphoma cells. For xenograft studies, 10 × 106 cells were injected into each flank of Nod/Scid mice. Tumor diameters were measured every other day with a shifting caliper by the same person followed by tumor volume calculation using the formula (length × width)/2. When xenotransplants reached a size of approximately 300 mm3, animals were treated with 1 mg/mL doxycycline (Dox) in 5% sucrose for 5 consecutive days. The control group received sucrose-containing water. At day 5, mice were euthanized and tumors collected for 3-dimensional measuring. Tumor volume was calculated using the formula (length × width × height)/2.
Plasmids and antibodies
Specific short hairpin RNA (shRNA) constructs targeting mouse Sae1 and Sae2 were ordered from Sigma MISSION (SAE1#1: TRCN0000040501, SAE1#2: TRCN0000040502, SAE2#1: TRCN0000040478, SAE2#2: TRCN0000040482). These plasmids are based on plko.1. The puromycinR gene was replaced by an eGFP complementary DNA. For interference with human SAE1 and SAE2, shRNA antisense sequences were taken from the Thermo Scientific Web site (SAE1: V3LHS_304633, SAE2: V2LHS_68112)24 and modified to fit the mi30 hairpin expression system.28 The sequences were synthesized by MWG-Biotech and cloned into the MSCV-LTRmiR30-SV40GFP (LMS) expression vector29 and the Dox-inducible TRE-dsRed-miR30/shRNA-PGK-Venus-IRES-NeoR plasmid.30 The antibodies used are listed in supplemental Table 1, available on the Blood Web site.
Viral infection
293T cells expressing the helper virus plasmids were transfected with the indicated plasmids (Lipofectamine 2000; Invitrogen). Lentiviral shRNA plasmids (Sigma) were cotransfected with lentiviral helper plasmids for virus production into 293T. Virus supernatants were used to infect lymphoma cells in the presence of 8 µg/mL polybrene (Sigma-Aldrich). Eµ-Myc cells were infected with lentivirus carrying specific shRNA targeting Sae1 or Sae2. BL cells bearing rtTA and eco receptor (Raji-Eco) were infected with retrovirus (LMS empty or shSAE1, shSAE2). At 96 hours postinfection, cells were sorted for green fluorescent protein (GFP) expression to a purity of >98%.
Cell viability and apoptosis assays
Proliferative indices were assessed by labeling cultured cells with 10 µM bromodeoxyuridine (BD Biosciences) for 5 minutes followed by staining according to the manufacturer’s protocol. For propidium iodide (PI) or 4′,6-diamidino-2-phenylindole (DAPI) cell-cycle analysis, cells were fixed in 70% ice-cold ethanol and stained in PI/DAPI staining solution (50 µg of PI/mL or 50 µg of DAPI/mL, 100 µg of RNase/mL, phosphate-buffered saline). To assess apoptosis, 5 × 105 cells were stained with PI/Annexin-V fluorescein isothiocyanate (Annexin-V-Fluos Kit; Life Technologies) and analyzed by flow cytometry. Apoptosis and cell-cycle distribution were measured by analysis of DNA content in the FL2 channel (linear mode, cell cycle) or FL3 channel (logarithmic mode, apoptosis). Cells with 1 log less or more below diploid DNA content were considered apoptotic (sub-G1).
RNA, expression profiling, and ChIP analyses
Expression profiling was performed using Affymetrix data31 or published data (GSE4475, BL; GSE40782, P493 low vs high Myc; GSE15907, ImmGen B-cell development) using GeneSpring and R-studio software. For quantitative PCR (qPCR), RNA was prepared using RNeasy (Qiagen). Complementary DNA was prepared using the Omniscript RT kit (Qiagen). qPCR was performed using a TaqMan cycler (Applied Biosystems) and the Platinum SYBR Green qPCR SuperMix-UDG kit (Invitrogen) and analyzed using the ΔΔCt method with control samples set as 1. For primer sequences, see supplemental Table 2. Chromatin immunoprecipitation sequencing (ChIP-seq) data files (GSE36354 and GSE30726) were visualized using IGV software (Broad Institute).
Immunoblotting
Protein extracts were electrophoretically separated on sodium dodecyl sulfate polyacrylamide gel electrophoresis gels, transferred to membranes (Immobilon-P; Millipore), and incubated with specific antibodies (supplemental Table 3).17
PET imaging
FDG (2-deoxy-2-[18F]fluoro-d-glucose) was synthesized as previously described.32 Imaging was performed using a micro–positron emission tomography system (Inveon, Siemens Preclinical Solutions). FDG was administered via tail vein injection (100 μL) at an activity dose of 5 to 10 MBq. Radiotracer accumulation was allowed for 60 minutes. Mice were imaged for a 15-minute static acquisition. Metabolic tumor volume was defined from the positron emission tomography (PET) images placing a semiautomated 3-dimensional region of interest of isocontour 50% around the maximum activity of the tumor.
Histology and immunohistochemistry
With institutional review board approval, and following informed consent, tumors from lymphoma patients were banked. Immunohistochemistry was performed as described previously.17 After incubation with antibody (supplemental Table 2), signals were visualized with peroxidase-conjugated secondary antibody (LSAB Kit; DAKO). Tissue sections were counterstained with Mayer hematoxylin solution. Staining intensity and quantity was evaluated by a certified pathologist analyzing high-power fields (×40) per sample (n = 3).
Statistical analyses
Statistical analyses were performed using GraphPad Prism. The bars represent the mean ± standard deviation (SD). All statistical analyses were Student t tests. P values <.05 were considered statistically significant.
