Key Points
Besides clustering, platelet factor 4/polyanion complexes require input of energy to become immunogenic.
Minute differences in chain length determine the induction of antigenicity of PF4.
Abstract
The chemokine platelet factor 4 (PF4) undergoes conformational changes when complexing with polyanions. This can induce the antibody-mediated adverse drug effect of heparin-induced thrombocytopenia (HIT). Understanding why the endogenous protein PF4 becomes immunogenic when complexing with heparin is important for the development of other negatively charged drugs and may also hint toward more general mechanisms underlying the induction of autoantibodies to other proteins. By circular dichroism spectroscopy, atomic force microscopy, and isothermal titration calorimetry we characterized the interaction of PF4 with unfractionated heparin (UFH), its 16-, 8-, and 6-mer subfractions, low-molecular-weight heparin (LMWH), and the pentasaccharide fondaparinux. To bind anti-PF4/heparin antibodies, PF4/heparin complexes require (1) an increase in PF4 antiparallel β-sheets exceeding ∼30% (achieved by UFH, LMWH, 16-, 8-, 6-mer), (2) formation of multimolecular complexes (UFH, 16-, 8-mer), and (3) energy (needed for a conformational change), which is released by binding of ≥11-mer heparins to PF4, but not by smaller heparins. These findings may help to synthesize safer heparins. Beyond PF4 and HIT, the methods applied in the current study may be relevant to unravel mechanisms making other endogenous proteins more vulnerable to undergo conformational changes with little energy requirement (eg, point mutations and post-translational modifications) and thereby predisposing them to become immunogenic.
Introduction
Heparin, widely used clinically as a parenteral anticoagulant,1-3 is a polyanion consisting of iduronic acid, glucuronic acid, and glucosamine residues carrying sulfate groups. Pharmaceutical heparin is obtained from porcine gut mucosa and, as biological material, is composed of polysaccharide chains with variable length. Degradation of unfractionated heparin (UFH) results in less polydisperse and smaller low-molecular-weight heparins (LMWHs). The pentasaccharide fondaparinux consists of the shortest sequence able to catalyze the activity of antithrombin. It was the first synthesized heparin approved for clinical use.4 Currently, several other heparin-based, synthesized polysaccharides are in preclinical development.5,6 Beside their anticoagulant activity, heparins have other biological effects including potential antitumor activity,7 which, however, differs depending on the chain length.7
Clinically, beside bleeding, heparin-induced thrombocytopenia (HIT) is the most important adverse effect of heparin.8 HIT is a life-threatening immune-driven adverse effect, which occurs in up to 3% of patients receiving UFH after major surgery.9 HIT is caused by antibodies that recognize platelet factor 4 (PF4), a CXC chemokine family protein, in ultralarge multimolecular complexes with heparin.10 Several of these pathogenic antibodies can bind to the multimolecular complexes of PF4 and heparin, forming immunocomplexes. When these PF4/heparin-immunoglobulin (Ig)G immunocomplexes bind to platelets, the Fc parts of the antibodies cross-link FcγIIa receptors on platelets, which induces platelet activation and aggregation.11 This results in a prothrombotic state and an increased risk for new thrombosis.12 Heparin-induced antibodies recognize an antigen exposed on PF4 at a certain PF4/heparin ratio,10 at which PF4 tetramers13 are forced into close approximation,14 accompanied by charge neutralization.14-16
Previously, we have shown, using circular dichroism (CD) spectroscopy in combination with an enzyme-linked immunosorbent assay (ELISA), that the antigenic site is a composite surface formed by ≥2 (or 3) PF4 monomers in a PF4 tetramer. Exposure of this antigenic site occurs when polyanions induce changes in the structure of PF4, resulting in an increase of the antiparallel β-sheet content in the PF4 secondary structure to more than ∼30%.17
Here we report the physicochemical characterization of complexes formed between PF4 and UFH, LMWH, or subfractions produced from unfractionated heparin with defined chain lengths (16- [HO16], 8- [HO08], and 6- [HO06] mer), as well as with synthetic fondaparinux (5-mer).4 We found that beside the conformational change of PF4 exposing >30% antiparallel β-sheets and formation of large PF4/heparin complexes, the enthalpy of binding (released heat) has to exceed a threshold value to provide the energy for the conformational change of PF4 required to expose the antigenic epitope. These findings may provide relevant aspects to understand the structure-function relationship for other biological functions of heparin derived drugs18 and may also underlie mechanisms making other endogenous proteins immunogenic.
Methods
Ethics
The use of human sera containing anti-PF4/heparin antibodies and human platelets and obtaining whole blood from healthy volunteers was approved by the Greifswald Ethics Board.
Reagents
We used the following reagents: lyophilized human PF4 isolated from platelets, Chromatec (Greifswald, Germany); UFH Heparin-Natrium-25000 (Ratiopharm, Ulm, Germany), fondaparinux (Arixtra; GlaxoSmithKline, London, UK); LMWH reviparin (Clivarin 1750; Abbott GmbH, Wiesbaden, Germany), hospital pharmacy; heparin oligosaccharides 6-mer (HO06), 8-mer (HO08) and 16-mer (HO16): Iduron Ltd. (Manchester, UK). These heparin fractions are obtained by partial heparin lyase digestion followed by high-resolution gel filtration and show a defined length as determined by the manufacturer. However, their interaction with antithrombin (ie, their anticoagulant capacity) is not defined. The smaller the oligosaccharides become, the more disruption on the pentasaccharide structure that is required for antithrombin binding to occur.
