Key Points
In ET, a CALR mutation correlates with a monoclonal X chromosome inactivation pattern, which differs from JAK2V617F mutant disease.
The presence of a CALR mutant is associated with suppression of wild-type myelopoiesis.
Abstract
Calreticulin mutations (CALRMUT) are found in a significant proportion of patients with essential thrombocythemia (ET) lacking JAK2V617F or MPL mutations. They are associated with substantially different hematological and clinical features and define a distinct subtype of ET. We show here that their presence is significantly correlated with a clonal X chromosome inactivation pattern (XCIP). Of 105 female ET patients investigated, 61 had an interpretable XCIP, and a clonal pattern was observed in 88% of CALRMUT patients compared with 26% of JAK2V617F (P = .0002) and 9% of JAK2V617F/MPL/CALR wild-type patients (P < .0001). Neutrophil CALRMUT level was significantly higher than JAK2V617F level (median, 50% vs 18%; P < .0001), and wild-type myelopoiesis was suppressed in CALRMUT but not JAK2V617F patients. These data are suggestive of truly monoclonal hematopoiesis in CALRMUT patients and provide further evidence that the biology associated with CALR mutations is markedly different from that of JAK2V617F mutations.
Introduction
Discriminating between a reactive thrombocytosis and the myeloproliferative neoplasm essential thrombocythemia (ET) can be a diagnostic challenge. In the absence of known clonal biomarkers, several groups, including our own, investigated clonality in female patients using X chromosome inactivation patterns (XCIPs) to distinguish between them on the premise that ET was a clonal disorder.1-4 However, these studies demonstrated considerable biological heterogeneity in ET patients, with 3 broad XCIP categories observed once constitutional and age-related skewing had been taken into account: monoclonal, oligoclonal, or polyclonal. Identification of JAK2V617F mutations provided the first molecular biomarker in ET, but this also led to a biological conundrum as their presence did not appear to relate to clonality status, with all 3 patterns observed in mutant-positive patients.5-8 Subsequent studies reported that the relative JAK2V617F mutant allele burden is generally lower than would be expected for a mutation driving clonal proliferation,8-13 and therefore in many mutant-positive patients, a significant proportion of the platelets are mutant negative and polyclonal in origin. Serial analysis of neutrophil samples has shown that JAK2V617F-mutated clones can be maintained as a stable subpopulation for many years, even in the absence of treatment.10-13 Furthermore, we demonstrated that in JAK2V617F cases, the observed neutrophil XCIP was very similar to that expected when the mutant level and constitutional (T-cell) XCIP were taken into account.10 Of note, in all these studies, a significant proportion of ET patients with clonal XCIPs were JAK2 wild type, implying the presence of other mutations leading to clonal expansion. MPL exon 10 mutations were later detected, but only account for 3% to 9% of ET patients lacking JAK2V617F mutations.14,15
Recently, calreticulin mutations (CALRMUT) have been reported in 49% to 71% of ET patients lacking JAK2V617F or MPL mutations that define a distinct subtype of ET with a substantially different hematological phenotype and clinical outcome from JAK2V617F-mutated patients.16-19 However, clonality status in these patients has not been reported. We therefore screened a cohort of 105 female patients with ET and known JAK2 and MPL genotype for CALR mutations and correlated the results with XCIPs in the 61 with interpretable patterns.
Study design
The median age at the first test for the 105 female patients investigated was 50 years (range, 10-92 years). Peripheral blood samples were collected between 1996 and 2010. The diagnosis was made according to the criteria for ET at the time of the first investigation (amended Polycythemia Vera Study Group or revised World Health Organization criteria). The studies were approved by the London Multi-centre Research Ethics Committee. Patient consent was obtained according to the Declaration of Helsinki.
XCIPs of purified neutrophil and CD3+ T-cell DNA were investigated using the human androgen receptor assay.10 As established in our previous studies, a clonal XCIP required skewed neutrophil X allele expression of >75% of one X allele, >20% difference between X allele expression in neutrophils and T cells (reflecting the hematopoietic stem cell pattern), and age ≤ 65 years at the time of test.20 Neutrophil JAK2V617F genotype and mutant allele level were determined as previously reported.10 Neutrophil DNA was screened for mutations in MPL exon 10 (primers 5′-GTGGGCCGAAGTCTGACCCT-3′, 5′-CGCTCTGTGACCCCAGATCTC-3′) and CALR exon 9 (primers 5′-GCCTGGTCCTGGTCCTGATGT-3′, 5′-AGGAGCGCTCAGGCCTCAGTC-3′) by heteroduplex analysis on the WAVE DNA Fragment Analysis System (Transgenomic Ltd., Glasgow, United Kingdom [UK]). Samples with abnormal chromatograms were directly sequenced. The relative CALR mutant level was quantified using the same polymerase chain reaction with a fluorescently labeled forward primer and analysis on the Beckman Coulter CEQ8000 Genetic Analysis System (Beckman Coulter, Fullerton, CA). Mutant level was expressed as a relative proportion of total CALR alleles.