Results
Myc induces the transcription of the SUMOylation machinery
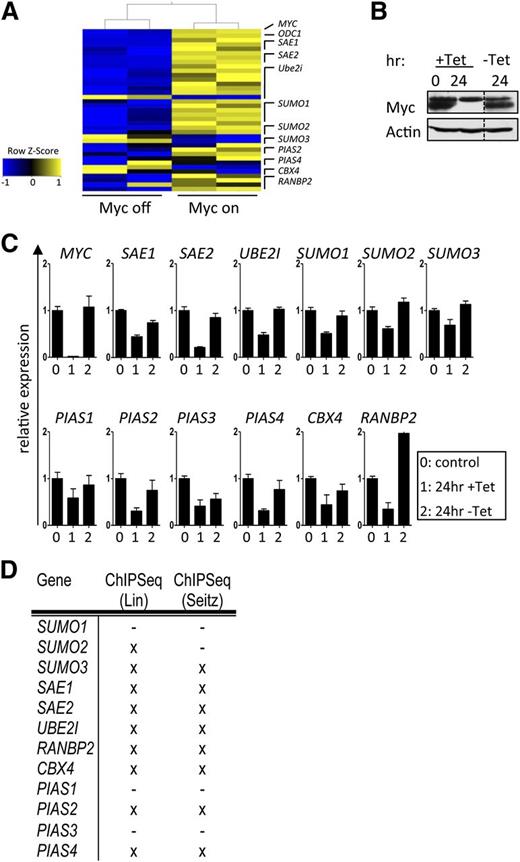
Myc plays essential roles in directing cell-cycle progression, cell growth (mass), and metabolism. Given the essential roles of protein SUMOylation in controlling the cell cycle and cell growth,10,11,33 we assessed if components and regulators of SUMOylation were Myc dependent. To initially test this, a gene expression data set (GSE40782)34 of P493-6 B lymphoma cells that carry a tetracycline (Tet)-repressible MYC transgene25 was analyzed. A striking upregulation of genes encoding components and regulators of the SUMOylation machinery was manifest in the Myc-on vs Myc-off state (Figure 1A). These data were confirmed in P493-6 cells cultured in media ±Tet in a Myc-on or Myc-off state (Figure 1B) using qPCR analyses (Figure 1C). Thus, Myc coordinates the control of SUMOylation gene expression in B cells.
Transcriptional regulation of the SUMOylation pathway by Myc is mainly direct. (A) P493-6 B cells carry a Tet-repressible MYC gene.25 Upon Tet treatment, the expression of Myc is reduced to nearly undetectable levels, and following withdrawal of Tet, Myc levels are restored within hours. Shown is the transcript expression of the indicated components of the SUMOylation pathway upon Myc activation (GSE40782). (B) Myc immunoblotting upon incubation of P493-6 cells with Tet (24 hr) and subsequent washout (-Tet 24 hr). (C) qPCR analysis of the indicated transcripts in P493-6 cells. Labeling of x-axes: 0, 0 hours Tet; 1, 24 hours Tet; 2, 24-hour Tet withdrawal (n = 3). (D) To assess direct Myc regulation, 2 publicly available quantitative ChIP-seq analyses were consulted.35,36 Shown is a summary of direct Myc binding to E-box sequences. X indicates direct Myc target genes.
Transcriptional regulation of the SUMOylation pathway by Myc is mainly direct. (A) P493-6 B cells carry a Tet-repressible MYC gene.25 Upon Tet treatment, the expression of Myc is reduced to nearly undetectable levels, and following withdrawal of Tet, Myc levels are restored within hours. Shown is the transcript expression of the indicated components of the SUMOylation pathway upon Myc activation (GSE40782). (B) Myc immunoblotting upon incubation of P493-6 cells with Tet (24 hr) and subsequent washout (-Tet 24 hr). (C) qPCR analysis of the indicated transcripts in P493-6 cells. Labeling of x-axes: 0, 0 hours Tet; 1, 24 hours Tet; 2, 24-hour Tet withdrawal (n = 3). (D) To assess direct Myc regulation, 2 publicly available quantitative ChIP-seq analyses were consulted.35,36 Shown is a summary of direct Myc binding to E-box sequences. X indicates direct Myc target genes.
To test if Myc directly binds to SUMOylation genes via identified E-boxes, we assessed 2 ChIP-seq data sets used to identify genome-wide Myc target genes.35,36 There was a marked and specific enrichment for Myc and Max binding to the promoter-regulatory regions in these SUMOylation genes in P493-6 B lymphoma cells, and in 5 different BL cell lines. Furthermore, Myc binding occurred at E-boxes in the promoter regions of the majority of these genes; thus, these are direct Myc target genes (Figure 1D and supplemental Figure 1). Collectively, these analyses show that critical regulators of SUMOylation are direct Myc transcription targets.
Activation of the SUMOylation pathway is a hallmark of Myc-induced lymphoma
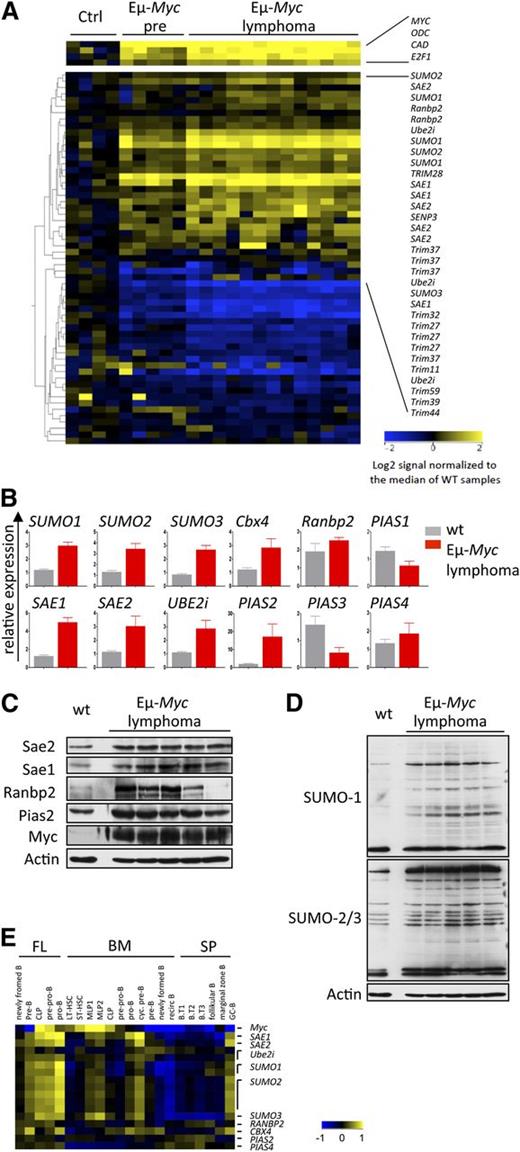
A powerful in vivo tool to dissect pathways that are downstream of Myc in transformation is afforded by the Eµ-Myc transgenic mouse,27 where one can assess direct effects of Myc in premalignant B cells and those events associated with neoplastic transformation.8,37 To assess the effects of Myc on the expression of established regulators of SUMOylation10,11,33 in B-cell lymphoma, expression profiling analysis was performed on B220+ B cells derived from 4- to 6-week-old wt vs Eµ-Myc littermates, as well as on Eµ-Myc lymphomas. A total of 36 of the 46 SUMOylation genes analyzed were differentially and significantly altered in their expression in premalignant Eµ-Myc B cells vs wt B cells (Figure 2A and supplemental Table 1). These changes included a marked increase in the expression of established positive regulators of SUMOylation, including E1 (Sae1, Sae2), E2 (Ube2i), and E3 ligases (Ranbp2, Cbx4, Pias2, Pias4), as well as SUMO1, SUMO2, and SUMO3. A number of genes often holding unclear roles in SUMOylation were also suppressed. Overall, the magnitude of the response was amplified in Eµ-Myc lymphomas (Figure 2A). These data were confirmed by qPCR and, for a select set of genes, by immunoblotting (Figure 2B-C). These analyses established a gross protein hyper-SUMOylation in Eµ-Myc lymphomas compared with wt B cells (Figure 2D).