ELISA
PF4/heparin ELISA was performed as described19 with 3 samples of human sera of patients known to contain anti-PF4/heparin IgG antibodies verified by PF4/heparin ELISA and the heparin-induced platelet activation test.19 UFH, LMWH, and the defined length heparins were added in rising concentrations to PF4 to form the complexes before they were coated on a microtiter plate (as indicated in the figures; UFH: 0-14 µg/mL, HO16 and HO08: 0-11.7 µg/mL, HO06: 0-15 µg/mL, fondaparinux: 0-30 µg/mL). To verify the reactivity pattern, we then coated the PF4/defined length heparin complexes at the optimal concentration determined by the titration experiment and tested them with an additional panel of 14 samples of characterized sera from patients with serologically confirmed HIT. LMWH was assessed by the same method.17
CD spectroscopy
Changes in the secondary structure of PF4 on interaction with heparins were studied by recording far-UV CD spectra (200-260 nm) using a Chirascan CD spectrometer (Applied Photophysics, Leatherhead, UK) as previously described.17 PF4 was dissolved in phosphate-buffered saline (PBS; 155 mM NaCl, 1.54 mM KH2PO4, 2.71 mM Na2HPO4-7 H2O, pH 7.2) to a final concentration of 80 µg/mL (2.5 µM). Complex formation was carried out at 20°C directly in the CD cuvette (Hellma, Müllheim, Germany) with a 5-mm path length. Each measurement started with a pure PF4 solution. Subsequently, increasing amounts of the heparins (UFH, HO16, HO08, HO06, and fondaparinux) were added to the cuvette, and CD spectra were recorded for each PF4/heparin ratio. Buffer baselines and baselines of each heparin concentration step (ie, without PF4 in the solution) were recorded. In the data analysis, the spectra of PF4 alone and of PF4/heparin complexes were corrected for the baselines, path length, number of amino acids, and concentration to obtain the wavelength-dependent mean residue delta epsilon values of the PF4/heparin complex. To estimate the secondary structure content of PF4, deconvolution of CD spectra was carried out with CDNN software using a database of 33 reference proteins.20 LMWH was assessed by the same method.17
Atomic force microscopy
To characterize the structural features of the PF4/heparin complexes, atomic force microscopy (AFM) imaging in liquid was carried out using a BioScope II scanning probe microscope from Digital Instruments (Santa Barbara, CA). NanoScope 7.3 software was used to control the AFM, to set the imaging parameters, and for flattening of the images. Preformed PF4/heparin complexes (at the optimal ratios found by isothermal titration calorimetry [ITC]) were incubated on freshly cleaved mica for 10 seconds, followed by washing the mica surface with ultrapure water. For imaging, a few drops of ultrapure water were added on the mica with adsorbed samples. All samples were imaged in tapping mode at room temperature using silicon nitride cantilevers DNP-S (Veeco, Camarillo, CA) with a drive frequency of 8 to 14 kHz in water and a nominal curvature radius of 10 nm. AFM images of the PF4/heparin complexes and of their constituents were recorded. All experiments were repeated ≥3 times.
The height distribution of the AFM-imaged features was analyzed using a MatLab script (MathWorks, Natick, MA).21,22 Briefly, the script scans over the images identifying the highest points within a moving 17 × 17 rectangle (supplemental Figure 1), giving the position and maximum height of the samples adsorbed to mica. To avoid cross-talk with measurement noise, a 0.9-nm threshold (the standard deviation of the background noise in the AFM images) was used in this study. Height values were then merged and plotted into a semilogarithmic point histogram. As the typically observed height values were on the order of the tip curvature radius, tip convolution led to a strong lateral broadening of the observed structures. We therefore did not further analyze the lateral dimensions of the detected structures.
ITC
The different heparins and PF4 were separately dialyzed against PBS buffer at pH 7.4. ITC measurements were carried out using an iTC200 calorimeter (GE Healthcare Life Sciences). A PF4 solution (500-2470 µg/mL [15.6-70 µM] in PBS) was added to the sample cell, and a solution of heparin (450-900 µg/mL [37-500 µM]) was loaded into the injection syringe. For each experiment, a 60-second delay was followed by 19 injections of 1 µL of the titrant solution, spaced 240 seconds apart. The sample cell was stirred at 1000 rpm throughout and maintained at 25°C. Control titration was performed by injecting heparin into PBS buffer and subtracted prior to data analysis. The area under each peak of the resulting heat profile was integrated, normalized by the concentrations, and plotted against the molar ratio of heparin to PF4 using an Origin script supplied with the instrument (Origin 7; OriginLab Corporation). The resulting binding isotherms were fitted by nonlinear regression using the single-site model. The stoichiometry of the interaction (n = cHeparin/cPF4, where c is the concentration in moles per liter), the equilibrium constant (KA), and the change in enthalpy (∆H) were obtained during the fitting of all titration data. Equilibrium dissociation constants (KD) were calculated as the reciprocal of KA. The Gibbs free energy change (ΔG) was calculated with the equation ΔG = −RT ln KA. All titrations were replicated to determine the experimental standard deviation for each parameter.