Results and discussion
CALR mutations were detected in 26 patients (25%): 9 (35%) were type 1 (p.L367Tfs*46), 11 (42%) were type 2 (p.K385Nfs*47), and 6 (23%) had other mutations. In all cases except 1, the predicted mutant would give rise to the common novel C-terminal peptide.16-18 The remaining patient had 2 mutations on the same allele (c.1137_1138insAG and 1227_1231del; p.E380Rfs*38) that replaced the final 20 amino acids of the mutant peptide with an alternative 7 amino acid sequence. In keeping with data reported by others,16-19 CALRMUT patients were significantly younger at diagnosis (P = .0002) and had significantly higher platelet counts than JAK2V617F patients (n = 43; P = .001; Table 1). There was no difference in age or platelet count at diagnosis between CALRMUT and the JAK2/MPL/CALR triple-negative patients (n = 34; Table 1). Two patients (2%) were MPLMUT and were excluded from further analysis. Cytogenetics were available from 36 patients (19 JAK2V617F, 9 CALRMUT, and 8 triple negative); all were normal except for 1 JAK2V617F-mutated patient with trisomy chromosome 8 and chromosome 9.
Characteristics of the CALRMUT patients studied compared with JAK2V617F and triple-negative patients
| Parameter . | Total . | JAK2V617F . | CALRMUT . | Triple negative . | P value for CALRMUT vs JAK2V617F . | P value for CALRMUT vs triple negative . |
|---|---|---|---|---|---|---|
| No. (% of cohort) | 105* | 43 (41%) | 26 (25%) | 34 (33%) | ||
| Age at diagnosis, years, median (range)† | 47 (10-84) | 54 (25-84) | 31 (10-71) | 40 (14-77) | .0002 | .2 |
| Platelet count at diagnosis, ×109/L, median (range)‡ | 807 (456-4875) | 800 (523-1140) | 1169 (500-4875) | 721 (456-2800) | .001 | .1 |
| Age at first test, years, median (range) | 50 (10-92) | 61 (25-92) | 37 (10-89) | 49 (16-89) | .003 | .8 |
| Months from diagnosis at first test, median (range)† | 18 (0-297) | 20 (0-217) | 40 (0-175) | 12 (0-297) | .5 | .3 |
| % Mutant, median (range) | 18% (5-100%)§ | 50% (22-56%) | <.0001 | |||
| % Mutant at or within 3 mo of diagnosis, median (range) | 17% (10-42%) | 46% (27-56%) | .0001 | |||
| XCIP status, clonal:polyclonal (%) | 22:39 (36%:64%) | 6:17 (26%:74%) | 14:2 (88%:12%) | 2:20 (9%:91%) | .0002¶ | <.0001¶ |
| Parameter . | Total . | JAK2V617F . | CALRMUT . | Triple negative . | P value for CALRMUT vs JAK2V617F . | P value for CALRMUT vs triple negative . |
|---|---|---|---|---|---|---|
| No. (% of cohort) | 105* | 43 (41%) | 26 (25%) | 34 (33%) | ||
| Age at diagnosis, years, median (range)† | 47 (10-84) | 54 (25-84) | 31 (10-71) | 40 (14-77) | .0002 | .2 |
| Platelet count at diagnosis, ×109/L, median (range)‡ | 807 (456-4875) | 800 (523-1140) | 1169 (500-4875) | 721 (456-2800) | .001 | .1 |
| Age at first test, years, median (range) | 50 (10-92) | 61 (25-92) | 37 (10-89) | 49 (16-89) | .003 | .8 |
| Months from diagnosis at first test, median (range)† | 18 (0-297) | 20 (0-217) | 40 (0-175) | 12 (0-297) | .5 | .3 |
| % Mutant, median (range) | 18% (5-100%)§ | 50% (22-56%) | <.0001 | |||
| % Mutant at or within 3 mo of diagnosis, median (range) | 17% (10-42%) | 46% (27-56%) | .0001 | |||
| XCIP status, clonal:polyclonal (%) | 22:39 (36%:64%) | 6:17 (26%:74%) | 14:2 (88%:12%) | 2:20 (9%:91%) | .0002¶ | <.0001¶ |
P values are the Student unpaired t test unless otherwise stated.
Two patients were MPLMUT.
Date of diagnosis was unknown in 10 patients: 2 JAK2V617F, 3 CALRMUT, and 5 triple negative.
Platelet count at diagnosis was unknown in 13 patients: 4 JAK2V617F, 3 CALRMUT, and 6 triple negative.
The 1 patient with a mutant level >50% had post-ET myelofibrosis at the time of analysis.
Two-sided Fisher's exact test.
