Abstract
Juvenile myelomonocytic leukemia (JMML) is a typically aggressive myeloid neoplasm of childhood that is clinically characterized by overproduction of monocytic cells that can infiltrate organs, including the spleen, liver, gastrointestinal tract, and lung. JMML is categorized as an overlap myelodysplastic syndrome/myeloproliferative neoplasm (MDS/MPN) by the World Health Organization and also shares some clinical and molecular features with chronic myelomonocytic leukemia, a similar disease in adults. Although the current standard of care for patients with JMML relies on allogeneic hematopoietic stem cell transplant, relapse is the most frequent cause of treatment failure. Tremendous progress has been made in defining the genomic landscape of JMML. Insights from cancer predisposition syndromes have led to the discovery of nearly 90% of driver mutations in JMML, all of which thus far converge on the Ras signaling pathway. This has improved our ability to accurately diagnose patients, develop molecular markers to measure disease burden, and choose therapeutic agents to test in clinical trials. This review emphasizes recent advances in the field, including mapping of the genomic and epigenome landscape, insights from new and existing disease models, targeted therapeutics, and future directions.
Introduction
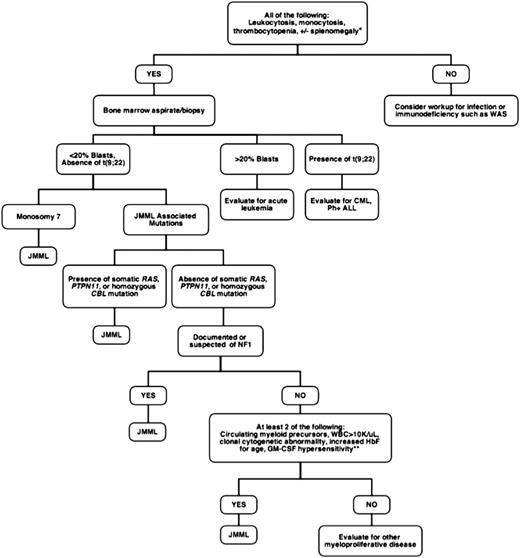
Juvenile myelomonocytic leukemia (JMML) is a rare but frequently lethal clonal myeloproliferative neoplasm (MPN) of early childhood characterized by the overproduction of myelomonocytic cells that infiltrate the spleen, lung, and intestines.1 It occurs with an estimated incidence of 1.2 cases per million annually, with a median age of diagnosis of 2 years.2,3 Patients usually present with fever, thrombocytopenia, failure to thrive, and splenomegaly. The sole curative option is hematopoietic stem cell transplantation (HSCT), which carries an event-free survival (EFS) at 5 years of only 52%.4 Treatment failure is typically a result of relapsed disease or transformation to acute myeloid leukemia (AML). Rarely, spontaneous resolution of this disorder can occur, but this is unpredictable.5-7 Historically, JMML has proven both difficult to diagnose and a challenge to treat. At presentation, circulating immature myeloid and nucleated red blood cells are found on the peripheral blood smear, and approximately half the patients present with an elevated hemoglobin F corrected for age.8 Figure 1 outlines a work-up for patients with suspected JMML based on currently accepted diagnostic criteria.
Diagnostic approach to suspected JMML. *Approximately 7% to 10% of JMML patients will not present with splenomegaly, although virtually all will develop splenomegaly within weeks to months of initial presentation. The mean age at diagnosis of JMML is 1.8 years, so that when encountering adolescents, alternative diagnoses such as chronic myelomonocytic leukemia should be considered. **GM-CSF in methylcellulose assays is not currently available in a Clinical Laboratory Improvement Amendments-approved setting. WAS, Wiskott-Aldrich syndrome; CML, chronic myelogenous leukemia; Ph+ ALL, Philadelphia-positive acute lymphoblastic leukemia; NF1, neurofibromatosis type 1.
Diagnostic approach to suspected JMML. *Approximately 7% to 10% of JMML patients will not present with splenomegaly, although virtually all will develop splenomegaly within weeks to months of initial presentation. The mean age at diagnosis of JMML is 1.8 years, so that when encountering adolescents, alternative diagnoses such as chronic myelomonocytic leukemia should be considered. **GM-CSF in methylcellulose assays is not currently available in a Clinical Laboratory Improvement Amendments-approved setting. WAS, Wiskott-Aldrich syndrome; CML, chronic myelogenous leukemia; Ph+ ALL, Philadelphia-positive acute lymphoblastic leukemia; NF1, neurofibromatosis type 1.