Results
Changes in the PF4 secondary structure by UFH, LMWH, and defined length heparins and binding of anti-PF4/heparin antibodies
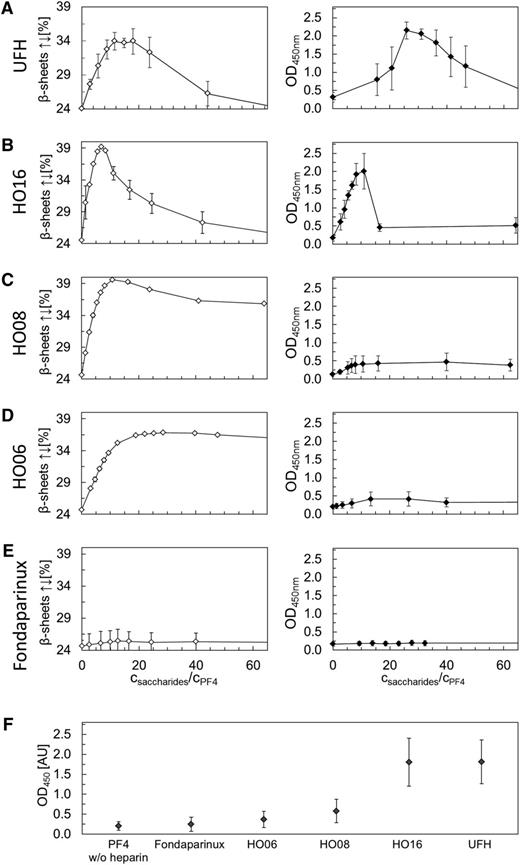
We compared the structural changes of PF4 (quantified by CD spectroscopy) with anti-PF4/heparin antibody binding to PF4 (quantified by optical density [OD] changes in an ELISA at the wavelength 450 nm), when PF4 was incubated with increasing concentrations of UFH, 16-, 8-, 6-mer heparins, and fondaparinux. The CD data are given in the left panels and the corresponding ELISA results in the right panels of Figure 1; data obtained with LMWH are given in supplemental Figure 2. We always normalized the heparin concentration by the PF4 tetramer concentration (number of heparin monomers per PF4 tetramer), taking into account that the PF4 concentration slightly changed by adding heparin in the titrating experiments. Moreover, we plotted in Figure 1 only the antiparallel β-sheet content of PF4, which is indicative for the overall changes of the PF4 structure17 (supplemental Figure 3).
Comparison of CD spectroscopy changes and ELISA results. Comparison of PF4 structural changes (antiparallel β-sheet content, determined by CD spectroscopy) and PF4/heparins antigenicity (OD obtained from ELISA measurements) for (A) UFH, (B) HO16, (C) HO08, (D) HO06, and (E) fondaparinux as a function of molar ratio monosaccharides/PF4. The open and filled squares represent average values of antiparallel β-sheet content and OD values, respectively. Error bars correspond to the standard deviation (calculated from the results of n = 3 experiments). (F) ELISA measurements for PF4 alone or in complex with fondaparinux, HO06, HO08, HO16, and UFH using a panel of 14 well-characterized sera containing anti-PF4/heparin antibodies. The filled squares represent average values of the maxima of the OD values. Error bars correspond to the standard deviation. Data for LMWH are given in supplemental Figure 2.
Comparison of CD spectroscopy changes and ELISA results. Comparison of PF4 structural changes (antiparallel β-sheet content, determined by CD spectroscopy) and PF4/heparins antigenicity (OD obtained from ELISA measurements) for (A) UFH, (B) HO16, (C) HO08, (D) HO06, and (E) fondaparinux as a function of molar ratio monosaccharides/PF4. The open and filled squares represent average values of antiparallel β-sheet content and OD values, respectively. Error bars correspond to the standard deviation (calculated from the results of n = 3 experiments). (F) ELISA measurements for PF4 alone or in complex with fondaparinux, HO06, HO08, HO16, and UFH using a panel of 14 well-characterized sera containing anti-PF4/heparin antibodies. The filled squares represent average values of the maxima of the OD values. Error bars correspond to the standard deviation. Data for LMWH are given in supplemental Figure 2.
Figure 1 shows 3 different patterns of the PF4/polyanion complexes: (1) reversible increase in antiparallel β-sheets of PF4, paralleled by a reversible increase in anti-PF4/heparin antibody binding (complexes with UFH and HO16, Figure 1A-B; LMWH, supplemental Figure 2); (2) nonreversible increase in antiparallel β-sheets of PF4 and minimal binding of anti-PF4/heparin antibodies (complexes with HO08 and HO06; Figure 1C-D); and (3) no increase in antiparallel β-sheets and no anti-PF4/heparin antibody binding (complexes with pentasaccharide fondaparinux; Figure 1E). We then tested a panel of 14 well-defined sera containing anti-PF4/heparin antibodies, with the PF4/defined length heparin complexes at the optimal concentrations determined by titration, which gave the same results (Figure 1F).
The maximum increase in antiparallel β-sheets (34% for UFH; 34% for LMWH; 39% HO16; 40% for HO08; and 36% for HO06) was observed at distinct concentrations of each heparin preparation. Due to the polydisperse (ie, variably sized) nature of UFH, it is impossible to define this on a molar basis. We therefore used an approach to take the mean MW of UFH 12 kDa, which corresponds to 39 monomers (saccharide monomers) per molecule. For consistency, the same approach was used for the heparin fragments.