SUMOylation pathway activation and hyper-SUMOylation in Myc-induced murine B-cell lymphoma. (A) Myc alters the expression of critical SUMOylation genes. Expression profiling of genes encoding critical components of the SUMOylation pathway is shown. B220+ B cells from wt mice (ctrl, n = 4), premalignant Eµ-Myc mice (Eµ-Myc pre, n = 5), and Eµ-Myc lymphomas (n = 13) were used. For a complete list of analyzed genes, see supplemental Table 1. All probe sets shown exhibit a >twofold change. (B) qPCR analysis of total RNA isolated from wt B220+ B cells (wt, n = 3) and Eµ-Myc lymphomas (n = 3). (C) Immunoblotting of the indicated proteins comparing pooled wt (n = 7) B220+ B cells and individual Eµ-Myc lymphomas. (D) Immunoblot analysis using SUMO1 and SUMO2/3 antibody in pooled wt (n = 7) vs individual Eµ-Myc lymphomas. (E) SUMOylation pathway gene expression during B-cell development assessed by Affymetrix analysis (GSE15907, ImmGen). B cells from fetal liver (FL), bone marrow (BM), and splenic (SP) origin were analyzed. B.T1-3, transitional B cells; CLP, common lineage progenitor; LT-HSC, long-term hematopoietic stem cell; MLP, multilineage progenitor; ST-HSC, short-term hematopoietic stem cell.
SUMOylation pathway activation and hyper-SUMOylation in Myc-induced murine B-cell lymphoma. (A) Myc alters the expression of critical SUMOylation genes. Expression profiling of genes encoding critical components of the SUMOylation pathway is shown. B220+ B cells from wt mice (ctrl, n = 4), premalignant Eµ-Myc mice (Eµ-Myc pre, n = 5), and Eµ-Myc lymphomas (n = 13) were used. For a complete list of analyzed genes, see supplemental Table 1. All probe sets shown exhibit a >twofold change. (B) qPCR analysis of total RNA isolated from wt B220+ B cells (wt, n = 3) and Eµ-Myc lymphomas (n = 3). (C) Immunoblotting of the indicated proteins comparing pooled wt (n = 7) B220+ B cells and individual Eµ-Myc lymphomas. (D) Immunoblot analysis using SUMO1 and SUMO2/3 antibody in pooled wt (n = 7) vs individual Eµ-Myc lymphomas. (E) SUMOylation pathway gene expression during B-cell development assessed by Affymetrix analysis (GSE15907, ImmGen). B cells from fetal liver (FL), bone marrow (BM), and splenic (SP) origin were analyzed. B.T1-3, transitional B cells; CLP, common lineage progenitor; LT-HSC, long-term hematopoietic stem cell; MLP, multilineage progenitor; ST-HSC, short-term hematopoietic stem cell.
Myc also plays essential roles in B-cell proliferation and development.38 To test whether the effects on SUMOylation genes were correlated with Myc expression, we assessed expression profiling data sets of the subsets of mouse progenitor immature and mature B cells from fetal liver, bone marrow, and spleen (GSE15907).39 These analyses established concordant expression of several SUMOylation genes with Myc during various stages of B-cell maturation (Figure 2E). Thus, the Myc-SUMOylation circuit is operational in both normal and cancer cells.
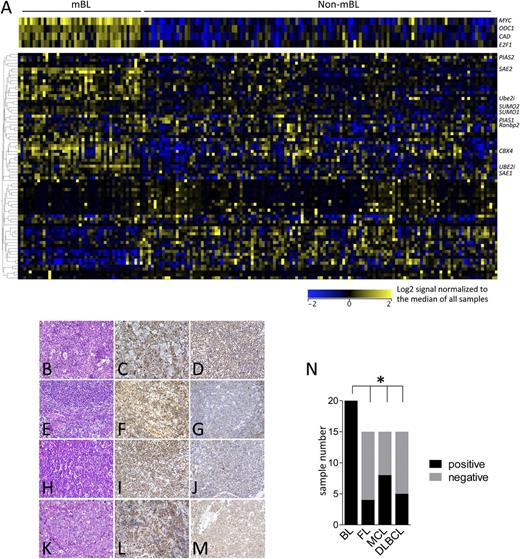
To investigate if Myc also induces the SUMOylation machinery in human B-cell lymphoma, publicly available gene expression data sets of aggressive human B-cell lymphomas were queried40 (GSE4475), including histopathologically defined BL samples, lymphomas with MYC/IG translocation, and those with non-MYC/IG status. These array data allow the distinction of molecular-defined Burkitt lymphoma (mBL) and non–Burkitt lymphomas (non-mBLs), which include diffuse large B cell lymphoma (DLBCL) and intermediate subtypes. A highly concordant upregulation of SUMOylation-related genes was observed in primary human mBL vs non-mBL (Figure 3A and supplemental Figure 2). To assess if the increased expression of SUMOylation regulators is manifest at the protein level, immunohistochemistry for SUMO1 and SUMO2/3 expression was performed on BL and various non-BL B-cell lymphomas. These analyses revealed that 100% of BL analyzed express high levels of SUMO2/3, whereas this was much less prevalent in other B-cell lymphoma subtypes analyzed (Figure 3B-M, summarized in Figure 3N). In contrast, SUMO1 expression was detected in all subtypes (supplemental Figure 3). Thus, alterations in SUMOylation in Myc-driven lymphomas in mice and humans are not due to transformation per se, and there are marked differences in this circuit in lymphomas with and without MYC involvement.