XCIP analysis could be interpreted in 61 of the 105 female patients (58%): 16 CALRMUT, 23 JAK2V617F, and 22 triple negative. Excluded patients either lacked a polymorphic marker (n = 6), had constitutional or age-related skewing (n = 36), or were MPLMUT (n = 2). There was a strong correlation between a clonal XCIP and presence of a CALR mutation: 14 (88%) of CALRMUT patients were clonal, 5 of 6 interpretable cases with a type 1 mutation, and 6 of 7 cases with a type 2 mutation. This was significantly different from JAK2V617F (26%; P = .0002) and triple-negative patients (9%; P < .0001; Table 1). The difference was not related to the length of time from diagnosis at first test, which did not differ between JAK2V617F and CALRMUT patients (P = .5). The CALR mutant level for all patients was significantly higher than the JAK2V617F level, both in samples available at or within 3 months of diagnosis (median, 46% vs 17%; P < .0001) and overall (median, 50% vs 18%; P < .0001; Table 1). No change in mutant level was observed in serial samples from 9 CALRMUT patients over 3 to 116 months of follow-up. The median difference was 1% (range, 0-15%), irrespective of treatment. Unlike the situation in JAK2V617F cases, this is therefore consistent with the presence of a heterozygous mutation in the majority of cells. This is supported by the XCIP data where 12 of the 14 clonal CALRMUT patients had monoclonal hematopoiesis with ≥95% expression of 1 allele. One of the 2 CALRMUT patients with a polyclonal XCIP had a lower mutant level of 30% with a borderline constitutively skewed XCIP (85%:15% expression in neutrophils, 72%:28% in T cells), and the observed neutrophil XCIP was only 4% different from the expected pattern of 89%:11% if the mutant-positive clone expressed the predominant X allele. This could not explain the polyclonal pattern in the remaining CALRMUT patient (42%:58% expression in neutrophils, 50%:50% in T cells; mutant level 36%). It is possible that this case is truly biclonal, as reported in a proportion of myeloproliferative neoplasm patients,21,22 with 2 major clones expressing different X alleles.
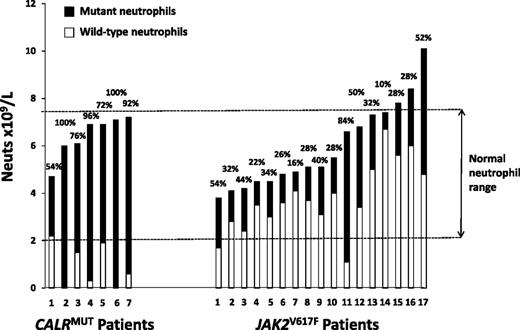
CALRMUT patients have significantly lower white cell counts than JAK2V617F patients,16,18,19 and immunohistochemistry studies have suggested that mutant CALR predominantly affects just the megakaryocytic lineage, with little/no expression in mature myeloid cells.23 To further investigate the difference in myelopoiesis, we used the mutant allele level (assuming heterozygosity) to calculate the absolute number of mutant and wild-type neutrophils in 24 mutant-positive patients tested prior to receiving any cytoreductive therapy. A CALR wild-type neutrophil count in the normal range for our institution (2.0-7.5 × 109/L) was observed in only 1 of 7 CALRMUT patients compared with 15 of 17 JAK2V617F patients (P = .001; Figure 1), which is indicative of suppression of wild-type myeloid cells at the hematopoietic stem cell stage in the CALRMUT but not JAK2V617F patients.
Absolute number and relative proportion of wild-type and mutant neutrophils in CALRMUT and JAK2V617F ET patients tested prior to receiving any cytoreductive therapy. Each bar represents an individual patient and shows the total neutrophil count at the time of testing, as well as the derived absolute number of mutant-positive and wild-type neutrophils. The latter are calculated from the relative proportion of mutant-positive neutrophils, as given above the bar, and assumes that mutant-positive cells were heterozygous for the mutation (ie, the proportion of mutated cells is double the percentage of mutant alleles).
Absolute number and relative proportion of wild-type and mutant neutrophils in CALRMUT and JAK2V617F ET patients tested prior to receiving any cytoreductive therapy. Each bar represents an individual patient and shows the total neutrophil count at the time of testing, as well as the derived absolute number of mutant-positive and wild-type neutrophils. The latter are calculated from the relative proportion of mutant-positive neutrophils, as given above the bar, and assumes that mutant-positive cells were heterozygous for the mutation (ie, the proportion of mutated cells is double the percentage of mutant alleles).
These results provide further evidence of the impact of CALR mutations on the hematopoietic stem cell with expansion of a dominant mutant-carrying clone, which leads to a distinct subtype of ET with a markedly different biology from that associated with JAK2V617F mutations.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by Leukaemia and Lymphoma Research, UK, and the UK Medical Research Council. The work was undertaken at University College London (UCL) Hospitals/UCL, which received a proportion of funding from the Department of Health's National Institute for Health Research Biomedical Research Centres funding scheme.
Authorship
Contribution: R.E.G. and D.C.L. designed the study; C.A., J.R.L., and R.E.G. performed experimental work and analyzed the data; and R.E.G. and D.C.L. wrote the manuscript, which was reviewed by all authors.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Rosemary E. Gale, Department of Haematology, UCL Cancer Institute, Paul O’Gorman Bldg, 72 Huntley St, London WC1E 6DD, UK; e-mail: rosemary.gale@ucl.ac.uk.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal