In this issue of Blood, Ostronoff et al report a low remission rate in acute myeloid leukemia (AML) patients coexpressing FLT3/ITD and cryptic translocation t(5;11)(q35;p15.5), known as NUP98/NSD1.1
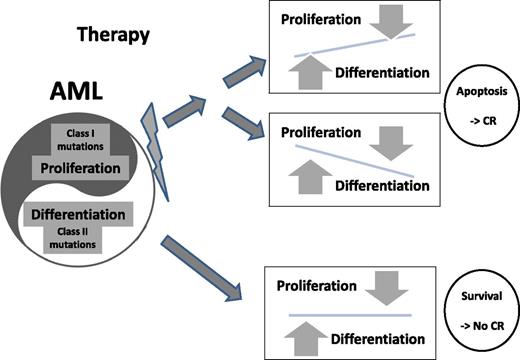
AML cells maintain a pathological homeostasis between aberrant proliferation and differentiation signals. When chemotherapy is administered, multiple signal pathways are affected. Apoptosis and, as a result, desired clinical response are induced in cases where the above-mentioned balance is impaired in either direction (upper panels). In cases illustrated in the lower panel, both signals are derived from powerful genetic aberrations and balance is maintained despite chemotherapy-induced stress resulting in the survival of the leukemic cells.
AML cells maintain a pathological homeostasis between aberrant proliferation and differentiation signals. When chemotherapy is administered, multiple signal pathways are affected. Apoptosis and, as a result, desired clinical response are induced in cases where the above-mentioned balance is impaired in either direction (upper panels). In cases illustrated in the lower panel, both signals are derived from powerful genetic aberrations and balance is maintained despite chemotherapy-induced stress resulting in the survival of the leukemic cells.
NUP98/NSD1 is more prevalent among pediatric patients2 associated with a normal karyotype and/or mutated FLT3/ITD and was earlier reported as a predictor of a poor prognosis.3 Ostronoff et al screened 1421 patients enrolled in 5 Children's Oncology Group (COG)/Children's Cancer Group (CCG) and Southwest Oncology Group (SWOG) studies. Focusing on patients presenting with a normal karyotype and FLT3/ITD, the authors provide convincing evidence that conventional induction is much less effective in patients expressing both NUP98/NSD1 and FLT3/ITD than in any other patient subpopulation. In their mostly pediatric cohort, the overall prevalence of NUP98/NSD1 was 3% (45/1421), but it was as high as 15% in patients presenting with FLT3/ITD. It is therefore worth questioning whether screening for NUP98/NSD1 fusion transcript in AML patients presenting with a normal karyotype and FLT3/ITD should be mandatory, particularly in pediatric population.
Recent progress in gene sequencing allows rapid coverage of a wide spectrum of genes at an affordable cost. In the very near future, comprehensive genetic data could become available at least for most AML patients diagnosed in developed countries. However, the capacity to identify all genetic aberrations at the DNA level, describe and quantify gene expression at the RNA level, and even analyze epigenetic profile, has not been widely adopted by clinicians. Unfortunately, the availability of such an enormous volume of data has not yet changed treatment algorithms for most AML patients. Several groups collected genetic data from a large number of AML patients, mostly those treated according to a uniform protocol, differentiated them based on their prognosis into genetically distinguishable groups and created a generic prediction model. Due to statistical constraints, in most models, the number of analyzed genetic parameters is limited to those that are most prevalent or considered to have the strongest clinical influence, and thus, most of available genetic data are excluded. Although several such models have been reported,4 none of them includes rare genetic aberrations such as NUP98/NSD1. Most models yield comprehensive risk stratification, yet they are mainly designed to support patient selection for allogeneic hematopoietic stem cell transplantation and neglect a preferred induction approach.
Ostronoff et al report that, within the group of patients diagnosed with normal karyotype AML, those presenting with both FLT3/ITD and NUP98/NSD1, FLT3/ITD alone, and NUP98/NSD1 alone achieved a complete remission rate of 37%, 67%, and 86%, respectively. Although based on data of only 11 patients, in cases where WT1 mutation was identified in addition to the former 2 aberrations, the complete response rate dropped to as low as 9%. Such a significant bedside finding should be taken back to the bench with an aim to explore mechanisms of interaction between these genes that antagonize chemotherapy effect. Understanding the biological mechanism underlying this exceptional synergistic effect is of particular interest because these 2 aberrations are not known to act within the same signaling pathway.
Twelve years ago, Gilliland and Griffin suggested that the FLT3/ITD mutation, whereas providing a strong proliferative signal, is not sufficient for leukemia initiation.5 According to this well-appreciated concept, mutations of 2 different classes, one that enhances proliferation and the other that impairs differentiation, are required for leukemia initiation. The model by Gilliland and Griffin highlights a fundamental take-home message in the biology of leukemia, ie, the significance of interaction between different genes. FLT3/ITD may coexist with many other mutations but only few specific gene pairs including the FLT3/ITD and NUP98/NSD1 combination are powerful enough to initiate leukemia.6
If, as suggested by Gilliland and Griffin, in all AML cells an aberrant proliferative signal is present along with impairment in differentiation process, it is interesting to decipher what makes cells harboring the FLT3/ITD and NUP98/NDS1 combination so resistant to chemotherapy. FLT3/ITD, is a strong driver for proliferation and beyond conventional cytogenetics, is the most common genetic aberration of definitive clinical significance in AML. The second player in this scene, NUP98/NDS1, is a fusion between a regulator of protein and RNA nucleo-cytoplasmic transport (NUP98) and histone methyltransferase (NDS1). In various hematological malignancies, 30 different genes including NSD1 have been identified to partner NUP98 for the creation of an oncogenic fusion transcript.7,8 NUP98 aberrant transcripts were shown to significantly impair differentiation, resulting in a variable biological effect correlating with the specific NUP98 partners. Patient specific prognosis is therefore determined by the level of biological interaction between fusion partners on the one side and the collaborative proliferation signal on the other.
Ostronoff et al also screened 80 acute promyelocytic leukemia (APL) patients: 30 (37.5%) were FLT3/ITD positive but none expressed NUP98/NSD1. Given that the presence of FLT3/ITD does not hamper prognosis of APL patients treated with therapy aimed to overcome differentiation blockade,9 the strength of the second hit seems to be a major component that differentiates APL from other types of AML. Following this line of thought, it may be suggested that in AML cells, apoptosis is induced by therapy, if chemotherapy significantly alters the inner cell balance between proliferation and differentiation signals (see figure). Coexpression of 2 powerful drivers such as FLT3/ITD and NUP98/NDS1 creates a stable pathological balance that drives leukemia forward but is very difficult to disrupt by regular chemotherapies.
One conclusion from the study by Ostronoff et al is that AML patients coexpressing FLT3/ITD and NUP98/NDS1 may require alternative therapies. In vitro data suggest that FLT3 inhibitors may be an option.6 Unfortunately, no clinical evidence favoring specific agents exist for patients presenting with both FLT3/ITD and NUP98/NSD1. In addition, because NUP98 can partner multiple other genes yielding oncogenic signals, it is worth exploring the clinical effect of other NUP98-related fusion transcripts. It might also be suggested that characterization of the interaction between proliferation and differentiation signals as a whole (may be by RNA expression patterns) may help progressing toward personalizing clinical decisions in AML. Leukemia patients presenting with a combination of a strong proliferation driver mutation and a dominant differentiation blocker are likely to require novel specific therapies.
Conflict-of-interest disclosure: The author declares no competing financial interests.