Activation of the SUMOylation pathways is a hallmark of human BL. (A) The expression of components of the SUMOylation pathway was analyzed in the gene expression data set GSE4475, comparing mBL with non-mBL samples.40 (B-M) Tissue microarrays of BL, DLBCL, mantle cell lymphoma (MCL), and follicular lymphoma (FL) were evaluated for SUMO2/3 expression using immunohistochemistry. (B) BL hematoxylin and eosin (H&E) staining. (C-D) BL SUMO2/3 positive. (E) FL H&E staining. (F) SUMO2/3-positive FL. (G) SUMO2/3-negative FL. (H) MCL H&E staining. (I) SUMO2/3-positive MCL. (J) SUMO2/3-negative MCL. (K) DLBCL H&E staining. (L) SUMO2/3-positive DLBCL. (M) SUMO2/3-negative DLBCL. Representative cases are shown. (N) Quantification of SUMO2/3 positivity in the indicated human B-cell lymphoma samples.
Activation of the SUMOylation pathways is a hallmark of human BL. (A) The expression of components of the SUMOylation pathway was analyzed in the gene expression data set GSE4475, comparing mBL with non-mBL samples.40 (B-M) Tissue microarrays of BL, DLBCL, mantle cell lymphoma (MCL), and follicular lymphoma (FL) were evaluated for SUMO2/3 expression using immunohistochemistry. (B) BL hematoxylin and eosin (H&E) staining. (C-D) BL SUMO2/3 positive. (E) FL H&E staining. (F) SUMO2/3-positive FL. (G) SUMO2/3-negative FL. (H) MCL H&E staining. (I) SUMO2/3-positive MCL. (J) SUMO2/3-negative MCL. (K) DLBCL H&E staining. (L) SUMO2/3-positive DLBCL. (M) SUMO2/3-negative DLBCL. Representative cases are shown. (N) Quantification of SUMO2/3 positivity in the indicated human B-cell lymphoma samples.
Hyper-SUMOylation is required for the proliferation of Myc-induced lymphomas
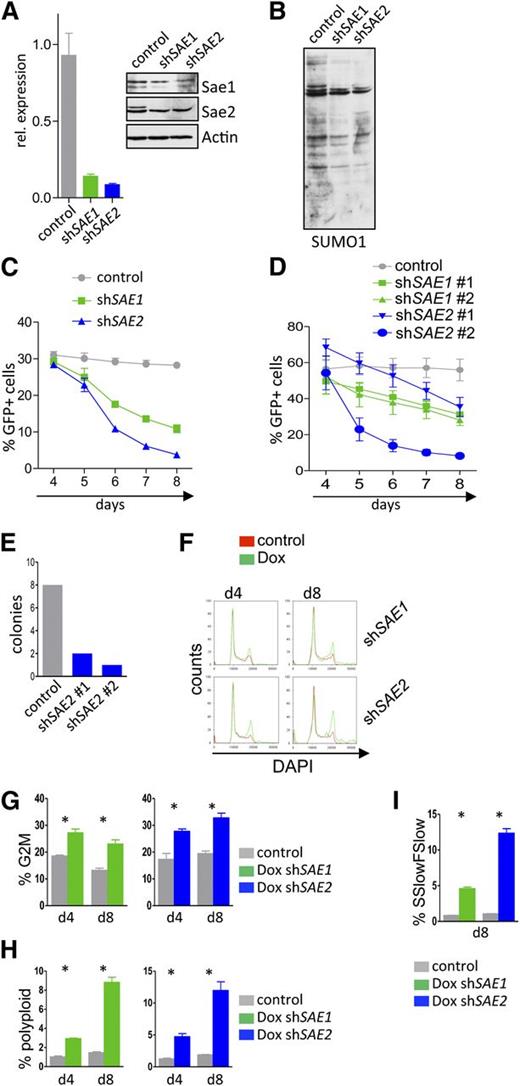
To address whether Myc-driven hyper-SUMOylation has biological significance, we evaluated the consequences of selective knockdown of the E1 SAE1/SAE2 complex, which is necessary for protein SUMOylation. A human BL cell line was stably infected with retroviral Mir30-based shRNA plasmids carrying an eGFP reporter. RNA interference effectively suppressed the messenger RNA and protein levels of both SAE1 and SAE2 (Figure 4A) and reduced overall protein SUMOylation (Figure 4B).
Interfering with the SUMOylation E1 ligase components SAE1 or SAE2 impairs proliferation and provokes G2/M arrest and polyploidy of Myc-induced lymphoma. (A) Human Raji BL cells carrying the ecotropic receptor were infected with retrovirus that expressed shRNAs directed against SAE1 or SAE2 and that also expressed a GFP reporter gene. Following GFP sorting for infected cells by flow cytometry, qPCR analyses (left) of the indicated transcripts and immunoblotting (right) of the indicated proteins were performed. (B) GFP-positive Raji cells from panel A were assessed for SUMO1 expression by immunoblotting. (C) Raji cells from panel A were followed for GFP expression in a competitive repopulation assay at the indicated time points (n = 3). (D) Eµ-Myc lymphoma cells were transduced with lentivirus expressing shRNAs targeting murine Sae1 and Sae2 or a scramble control shRNA. Cells were then followed for GFP positivity in a competitive repopulation assay by flow cytometry at the indicated time points (n = 3). (E) Colony-forming unit assay of GFP-sorted Eµ-Myc cells after knockdown of Sae2. A total of 2 × 104 cells were plated and colony number determined at day 10. (F-I) Human Raji BL cells engineered to express the ecotropic receptor and rtTA were transduced with retroviruses expressing Dox-inducible shRNAs targeting human SAE1 and SAE2 and the dsRed reporter gene, as well as a constitutively expressed GFP reporter. GFP-sorted cells were incubated for the indicated intervals with 1 µg/mL Dox. (F) Flow cytometric cell-cycle analysis measuring DAPI uptake (n = 3). (G) Quantification of polyploid cells (n = 3). (H) Quantification of cells in G2/M. (I) Forward scatter low/side scatter low as a measure of pycnotic cell death is shown (n = 3).