AFM morphological characterization of the complexes formed by PF4 with heparins of different chain length
The differences in anti-PF4/heparin antibody binding raised the question of whether the smaller heparins, albeit causing an increase in antiparallel β-sheets, form large complexes with PF4. We used tapping mode liquid AFM imaging to characterize the dependency between length of heparin and the size of the complexes formed with PF4.
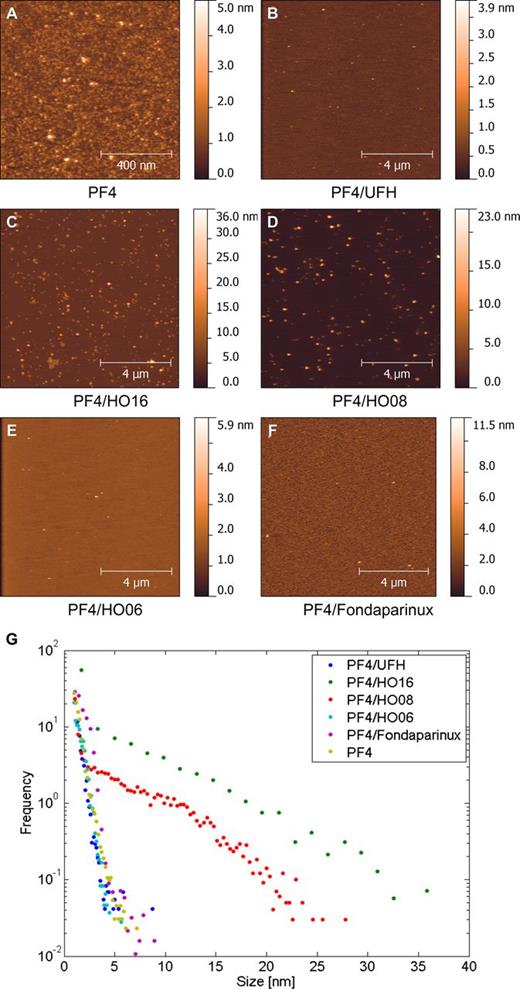
We show the complexes that PF4 forms with 16, 8, 6-mer heparins and fondaparinux in Figure 2. PF4 forms ultralarge complexes (>20 nm) with the 16-mer (HO16) and the 8-mer (HO08) heparins (Figure 2C-D). The complexes formed by PF4 with HO16 and HO08 have a broad height distribution reaching up to 35 and 25 nm, respectively (Figure 2G). The 6-mer (HO06) and the 5-mer (fondaparinux) heparins formed small, if any, complexes with PF4 with heights in the range of 1 to 5 nm (Figure 2E-G). The complexes were similar to PF4 alone, which formed structures with a large height distribution (from 1 to 7 nm; supplemental Figure 4A,G).
Assessment of PF4/heparin complexes by AFM. Representative liquid AFM tapping mode images of (A) PF4 alone and (B) PF4/UFH, (C) PF4/HO16, (D) PF4/HO08, (E) PF4/HO06, and (F) PF4/fondaparinux complexes on mica; the corresponding height histograms derived from all experiments are shown in G.
Assessment of PF4/heparin complexes by AFM. Representative liquid AFM tapping mode images of (A) PF4 alone and (B) PF4/UFH, (C) PF4/HO16, (D) PF4/HO08, (E) PF4/HO06, and (F) PF4/fondaparinux complexes on mica; the corresponding height histograms derived from all experiments are shown in G.
When we tried to measure PF4/UFH complexes, we could not stably immobilize them on the mica surface. Most likely they were so large that they detached when we rinsed the surface. Consistently, in additional experiments with 300-second incubation time and gentle dipping instead of washing we found several large PF4/UFH complexes (data not shown).
We show the control experiment with UFH, 16, 8, 6-mer heparins, and fondaparinux adsorbed on mica surface alone in supplemental Figure 4B-F, where we found grains with average height of 1 to 2 nm.
Binding interaction between PF4 and UFH, LMWH, and defined chain length heparins: ITC study
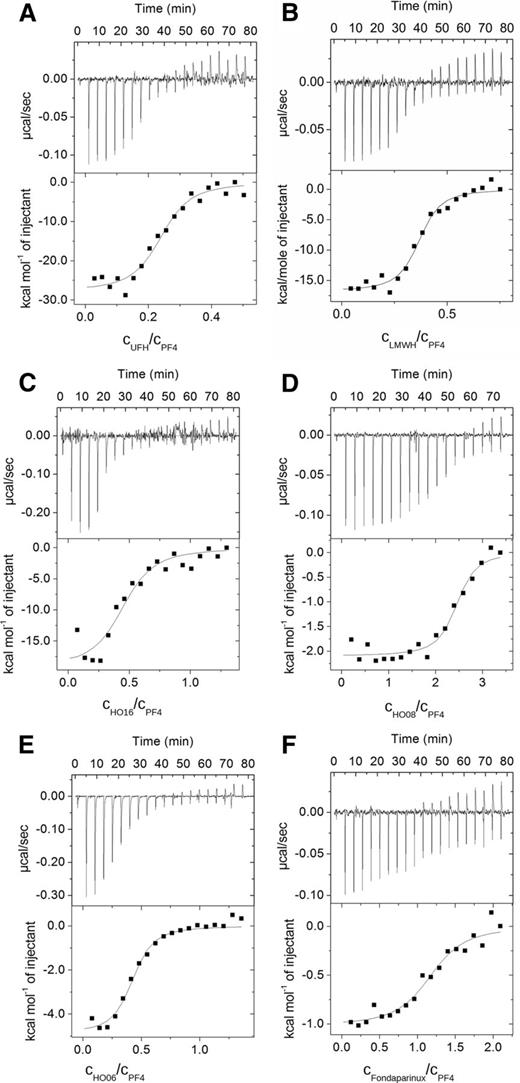
We assessed the energetic characteristics of the interaction of PF4 with UFH, LMWH, and defined chain length heparins by ITC, which directly measures changes in heat that occur during complex formation.26,27 The upper panels of Figure 3 show the sequence of the titration, with each peak corresponding to the injection of the solution in the syringe, whereas the lower panels show the integrated heat plot as a function of heparin/PF4 tetramer ratio. The thermodynamic parameters are given in Table 1.
Representative binding isotherms for the titration of PF4 with defined chain length heparins. (Upper) Raw titration data and (lower) integrated heats as a function of the molar ratio of heparin/PF4 for (A) UFH, (B) LMWH, (C) HO16, (D) HO08, (E) HO06, and (F) fondaparinux.
Representative binding isotherms for the titration of PF4 with defined chain length heparins. (Upper) Raw titration data and (lower) integrated heats as a function of the molar ratio of heparin/PF4 for (A) UFH, (B) LMWH, (C) HO16, (D) HO08, (E) HO06, and (F) fondaparinux.
Thermodynamic parameters (calculated per mole PF4 tetramer) for the interaction of PF4 with UFH, HO16, LMWH, HO08, HO06, and fondaparinux at 25°C
| Complexes . | Enthalpy, ∆H (cal/mol) . | Dissociation constant, KD (µM) . | Stoichiometry, n . | Gibbs free energy, ∆G (cal/mol) . | Entropy, ∆S (cal/mol/K) . |
|---|---|---|---|---|---|
| PF4/UFH | −6682 ± 1150 | 0.14 | 0.24 ± 0.03 | −2240 ± 80 | −15.6 ± 4.1 |
| PF4/HO16 | −7260 ± 1365 | 0.05 | 0.42 ± 0.1 | −3865 ± 489 | −11.4 ± 6.2 |
| PF4/LMWH | −6663 ± 906 | 0.09 | 0.36 ± 0.01 | −3515 ± 136 | −11.5 ± 3.8 |
| PF4/HO08 | −4240 ± 1467 | 0.20 | 1.93 ± 0.42 | −17 148 ± 635 | 46.1 ± 23.3 |
| PF4/HO06 | −2071 ± 35 | 2.50 | 0.42 ± 0.04 | −3190 ± 151 | 3.7 ± 0.5 |
| PF4/fondaparinux | −1333 ± 57 | 1.25 | 1.30 ± 0.15 | −10 415 ± 404 | 30.4 ± 1.4 |
| Complexes . | Enthalpy, ∆H (cal/mol) . | Dissociation constant, KD (µM) . | Stoichiometry, n . | Gibbs free energy, ∆G (cal/mol) . | Entropy, ∆S (cal/mol/K) . |
|---|---|---|---|---|---|
| PF4/UFH | −6682 ± 1150 | 0.14 | 0.24 ± 0.03 | −2240 ± 80 | −15.6 ± 4.1 |
| PF4/HO16 | −7260 ± 1365 | 0.05 | 0.42 ± 0.1 | −3865 ± 489 | −11.4 ± 6.2 |
| PF4/LMWH | −6663 ± 906 | 0.09 | 0.36 ± 0.01 | −3515 ± 136 | −11.5 ± 3.8 |
| PF4/HO08 | −4240 ± 1467 | 0.20 | 1.93 ± 0.42 | −17 148 ± 635 | 46.1 ± 23.3 |
| PF4/HO06 | −2071 ± 35 | 2.50 | 0.42 ± 0.04 | −3190 ± 151 | 3.7 ± 0.5 |
| PF4/fondaparinux | −1333 ± 57 | 1.25 | 1.30 ± 0.15 | −10 415 ± 404 | 30.4 ± 1.4 |
The stoichiometry n is the molar ratio heparin molecule/PF4.
For all investigated heparins, the reaction with PF4 was exothermic (heat release; Figure 3), but 2 distinct reaction patterns occurred: (1) reactions with large heat release (enthalpy change) and (2) reactions with little heat release. PF4/UFH, PF4/LMWH, and PF4/HO16 complexes showed the largest heat release (Table 1; Figure 3A-C), whereas PF4/HO08 (Table 1; Figure 3D), PF4/HO06 (Table 1; Figure 3E), and PF4/fondaparinux complexes showed ∼60%, 30%, and <20% of the heat release of PF4/UFH complexes, respectively (Table 1; Figure 3F, please note the different scales in Figure 3).
We calculated relatively low values of equilibrium dissociation constant KD for the complexes formed by PF4 with HO16 heparin (0.05 μM), LMWH (0.09 µM), and UFH (0.14 μM), indicating strong binding compared with the KD for the complexes formed with short chain length heparins (1.25 μM for PF4/fondaparinux complexes; 2.5 μM for PF4/HO06 complexes, and 0.2 μM for PF4/HO08 complexes).
The key finding of the thermodynamic studies is the change in randomness of the system (entropy, ∆S; Table 1), which correlates very well with the binding capacity of the resulting PF4/heparin complexes for anti-PF4/heparin antibodies (as shown by ELISA). We give the enthalpy per PF4 molecule because this is the constant reaction partner in our experiments.
The calculated ∆S for the PF4/UFH complex and PF4/LMWH complex showed a negative value (−15.6 ± 4.1 and −11.5 ± 3.8 cal/mol/K, respectively, with respect to 1 PF4 molecule), ie, a considerable amount of energy released after PF4 binding was consumed by the conformational changes of the complexes (an alternative explanation is that hydrophobic functional groups move to the surface of PF4 coming into direct contact with water). A similar pattern (negative change in entropy, ∆S = −11.4 ± 6.2 cal/mol/K with respect to 1 mol PF4) was found for PF4/HO16 complexes. In contrast, complexes formed by PF4 with HO08, HO06, and fondaparinux showed a positive entropy change, ie, binding of these heparins does not result in conformational changes of the PF4/heparin complexes that require additional energy.
Discussion
Our studies combining physicochemical characterization with assessment of anti-PF4/heparin antibody binding provide new insights into the interaction mechanism between PF4 and heparins. Using heparin fragments with defined length, we found that PF4/heparin complexes require the following characteristics to bind anti-PF4/heparin antibodies: induction of an increase in antiparallel β-sheets in PF4 exceeding ∼30%; formation of multimolecular complexes; and an amount of energy larger than −4000 cal/molPF4. This energy is required for the conformational changes in PF4 needed to expose the antigenic epitopes and is provided when heparins ≥11-mers bind to PF4.
Although we confirm in this study that an increase in the antiparallel β-sheet content of PF4 >30% is a requirement17 for binding of anti-PF4/heparin antibodies, it was a major surprise when we found that in complexes with HO08 and HO06, the antiparallel β-sheet content of PF4 increased clearly >34% (ie, even more than in PF4/UFH complexes). It is well known that a critical heparin chain length of ∼12 saccharide units is required to form PF4/polyanion complexes that express the antigen to which anti-PF4/heparin antibodies bind28 and induce subsequent platelet activation.24 10-mer heparin fragments induce only weak recognition, and 8- and 6-mer heparin fragments are even less29,30 or nonreactive.28 Consistent with these findings, anti-PF4/heparin antibodies did not (or only minimally) bind to PF4/HO08 and PF4/HO06 complexes in our study. Only fondaparinux neither increased the antiparallel β-sheet structures of PF4 nor facilitated binding of anti-PF4/heparin antibodies when complexed with PF4 (Figure 1). However, fondaparinux still binds to PF4 as shown by ITC, where interaction of PF4 with fondaparinux results in an exothermic reaction (ie, the enthalpy change is negative; Table 1; Figure 3E).
CD spectroscopy already gave a first hint that HO08 and HO06 form with PF4 different complexes than longer heparins. The typical reversible changes in the secondary structure of PF4 at high heparin concentrations were only seen for UFH, LMWH, and HO16, but not for HO08 and HO06. This feature, can, however, not be the only feature differentiating between PF4/polyanion complexes that bind anti-PF4/heparin antibodies and those that do not. Dextran sulfate also induces irreversible changes in the PF4 secondary structure but at the same time, anti-PF4/heparin antibodies bind strongly to PF4/dextran sulfate complexes.17,31
We assumed that formation of multimolecular complexes is the requirement for anti-PF4/heparin antibody binding as already shown by us14,24,32 and others10 and that at minimum a 12-mer is required for formation of such large complexes.28 Accordingly, we found by AFM that the size of PF4/fondaparinux and PF4/HO06 complexes did not differ largely from the size of PF4 alone (Figure 2). However, PF4/HO08 complexes were as large as PF4/HO16 complexes, and still anti-PF4/heparin antibodies did not bind to them. Thus, exposure of the antigen allowing binding of anti-PF4/heparin antibodies must require more than formation of multimolecular complexes between PF4 and a polyanion even if this induces a change in antiparallel β-sheets of PF4 >30%.
These puzzling observations were further clarified by ITC, which measures the thermodynamic changes when PF4/polyanion complexes are formed. For all investigated heparins, heat was released on binding to PF4. However, longer heparins induced a higher heat release (negative change in enthalpy) compared with shorter heparins. Normalized per mole PF4 tetramers, the largest heat release (the highest negative values for enthalpy) was measured for the complexes formed by PF4 with UFH, LMWH, and HO16, whereas HO08, HO06, and fondaparinux (Tables 1 and 2) induced much less heat release when complexed with PF4. This is an unexpected finding for a mainly electrostatically mediated interaction.33 The smaller heparin molecules should have been able to pack closer to the PF4 tetramers, thereby displacing more water molecules and consequently leading to an increased heat release.
Thermodynamic energies for the interaction of PF4 with UFH, LMWH, HO16, HO08, HO06, and fondaparinux at 25°C, calculated per mole of PF4 tetramers
| Complexes . | Enthalpy, ∆H (cal/mol) . | Comparison* . | Required enthalpy, ∆Hreq (cal/mol)† . |
|---|---|---|---|
| PF4/UFH | −6682 ± 1150 | > | −4673 ± 1238 |
| PF4/HO16 | −7260 ± 1365 | > | −3415 ± 1865 |
| PF4/LMWH | −6663 ± 906 | > | −3450 ± 1140 |
| PF4/HO08 | −4240 ± 1467 | ≈ | −3840 ± 717‡ |
| PF4/HO06 | −2071 ± 35 | < | −3840 ± 717‡ |
| PF4/fondaparinux | −1333 ± 57 | < | −3840 ± 717‡ |
| Complexes . | Enthalpy, ∆H (cal/mol) . | Comparison* . | Required enthalpy, ∆Hreq (cal/mol)† . |
|---|---|---|---|
| PF4/UFH | −6682 ± 1150 | > | −4673 ± 1238 |
| PF4/HO16 | −7260 ± 1365 | > | −3415 ± 1865 |
| PF4/LMWH | −6663 ± 906 | > | −3450 ± 1140 |
| PF4/HO08 | −4240 ± 1467 | ≈ | −3840 ± 717‡ |
| PF4/HO06 | −2071 ± 35 | < | −3840 ± 717‡ |
| PF4/fondaparinux | −1333 ± 57 | < | −3840 ± 717‡ |
A comparison of the minimum required enthalpy with the measured enthalphy indicates if an antigenicity inducing conformational change (similar to UFH, LMWH, and HO16) is thermodynamically allowed (ΔH > ΔHreq), unlikely (ΔH ≈ ΔHreq), or forbidden (ΔH < ΔHreq).
The lower limit for the enthalpy required to drive antigenicity inducing conformational changes was estimated using TΔS by taking either the measured ΔS values (UFH, LMWH, HO16) or assuming ΔS = −12.8 cal/mol/K (for heparins labeled with ‡), which is the average of the entropy changes for UFH and HO16.
By comparing the ELISA data with the ITC data, it became obvious that the heat release has to be stronger than approximately –4000 cal/molPF4 to allow expression of the binding site for anti-PF4/heparin antibodies. In other words, this energy is needed to drive the structural changes in PF4 required for exposure of the neoantigens. This is hardly fulfilled by HO08, but not by HO06 and fondaparinux (Table 2).
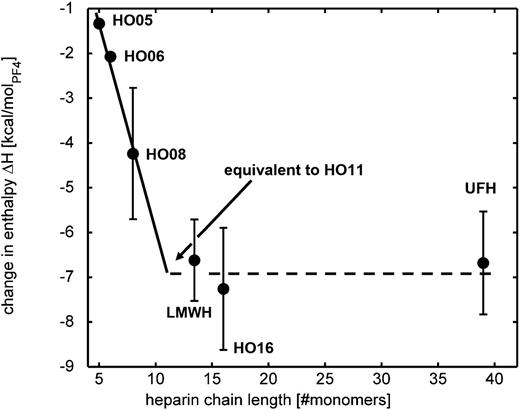
We then extrapolated the change in enthalpy (heat release) of the different heparins, as shown graphically in Figure 4. The required change in enthalpy approaches the values of UFH and HO16 at a chain length of ∼11 monosaccharide units. This excellently matches with empirically observed interaction patterns of PF4 with different heparins showing that ≥12 monosaccharides are necessary to induce anti-PF4/heparin antibody binding in vitro.28-30 This critical heparin chain length can therefore also be interpreted by the minimum chain length that is necessary to release enough energy to drive the PF4 conformational changes, which finally lead to epitope exposure.16
Dependence between change in enthalpy and heparin chain length. The change in enthalpy (circles, values taken from Table 2) increases with chain length and reaches the values of HO16 and UFH at a heparin chain length around 11 monosaccharides, which is close to the critical heparin chain length (=12) that has to be exceeded to form antigenic PF4/heparin complexes. The error bars for HO05 and HO06 are so small that they are hidden by the circles.
Dependence between change in enthalpy and heparin chain length. The change in enthalpy (circles, values taken from Table 2) increases with chain length and reaches the values of HO16 and UFH at a heparin chain length around 11 monosaccharides, which is close to the critical heparin chain length (=12) that has to be exceeded to form antigenic PF4/heparin complexes. The error bars for HO05 and HO06 are so small that they are hidden by the circles.
The CD experiments showed different patterns of the antiparallel β-sheet content of PF4 (Figure 1), which was reversible at higher concentrations when UFH, LMWH, and HO16 were added, but irreversible with HO08 and HO06. Likely, shorter heparins (HO08 and HO06, and presumably all heparins <11-mers) pack closely around PF4 tetramers; thereby, each negative charge of the heparin chain finds positive binding partners on PF4. Larger heparins, however, are too long to just bind to one PF4 tetramer. Therefore, they likely bridge between two PF4 tetramers to find maximal binding partners to neutralize their negative charges. This requires close approximation and a conformational change that consumes energy. By adding more heparin, the long heparin molecules all compete for the positive binding sites on PF4. As already proposed in earlier studies,24 this then results in disruption of the multimolecular complexes and reversal of the conformational change. In contrast, fondaparinux, HO06, and HO08 already form energetically favorable complexes with PF4, which will not be reversed by the addition of more fondaparinux, HO06, or HO08.
Our study has some limitations. We determined the structural changes of the PF4 molecule associated with exposure of the antibody binding site. Whether these changes are causative or indirectly related can only be determined when the structure of the binding site(s) for PF4/heparin antibodies is identified. This will require crystallization of PF4/heparin complexes together with an anti-PF4/heparin antibody. Although we did use typically reacting human anti-PF4/heparin antibodies, the panel of 14 human sera may not cover the entire spectrum of binding characteristics of human anti-PF4/heparin antibodies. In addition, the defined heparin fragments that we used are not characterized for their anticoagulatory potency. They are model substances and might show different physical characteristics, especially depending on the number of their sulfate groups. In the AFM experiments, we had technical difficulties to immobilize PF4/UFH complexes on the mica surface in the fluid phase, in contrast to previous experiments, in which we assessed PF4/heparin complexes by AFM on dried mica.14 Most likely, the PF4/UFH complexes are so large10 that they had been flushed away in the present fluid phase experiments. Consistent with this hypothesis, we found large PF4/UFH complexes in additional experiments (300-second incubation time and gentle dipping instead of washing; data not shown). In addition, the PF4/polyanion complexes we observed in the fluid phase were predominantly globular, similar to the complexes seen in transmission electron microscopy images of Rauova et al.10 We did not find ridge-like structures as previously described,14 which had a height of only about 2.9 nm and a length of up to 200 nm. These differences can be explained by the fact that measurements were carried out in liquid and tip convolution is much larger here than in the previous work. We also did not exclude the interaction of several polyanions with PF4, eg, endothelial cell heparin sulfate and the pentasaccharide, which may together induce conformational changes that likely happen in vivo and may be the explanation why fondaparinux can induce anti-PF4/heparin antibodies.34
Our study shows for the first time that besides clustering, conformational changes of PF4 by a polyanion do not necessarily lead to the expression of the binding site for anti-PF4/heparin antibodies. This only occurs if binding of a polyanion to PF4 results in a conformational change that requires input of energy. Only then does PF4 expose structures to which the immune system reacts and that can therefore be seen as danger signal, eg, for labeling bacteria.35 Thus, our biophysical methods may be applied to guide the development of synthetic heparins and other polyanion-based drugs, eg, aptamers,36 that do not lead to expression of these danger signals, which results in an increased risk for heparin-induced thrombocytopenia.
Beyond HIT, understanding the conformational changes making PF4 immunogenic may be relevant for mechanisms underlying other autoimmune blood disorders and immune reactions to human recombinant proteins used as biotherapeutics. Many proteins that are the target of autoantibodies in hematology tend to cluster, eg, PF4 clusters with polyanions like heparin; platelet glycoprotein IIbIIIa (target in immune thrombocytopenia) clusters in rafts37 ; and ADAMTS13 (target in thrombotic thrombocytopenic purpura) may cluster on von Willebrand factor.38 If in addition to clustering, these proteins undergo conformational changes, they may also trigger an immune response. In this regard, it is of interest that GPIIbIIIa and ADAMTS13 have in common that their genes show many polymorphisms. Potentially, certain point mutation or post-translational modifications allow conformational changes critical for the immune system with less energy input.
In summary, biophysical methods allowed us to characterize the conformational changes, which the endogenous protein PF4 undergoes when it forms complexes with well-defined polyanions. Our findings may help to synthesize safer heparins. Beyond HIT, these methods may be relevant to unravel mechanisms that predispose other endogenous proteins relevant in autoimmunity to become immunogenic.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Marco Marenchino (GE Healthcare) for valuable discussion and help with ITC data analysis and interpretation. This work was supported by the German Ministry of Education and Research within project FKZ 03Z2CN11. K.K. was supported by the Volkswagen Stiftung (Lichtenberg Professorship to Hansjörg Schwertz).
Authorship
Contribution: M.K. designed and carried out the ITC and AFM experiments, analyzed and interpreted the results, and wrote and reviewed the manuscript; S.B. designed and carried out the ELISA and CD measurements, analyzed and interpreted the results, and reviewed the manuscript; K.K. discussed the results and reviewed the manuscript; S.B. supported the AFM experiments and participated in the thermodynamical interpretation of the data; C.A.H. discussed the results critically and reviewed the manuscript; W.W. discussed the results and reviewed the manuscript; A.G. provided the conceptual design of the experiments, provided critical review and discussion of results; and wrote the manuscript; M.D. provided the conceptual design of biophysical characterization of PF4-defined heparins complexes, designed the experiments, interpreted the results, and wrote and reviewed the manuscript; and all authors reviewed and approved the final version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Andreas Greinacher, Institut für Immunologie und Transfusionsmedizin, Sauerbruchstrasse, 17475 Greifswald, Germany; e-mail: greinach@uni-greifswald.de; or Mihaela Delcea, ZIK HIKE-Zentrum für Innovationskompetenz, Humorale Immunreaktionen bei kardiovaskulären Erkrankungen, Ernst-Moritz-Arndt-Universität Greifswald, Fleischmannstrasse 42-44, 17489 Greifswald, Germany; e-mail: delceam@uni-greifswald.de.