Interfering with the SUMOylation E1 ligase components SAE1 or SAE2 impairs proliferation and provokes G2/M arrest and polyploidy of Myc-induced lymphoma. (A) Human Raji BL cells carrying the ecotropic receptor were infected with retrovirus that expressed shRNAs directed against SAE1 or SAE2 and that also expressed a GFP reporter gene. Following GFP sorting for infected cells by flow cytometry, qPCR analyses (left) of the indicated transcripts and immunoblotting (right) of the indicated proteins were performed. (B) GFP-positive Raji cells from panel A were assessed for SUMO1 expression by immunoblotting. (C) Raji cells from panel A were followed for GFP expression in a competitive repopulation assay at the indicated time points (n = 3). (D) Eµ-Myc lymphoma cells were transduced with lentivirus expressing shRNAs targeting murine Sae1 and Sae2 or a scramble control shRNA. Cells were then followed for GFP positivity in a competitive repopulation assay by flow cytometry at the indicated time points (n = 3). (E) Colony-forming unit assay of GFP-sorted Eµ-Myc cells after knockdown of Sae2. A total of 2 × 104 cells were plated and colony number determined at day 10. (F-I) Human Raji BL cells engineered to express the ecotropic receptor and rtTA were transduced with retroviruses expressing Dox-inducible shRNAs targeting human SAE1 and SAE2 and the dsRed reporter gene, as well as a constitutively expressed GFP reporter. GFP-sorted cells were incubated for the indicated intervals with 1 µg/mL Dox. (F) Flow cytometric cell-cycle analysis measuring DAPI uptake (n = 3). (G) Quantification of polyploid cells (n = 3). (H) Quantification of cells in G2/M. (I) Forward scatter low/side scatter low as a measure of pycnotic cell death is shown (n = 3).
Infected cells were followed for GFP positivity in a competitive repopulation assay, analyzing the effects of silencing SAE1 or SAE2 on BL cell growth and survival. SAE1 or SAE2 knockdown led to a highly significant reduction in BL cell proliferation (Figure 4C), in accord with marked reduction in bromodeoxyuridine incorporation (supplemental Figure 4A). Knockdown of Sae1 or Sae2 also impaired the proliferation of primary Eµ-Myc lymphomas stably infected with lentivirus expressing shRNAs that selectively silence the expression of either Sae1 or Sae2 (Figure 4D and supplemental Figure 4B). Moreover, knockdown of Sae2 abolished the ability of Eµ-Myc lymphoma cells to form colonies in methylcellulose (Figure 4E).
Stable knockdown via RNA interference can mask the roles of proteins through the selection of secondary events. The effects of acute, inducible knockdown of SAE1 and SAE2 were evaluated using a vector that harbors a Venus reporter and that expresses the shRNA under control of a Tet-response element that also induces a dsRed reporter.30 This system thus allows the selection of infected cells and then evaluation of the effects of inducible knockdown. Raji BL cells engineered to express the ecotropic receptor and rtTA were infected with this expression vector and sorted for Venus expression. Treatment of Venus+ cells with Dox efficiently reduced the levels of SAE1 or SAE2 transcripts and proteins (supplemental Figure 4C). Acute knockdown of SAE1 or SAE2 markedly impaired BL cell growth (supplemental Figure 4D), and growth arrest was associated with an accumulation of cells in the G2/M phase, as well as with an accumulation of polyploid cells (Figure 4F-H) and cell death (Figure 4I). Thus, blocking SUMOylation impairs the proliferation of both human and mouse Myc-driven B-cell lymphoma, disrupts the G2/M transition, and leads to aneuploidy.
Pharmacologic inhibition of hyper-SUMOylation triggers synthetic lethality in Myc-driven lymphoma
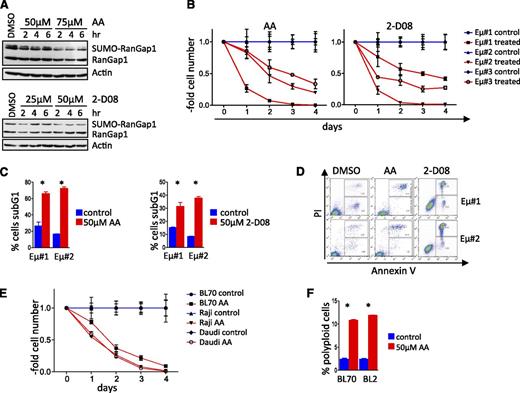
To assess if pharmacologic targeting of SUMOylation has potential therapeutic significance for lymphomas with MYC involvement, we evaluated the in vitro effectiveness of the available SUMOylation inhibitors (SUMOi) anacardic acid (AA) and 2-D08. AA blocks the first step in the SUMOylation process, specifically the formation of the SAE1 and SAE2 complex41 to abolish protein SUMOylation, whereas 2-D08 inhibits SUMOylation of Ube2i and the subsequent transfer of SUMO to target proteins.42 Both compounds reduced SUMOylation of RanGap1, an established SUMOylation target,43,44 in a dose-dependent fashion (Figure 5A). AA or 2-D08 treatment triggered rapid growth arrest of Eµ-Myc lymphomas (Figure 5B), followed by apoptotic cell death (Figure 5C-D). All human BL cell lines tested also underwent growth arrest and apoptosis following AA or 2-D08 treatment (Figure 5E and supplemental Figure 5A-B). Growth arrest was again associated with the induction of polyploidy (Figure 5F). Thus, SUMOylation inhibitors have potent cytostatic and cytotoxic effects on lymphomas with MYC involvement.
SUMOylation inhibitors compromise the growth and survival of Myc-driven lymphoma. (A) AA and 2-D08 are recently identified pharmacologic SUMOi that block SUMOylation of RanGap1. Immunoblot analyses of HEK293T cells treated with AA (top). Immunoblot analyses of Eµ-Myc lymphoma cells treated with 2-D08 (bottom). (B) Three different Eµ-Myc lymphoma cell lines were incubated with 50 µM AA (left) or 75 µM 2-D08 (right) and were counted at the indicated intervals (n = 3). Cell count was normalized to that of untreated control cells for each time point. (C) Cell death (sub-G1 cell fraction determined PI staining) following treatment with AA (left panel) or 2-D08 (right panel) (n = 3). (D) Primary Eµ-Myc lymphoma cells were treated with the indicated dose of AA or 2-D08. Annexin-V/fluorescence-activated cell sorter analyses was used to assess apoptosis. A representative experiment is shown. (E) Human BL cell lines were incubated with 50 µM AA and counted at the intervals indicated. Cell count was normalized to untreated controls for each time point (n = 3). (F) Human BL cell lines were treated with 50 µM AA for 48 hours and assessed by PI flow cytometry for DNA content (n = 3). DMSO, dimethylsulfoxide.
SUMOylation inhibitors compromise the growth and survival of Myc-driven lymphoma. (A) AA and 2-D08 are recently identified pharmacologic SUMOi that block SUMOylation of RanGap1. Immunoblot analyses of HEK293T cells treated with AA (top). Immunoblot analyses of Eµ-Myc lymphoma cells treated with 2-D08 (bottom). (B) Three different Eµ-Myc lymphoma cell lines were incubated with 50 µM AA (left) or 75 µM 2-D08 (right) and were counted at the indicated intervals (n = 3). Cell count was normalized to that of untreated control cells for each time point. (C) Cell death (sub-G1 cell fraction determined PI staining) following treatment with AA (left panel) or 2-D08 (right panel) (n = 3). (D) Primary Eµ-Myc lymphoma cells were treated with the indicated dose of AA or 2-D08. Annexin-V/fluorescence-activated cell sorter analyses was used to assess apoptosis. A representative experiment is shown. (E) Human BL cell lines were incubated with 50 µM AA and counted at the intervals indicated. Cell count was normalized to untreated controls for each time point (n = 3). (F) Human BL cell lines were treated with 50 µM AA for 48 hours and assessed by PI flow cytometry for DNA content (n = 3). DMSO, dimethylsulfoxide.
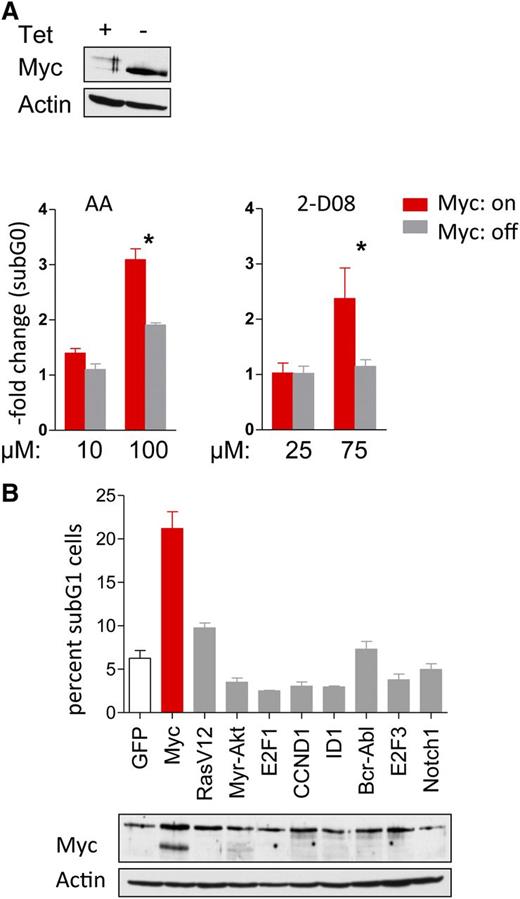
For a cancer drug to move forward to the clinic, there must be a suitable therapeutic window, where there is activity toward the tumor cell but not normal cells or tissues. Using synthetic lethal approaches, one can specifically target cells with certain genotypes/phenotypes to enhance the tumor therapeutic index.17,45 To address whether the effects of SUMO inhibition were Myc specific, P493-6 B lymphoma cells cultured with (Myc-off) or without (Myc-on) Tet were treated with AA and 2-D08. Only cells in the Myc-on state were sensitive to the AA or 2-D08 SUMOylation inhibitors (Figure 6A). To confirm these data, and to test if there is a Myc-specific susceptibility to SUMO inhibition, Rat-1 fibroblasts that stably overexpress a number of well-established oncoproteins (activated Ha-Ras, Akt, E2F1, E2F3, cyclin D1, BCR-ABL, Notch1, and ID1)17,46 were treated with AA. Only Myc-overexpressing fibroblasts were sensitive to the inhibition of SUMOylation (Figure 6B), and this selectivity was independent of the percentages of cells in S phase.17 Thus, there are selective synthetic lethal effects of inhibition of SUMOylation in Myc-driven cancer cells.
Synthetic lethality of Myc overexpression and pharmacologic interference with SUMOylation. (A) P493-6 cells treated with or without tetracycline (Tet) were incubated with the indicated doses of AA and 2-D08 for 24 hours. Cells were stained with PI and DNA content was determined by flow cytometry. The fold change of the sub-G1 fraction as measure for cell death is shown (n = 3). (B) Rat-1 fibroblasts stably infected with indicated oncogenes were incubated with 25 µM AA for 48 hr (top). Cells were PI stained and DNA content analyzed by flow cytometry. The sub-G1 cell fraction is shown (n = 3). Immunoblotting for Myc expression (bottom).
Synthetic lethality of Myc overexpression and pharmacologic interference with SUMOylation. (A) P493-6 cells treated with or without tetracycline (Tet) were incubated with the indicated doses of AA and 2-D08 for 24 hours. Cells were stained with PI and DNA content was determined by flow cytometry. The fold change of the sub-G1 fraction as measure for cell death is shown (n = 3). (B) Rat-1 fibroblasts stably infected with indicated oncogenes were incubated with 25 µM AA for 48 hr (top). Cells were PI stained and DNA content analyzed by flow cytometry. The sub-G1 cell fraction is shown (n = 3). Immunoblotting for Myc expression (bottom).
Interfering with SUMOylation impairs the maintenance of Myc-induced lymphoma
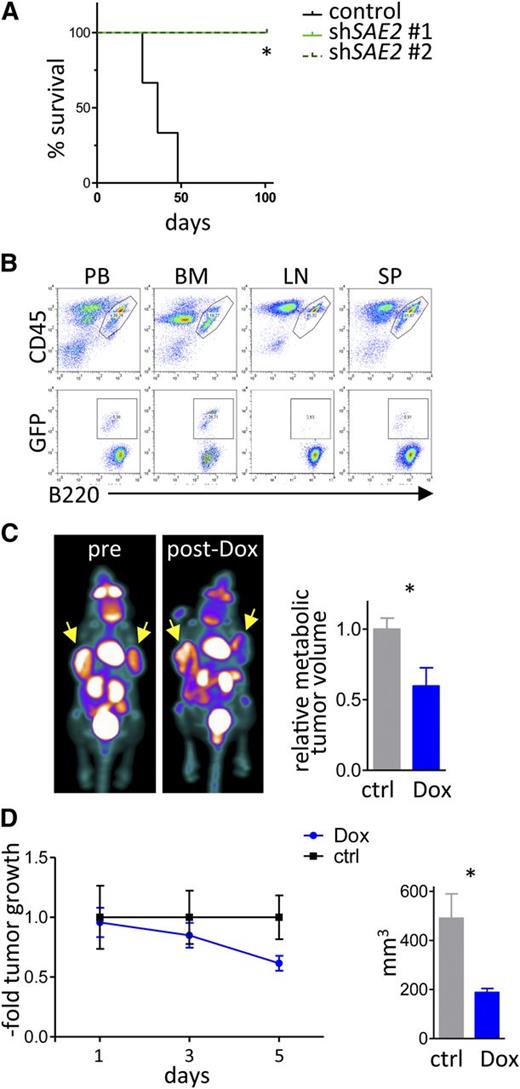
Collectively, these findings suggested that SUMOylation would be necessary for the maintenance of Myc-driven lymphoma. To test this, we assessed the effects of isogenic GFP-positive Eµ-Myc lymphoma cells engineered to express 2 different shRNAs that selectively knock down Sae2 expression vs a control scrambled shRNA. These isogenic lymphomas were transplanted IV (via the tail vein) into syngeneic C57Bl/6 mice, and recipient mice were followed for disease onset. Strikingly, only mice transplanted with the control lymphoma succumbed to disease, whereas those transplanted with lymphomas expressing the Sae2 targeting shRNA did not die within the observation period (Figure 7A). Flow cytometry analyses confirmed that all recipient mice transplanted with lymphomas expressing the control shRNA developed GFP+B220+ tumors in blood, bone marrow, lymph nodes, and spleen, indicating death was due to lymphoma. In contrast, basically no GFP+B220+ lymphoma cells were detected in recipients transplanted with lymphomas expressing Sae2 shRNA (supplemental Figure 6).
Sae2 is required for maintenance of Myc-induced lymphoma in vivo. (A) Eµ-Myc lymphoma cells engineered to express shRNAs targeting Sae2 or a control shRNA as well as GFP were injected (20 000 cells IV) into syngeneic recipients (n = 3 control mice, n = 4 mice for each Sae2 targeting shRNA). Mice were followed for lymphoma onset. Shown is the disease-free survival. (B) After lymphoma onset, mice were euthanized and peripheral blood (PB), bone marrow (BM), lymph nodes (LN), and spleen tissue (SP) were harvested and analyzed for the indicated surface markers using flow cytometry. (C-D) Raji BL cells expressing the ecotropic receptor and rtTA were transduced with a retrovirus expressing Dox-inducible, miR30-based shRNA to silence SAE2 expression. A total of 10 × 106 cells were injected into the flanks of immunocompromised Nod/Scid mice (n = 8). Mice were followed for tumor onset, and a group of 4 mice was treated with Dox to induce the expression of the SAE2 shRNA. (C) FDG-PET imaging of human BL xenografts showing representative images (left) and quantification of metabolic tumor volume by FDG uptake (right) (n = 4 tumors for control, n = 6 tumors from mice treated with Dox). (D) Assessment of xenograft tumor growth (left) and size at end of experiment (right) during a 5-day ±Dox treatment period (n = 6 tumors for control mice, n = 8 tumors from mice treated with Dox).
Sae2 is required for maintenance of Myc-induced lymphoma in vivo. (A) Eµ-Myc lymphoma cells engineered to express shRNAs targeting Sae2 or a control shRNA as well as GFP were injected (20 000 cells IV) into syngeneic recipients (n = 3 control mice, n = 4 mice for each Sae2 targeting shRNA). Mice were followed for lymphoma onset. Shown is the disease-free survival. (B) After lymphoma onset, mice were euthanized and peripheral blood (PB), bone marrow (BM), lymph nodes (LN), and spleen tissue (SP) were harvested and analyzed for the indicated surface markers using flow cytometry. (C-D) Raji BL cells expressing the ecotropic receptor and rtTA were transduced with a retrovirus expressing Dox-inducible, miR30-based shRNA to silence SAE2 expression. A total of 10 × 106 cells were injected into the flanks of immunocompromised Nod/Scid mice (n = 8). Mice were followed for tumor onset, and a group of 4 mice was treated with Dox to induce the expression of the SAE2 shRNA. (C) FDG-PET imaging of human BL xenografts showing representative images (left) and quantification of metabolic tumor volume by FDG uptake (right) (n = 4 tumors for control, n = 6 tumors from mice treated with Dox). (D) Assessment of xenograft tumor growth (left) and size at end of experiment (right) during a 5-day ±Dox treatment period (n = 6 tumors for control mice, n = 8 tumors from mice treated with Dox).
To test if inducible knockdown of SAE2 was sufficient to provoke human B lymphoma regression in vivo, Raji BL cells expressing Dox-inducible SAE2 targeting shRNA were injected into the flank of immune-compromised Nod/Scid mice. After the formation of lymphomas was detectable by FDG-PET, a cohort of these mice were treated with Dox (1 mg/mL in 5% sucrose in their water supply) to induce knockdown of SAE2. Notably, the lymphomas in this cohort showed marked growth reduction following silencing of SAE2, whereas tumors present in recipient mice on sucrose only continued to rapidly expand (Figure 7C-D). Thus, SUMOylation is necessary for maintenance of the malignant state and possibly lymphomagenesis, and SUMO inhibition is an attractive strategy for treating lymphomas with MYC involvement.
Discussion
Myc is known to bind to and stimulate the transcription of a large cast of genes.35 Given this, one is confronted with redundancy when trying to target individual genes or gene products.7,47-49 Nevertheless, Myc target genes with metabolic functions such as Odc31 and ribosomal L2421 are essential for Myc-driven lymphomagenesis. This raises the possibility that some metabolic or signaling nodes exist whose inhibition is synthetic lethal with Myc overexpression. Here, we show that the SUMO pathway may constitute such a node. Our data establish that Myc directly regulates the transcription of many components of the SUMOylation machinery in both B-cell development and in B-cell lymphoma and that high levels of Myc drive a hyper-SUMOylation phenotype in premalignant and neoplastic mouse and human B-cell lymphoma. Further, genetic and pharmacologic approaches revealed that both mouse and human lymphomas with Myc involvement require the hyper-SUMOylation phenotype to maintain the malignant state. Elevated levels of Myc and of SUMOylation connote poor prognosis in various cancers.2,4,14 Importantly, the hyper-SUMOylation phenotype is druggable, and we show that agents that disable SUMOylation are selective for malignancies with MYC involvement.
One striking finding of our analyses is the nearly full coverage of central regulators of SUMOylation as direct Myc target genes, which has previously only been shown for SAE1.50 This observation points to other meaningful links between Myc and SUMOylation, where the concordant expression of c-Myc and SUMO regulators in B-cell subsets suggests developmental roles of the Myc-SUMOylation circuit, which could also include roles in stem cells where Myc plays key roles in self renewal.51,52 While Myc oncoproteins are targets of SUMOylation that bear a highly conserved SUMOylation site, extensive studies of mutant Myc that cannot be SUMOylated did not show any effects on cell growth, cell-cycle control, or apoptosis.53 We conclude that while Myc drives the hyper-SUMOylation phenotype, and thus facilitates its own SUMO modification in a feed-forward loop, the therapeutic outcome of SUMOi is unlikely caused by inhibition of Myc SUMOylation.
Further, because Myc oncogenes are activated in a broad spectrum of human cancers,1,2 and because Myc plays essential roles downstream of other oncogenic drivers,54 targeting the SUMOylation pathway might have a benefit in other highly proliferative or resistant malignancies and/or be important for eradicating cancer-initiating cells that are particularly resistant to therapy. Specifically interesting in this scenario is that MYC translocations or MYC overexpression by other means is associated with poor survival, in particular in DLBCLs that also overexpress BCL2.55,56 For such double-hit DLBCL patients, and also in DLBCL patients with just high MYC levels, SUMOi could be an attractive approach.
Myc coordinates the control of translation via activation of mTOR, and this represents a therapeutic vulnerability for Myc-driven malignancies.5,20 Further, Myc can control aspects of the UPS. For example, Myc activates the E3 ubiquitin ligase SCFSkp2 by upregulating the allosteric regulator Cks1, and this SCF complex directs ubiquitylation and destruction of p27Kip1 to augment cell proliferation.7,8,57 Inhibition of the UPS by bortezomib or second-generation proteasome inhibitors has been highly successful in the clinic in multiple myeloma, in large part due to the acute sensitivity of myeloma cells to proteotoxic stress and the disruption of key survival pathways.58 Our studies show that the regulation of protein SUMOylation by Myc provides an additional level of control of proteins as well as new therapeutic vulnerability. Specifically, our studies suggest that inhibiting SUMOylation is a therapeutic option that should be considered for the treatment of tumors, such as those driven by Myc, that are characterized by a high load of SUMOylated proteins. Furthermore, one prediction is that the synthetic lethality of inhibitors of mTOR could be augmented by adding agents that disable SUMOylation.
Considering the magnitude of hyper-SUMOylation manifest in Myc-activated lymphoma, it is difficult to define a select SUMOylated protein that is the key target within this Myc cancer phenotype. Because defining features of SUMO inhibition are G2/M arrest and polyploidy, one potential candidate is the Aurora family of kinases.59 The function of Aurora kinases relies on their SUMOylation, and treatment of Myc-driven malignancies with Aurora inhibitors, or disrupting their SUMOylation motifs, mimics the effects of inhibiting SUMOylation, including defects in mitosis, polyploidy, and Myc-dependent synthetic lethal interactions.17,18,60,61 It is, however, likely that the effectiveness of SUMO inhibition is due to effects on a broader cast of targets necessary for tumor growth and survival. Given that direct targeting of Myc transcription functions is difficult, the data establish that inhibition of SUMOylation is an attractive means to selectively disable Myc-driven cancer, where treatment of Myc-induced lymphomas with 2 SUMOylation inhibitors that block different stages of SUMOylation41,42 compromise the maintenance of lymphoma. Although the potency of the AA and 2-D08 agents is an issue, because the 50% inhibition/inhibitory concentration values are in the low-µM concentration, the fact that both drugs act on different targets in the SUMOylation pathway and effectively block SUMOylation in our system supports the notion that one can effectively target this pathway for cancer therapy.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
Scott Lowe and Cornelius Miething (Memorial Sloan Kettering Cancer Center, NY) provided the TRMPV construct and RIEP-transfected cell lines. This work benefitted from data assembled by the ImmGen consortium.39 Jolanta Slawska and Viktoriya Tomiatti helped with cell injection and PET imaging.
This work was supported by the Deutsche Forschungsgemeinschaft (SFB824, KE 222/7-1) (U.K.), Deutsche Jose Carreras Leukämie Stiftung (R11/18) (U.K.), a Leukemia Lymphoma Society Scholar Award (A.G.), National Institutes of Health, National Cancer Institute grant CA100603 (J.L.C.), monies from the State of Florida to Scripps Florida (J.L.C.), the Swedish Cancer Society (J.A.N.), BioCARE (J.A.N.), the Swedish Research Council (J.A.N.), and the German Cancer Consortium (DKTK; U.K. and C.P.).
Authorship
Contribution: A.H. and U.K. initiated the research and planned the experiments; A.H., S. Schoeffmann, S. Steidle, F.X.S., L.A.N., M.R., and I.L., performed the experiments; M.F. performed the bioinformatics analyses; A.G. provided crucial reagents; A.H., C.P., J.A.N., J.L.C., and U.K. wrote the manuscript; and all authors reviewed the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for J.L.C. and F.X.S. is Department of Tumor Biology, The Moffitt Cancer Center & Research Institute, Tampa, FL.
Correspondence: Ulrich Keller, Ismaninger Str 15, Munich 81675, Germany; e-mail: ulrich.keller@lrz.tum.de.