Major progress in understanding the pathogenesis of JMML has been achieved by deciphering the genetic lesions that initiate the disease, the majority of which encode proteins that signal in the RAS/MAPK pathway. This finding was made possible largely as a result of clinical observations in patients with inherited disorders that predispose to JMML and illustrates how insights from the bedside can be powerful forces in advancing discoveries at the bench.
Defining the molecular genetics of JMML: insights from the bedside
Developmental syndromes implicate hyperactive Ras in leukemogenesis
Delineating the molecular basis of inherited cancer predispositions identifies genes that, when mutated, play a key role in tumorigenesis. Indeed, initial observations from children with neurofibromatosis type 1 (NF1) and Noonan syndrome (NS), both developmental disorders that predispose to MPN, enhanced our understanding of the fundamental pathway involved in myeloid proliferation and maturation. Defining aberrant Ras signaling as the molecular mechanism linking NF1, NS, and other Rasopathies to JMML and a transient JMML-like disease also provided new insights into leukemogenesis. In particular, it is now clear that JMML is fundamentally a disease of hyperactive Ras signaling, with somatic mutations (superimposed on germline lesions in some instances) in the NF1, NRAS, KRAS, PTPN11, and CBL genes found in more than 90% of cases (Figure 2).8,9 The vast majority of these mutations are mutually exclusive in individual patients, which suggests that these proteins act as drivers, encoding components of a conserved biochemical network that regulates the proliferation, differentiation, and survival of HSCs and their progeny.
The Ras-MAPK pathway and gene mutations contributing to the neuro-cardio-facial-cutaneous syndromes, a Cbl germline syndrome, RALD, and JMML. NLSML, NS with multiple lentigines; CFC, cardio-facio-cutaneous; RALD, Ras-associated lymphoproliferative disease.
The Ras-MAPK pathway and gene mutations contributing to the neuro-cardio-facial-cutaneous syndromes, a Cbl germline syndrome, RALD, and JMML. NLSML, NS with multiple lentigines; CFC, cardio-facio-cutaneous; RALD, Ras-associated lymphoproliferative disease.
Ras proteins are key regulators of cell growth, survival, and differentiation in response to many extracellular stimuli, including granulocyte macrophage colony-stimulating factor (GM-CSF) and other hematopoietic growth factors.10 On ligand binding, the GM-CSF receptor (GMR) activates nonreceptor tyrosine kinases Janus kinase 2 (JAK2) and Src family kinases Src and Lyn, which subsequently phosphorylate the β-subunit of GMR.11 Activated GMR serves as a docking site for adaptors and signaling molecules such as SHP-2, which results in activation of downstream Ras signaling pathways.
Ras proteins function as molecular switches by cycling between active guanosine triphosphate (GTP)-bound and inactive guanosine diphosphate-bound conformations. This process is largely controlled by 2 competing classes of regulatory proteins: guanosine nucleotide exchange factors and GTPase activating proteins (GAPs).10 Guanosine nucleotide exchange factors induce the dissociation of guanosine diphosphate to allow binding of GTP, thus allowing Ras to turn “on.” GTP-bound Ras interacts with effector molecules including RAF, phosphatidylinositol 3-kinase (PI3K), and Ral-GDS to activate downstream kinases. GAPs such as p120GAP and neurofibromin terminate RAS signaling by accelerating the rate of GTP hydrolysis to guanosine diphosphate, augmenting this intrinsic “off” reaction.10
NF1
In 1958, a paper describing an unusual MPN in a young child with café au lait macules and xanthomatous skin lesions suggested a potential association between NF1 and hematologic malignancies.12 This report proved prescient, as subsequent research confirmed that the incidence of this aggressive MPN, now known as JMML, is increased 200- to 500-fold in children with NF1.13,14 Interestingly, children (but not adults) are at particular risk of developing JMML and other myeloid malignancies.14,15 The NF1 gene contains a GAP domain with a conserved arginine finger motif.16 Taken together, the facts that clinical NF1 is a dominantly inherited cancer predisposition syndrome, NRAS and KRAS are frequently mutated in human cancers, and neurofibromin is a Ras GAP16,17 suggested that NF1 was a bona fide tumor suppressor gene. Indeed, genetic analysis of JMML bone marrows revealed somatic loss of heterozygosity at the NF1 locus in multiple children with NF1, which invariably involved deletion of the normal parental allele in familial cases with duplication of the mutant gene, a phenomenon known as acquired uniparental isodisomy.18,19 Interestingly, a similar genetic mechanism is operative in CMML and JMML patients with CBL mutations.20-22 Importantly, leukemia cells from children with NF1 exhibited reduced neurofibromin-specific GAP activity, elevated levels of Ras-GTP, and aberrant activation of the canonical downstream Raf/mitogen-activated protein kinase (MEK)/extracellular signal-regulated kinase (ERK) effector pathway.23 As described here (targeted therapy), recent preclinical studies in genetically accurate animal models of JMML strongly support this hypothesis and provide a rationale for implementing clinical trials of MEK inhibition.
PTPN11
Germline PTPN11 mutations are found in approximately 50% of patients with NS, a developmental disorder that shares clinical features with NF1.24,25 As in NF1, clinical reports describing a spectrum of hematologic abnormalities including JMML and a JMML-like transient myeloproliferative disorder in children with NS26-28 led investigators to screen nonsyndromic patients with JMML for somatic mutations in PTPN11. Somatic mutations in PTPN11 were identified in 35% of nonsyndromic JMML cases.29,30
PTPN11 encodes SHP-2, a nonreceptor protein tyrosine phosphatase (PTP) involved in relaying signals from activated GMR complexes, to regulate proliferation, differentiation, and migration.31,32 SHP-2 contains 2 N-terminal Src homology 2 domains (N-SH2), 1 PTP domain, and a C-terminal tail.31 In its basal state, the N-SH2 domain blocks the catalytic site in the PTP domain. Ligand binding disrupts this autoinhibitory interaction, resulting in PTP activation and enhanced Ras/MAPK pathway activation.33 Most PTPN11 mutations in JMML patients disrupt the interface between the N-SH2 and PTP domains, thus preventing basal autoinhibition and leading to gain of function by allowing constitutive access to the catalytic site of the enzyme.34 The Ras/MAPK pathway is the primary cascade regulated by Shp2,31 although numerous reports have also linked Shp2 to modulation of the JAK/Stat pathway,35 PI3K/Akt pathway,36 and focal adhesion kinase pathway37 in various cellular contexts.
A comparison of the PTPN11 mutations in de novo versus syndromic JMML (NS) reveals that many, but not all, of the same codons in exons 3, 4, and 13 are affected.38 However, the precise codon substitutions are generally different in NS than in JMML, leading investigators to hypothesize and demonstrate that the transforming ability of mutations in NS are “weaker,” and thus better tolerated as germline events. Indeed, the most common PTPN11 mutation in de novo JMML is the c. 226G > A, resulting in E76K, an alteration that has never been documented as a germline lesion in NS. Biochemical and cell biologic studies have shown that the somatic leukemia-associated mutations encode stronger gain of function compared with the germline mutations found in NS.39,40 This “weaker” biochemical phenotype found in germline mutations likely explains the observation that many of the myeloproliferative disorders in NS infants are transient, but it remains puzzling as to why they spontaneously resolve over time.
Although SHP2 phosphatase activity is required for most known signaling functions, the key substrates that are required to activate the Ras–Raf–MEK–ERK pathway remain uncertain.32 RUNX1 has recently been identified as a direct interacting partner of Shp2 in megakaryocytes.41 Specifically, RUNX1 is negatively regulated by src-family kinase-mediated tyrosine phosphorylation, and Shp2 contributes to RUNX1 tyrosine dephosphorylation in this cellular context. A recent report describes the potential role of RUNX1 as a downstream target of activated Shp2 in the pathogenesis of JMML.42 Overexpression of a mutant RUNX1 (“RUNX1-5F”) that mimics constitutive dephosphorylation by Shp2 in murine Lin− Sca1+ ckit+ (LSK) bone marrow cells renders them resistant to src-family kinase-mediated tyrosine phosphorylation, resulting in dramatic colony-forming unit (CFU)-GM expansion, decreased apoptosis, and reduced cytokine dependence, which closely phenocopies mutant Shp2 knock-in mice with MPN.
RAS
Discoveries of oncogenic Ras mutations in JMML patients further implicated hyperactive Ras signaling in JMML pathogenesis. Somatic point mutations in NRAS and KRAS genes occur in about 25% of JMML cases, with the most common amino acid substitutions occurring at codons 12, 13, and 61.43,44 The resultant proteins accumulate in the GTP-bound conformation because of both defective intrinsic GTPase activity and resistance to GAPs. Animal models have subsequently revealed that Kras mutant mice also develop fatal myeloid disorders that resemble JMML in vivo and in vitro.45,46
Doisaki et al47 recently reported 2 patients with nonsyndromic JMML who were found to have somatic mosaicism for oncogenic NRAS G12D and G12S mutations, with mutated allele frequencies ranging from 3% to 50% in bone marrow and other somatic tissue (buccal, hair, nails). Both patients demonstrated spontaneous resolution of the disease without HSCT, although it is worth noting that the patient with the NRAS G12D mutation developed severe thrombocytopenia, refractory anemia, and trilineage dysplasia in the marrow with a low blast count 11 years after his JMML diagnosis.47 An analogous case of a nonsyndromic JMML patient with mosaicism for KRAS G13D experienced an indolent course for 1 year, followed by fatal malignant transformation. Notably, at time of progression, this patient was found to have acquired uniparental isodisomy at the 12p locus.48 Further studies are needed to evaluate the prognostic implications of mosaicism for oncogenic RAS mutations, especially given the potential for delayed transformation.
The link between JMML and other developmental syndromes implicates Ras signaling as a fundamental deregulated pathway in other hematopoietic and immunologic diseases. Recent reports describe somatic RAS mutations in patients with a new class of lymphoproliferative disease called Ras-associated lymphoproliferative disease.49-51 Patients present with crossover features of JMML and autoimmune lymphoproliferative syndrome including cytopenias, lymphadenopathy, and splenomegaly. Takagi et al49 demonstrated a somatic KRAS G13D mutation in multiple populations of hematopoietic cells, implying that the initial mutation occurred in an early multipotent progenitor or HSC. Niemela et al and Oliveira et al50,51 found that KRAS and NRAS mutant T cells deprived of interleukin 2 failed to induce the proapoptotic protein BIM and exhibited increased proliferation. It is thus unclear whether Ras-associated lymphoproliferative disease is a distinct entity from JMML, but differences in the clinical phenotype manifested from a RAS mutation likely depend in part on the type of cell in which the mutation originates, potential acquisition of cooperating mutations, existing modifier genes, and the degree of biochemical activity conferred by the specific RAS mutation.
CBL
We and others previously identified homozygous Casitas B-lineage lymphoma (CBL) mutations in approximately 10% to 15% of JMML cases.20,52 Recent reports had uncovered similar lesions in adults with MPN and myelodysplastic syndromes.22,53,54 In collaboration with investigators from the European Working Group on Myelodysplastic Syndromes (EWOG-MDS), we subsequently detected a total of 27 CBL mutations in a group of 68 JMML patients with no other known genetic lesions.20 The most common mutation was the c.1111T > C transversion, resulting in the substitution of a histidine for a key tyrosine residue at codon 371 in the linker region of the Cbl protein. Others have since confirmed these findings.52,55 We also have demonstrated that the initial CBL lesion in the majority of JMML patients occurs as a germline event, either inherited in an autosomal dominant fashion or arising spontaneously, similar to NF1 and NS.21 In nearly all patients with JMML and CBL mutations, the mutant allele is duplicated via acquired uniparental isodisomy. This results in the loss of both wild-type (WT) tumor suppressor alleles and the gain of an oncogenic mutation. A hemizygous mutant allele with a WT deletion is rarely seen and is understood to be the critical event driving leukemogenesis.
Cbl is an E3 ubiquitin ligase that is known to mark activated receptor and nonreceptor tyrosine kinases and other proteins for degradation by ubiquitination but also retains important adaptor functions (reviewed in Schmidt and Dikic).56 We and others showed that defective E3 ligase activity results from specific substitutions at this location (Y371H, Y371S) only in the absence of the WT protein,21,22 and that these homozygous mutations can also lead to cytokine-independent growth. Multiple mechanisms of Cbl-driven oncogenesis have been proposed that invoke both a dominant negative function of mutant CBL on other Cbl proteins as well as a gain of function via activated receptor and nonreceptor tyrosine kinases that can no longer be ubiquitinated.57 To further investigate the functional consequences of CBL in GM-CSF signaling, Javadi et al58 expressed WT CBL, linker region mutant CBL-Y371H, and RING finger mutant CBL-C384R in TF-1 hematopoietic cells, which expresses the endogenous GM-CSF receptor.58 On cytokine stimulation, expression of CBL mutants resulted in enhanced phosphorylation of GM-CSF, which occurred concurrently with elevated expression of JAK2 and LYN, as well as prolonged survival. Pharmacologic inhibition of either Src or JAK2 delineated which proximal GM-CSF-dependent pathways are modulated through JAK2 and LYN activation and provide a rationale for potential targeted therapy in CBL-associated JMML.
One striking phenomenon about patients with JMML and homozygous CBL mutations that first arise as a germline event is the high rate of spontaneous resolution of disease.6,21,59 Indeed, in our series, 6 children did not receive HSCT for various reasons, and only 1 died of progressive JMML. However, 4 of the remaining patients developed signs consistent with serious vasculopathies by the end of their second decade of life, as evidenced by optic atrophy, hypertension, cardiomyopathy, or arteritis.21 Recently, Strullu et al60 and Muramatsu et al61 performed screens for CBL mutations in 2 independent JMML cohorts. Although the majority of mutations were homozygous, as expected, both groups identified heterozygous CBL mutations. These consisted of splice site mutations and deletions and, in some cases, were present alongside another Ras-activating lesion, suggesting that CBL mutations can occur as secondary events in JMML. Both groups reported clinical heterogeneity in terms of outcomes. Muramatsu et al61 noted a trend toward improved survival in the CBL mutant subgroup compared with CBL WT, although serious vascular events such as Moyamoya were seen.
Additional genetic and epigenetic lesions in JMML.
In contrast to other MPNs of adulthood, JAK2, RUNX1, and TET2 mutations have not been found in JMML.52,62 Other mutations that have been reported to occur less frequently in JMML include ASXL163 and FLT3.64 Recently, investigation of spliceosomal gene aberrations uncovered rare mutations in SRSF2 in JMML.65
Whole-exome strategies have also identified secondary mutations in SETBP1 and JAK3 occurring in up to 17% of JMML patients and conferring a poorer prognosis, although the therapies delivered were variable.66 As these mutations were primarily found in a subpopulation of leukemic clones, they are thought to drive progression of disease rather than initiation; however, recent data may suggest that subclonal mutations are present at diagnosis (M.L.L., unpublished data, August 2014). SETBP1 has been recently described in 15% to 25% of adult cases of atypical CML, CMML, secondary AML, and unclassified MDS/MPNs.66,67 It is worth noting that recurrent germline SETPBP1 mutations (p.Gly870Ser) were initially reported in atypical CML patients67 and later found to be identical to those causing a rare but devastating autosomal recessive syndrome called Schinzel-Giedion syndrome, a neurodegenerative disorder characterized by midface retraction syndrome and cardiac, renal, and skeletal anomalies. If patients survive beyond infancy, they have a predisposition to neuroepithelial tumors but, curiously, not myeloid malignancies.68 Evidence suggests that SETBP1 is a protooncogene with overexpression in myeloid progenitors leading to enhanced self renewal.69 SETBP1 binds SET nuclear oncoprotein and inhibits protein phosphatase type 2a, a tumor suppressor.70 SETBP1 may also be involved in the direct transcriptional activation of HOXA9 and HOXA10 genes in myeloid progenitors.69 Its strict role in leukemogenesis remains unclear. Importantly, however, several studies have shown SETBP1 to be a poor prognostic marker in myeloid diseases.66,67,71
Alterations in microRNA (miR) expression are involved in the initiation, progression, and metastasis of human tumors, and germline mutations in specific miR clusters are thought to be associated with inherited cancer predispotions.72 The let-7 miR family is known to target Ras, and a SNP in a let-7 binding site in the KRAS 3′ untranslated region yielded KRAS overexpression and an increased risk for nonsmall cell lung cancer.73 Steineman et al74 found no evidence of mutations of let-7 or let-7 binding sites in a study of 10 JMML patients without known classic JMML mutations, although other mIRs known to bind NRAS or KRAS untranslated regions may play a role in JMML pathogenesis. Liu et al75 recently demonstrated overexpression of mIR-183 in JMML. They also identified significant reductions in levels of cyclic adenosine monophosphate-response-element-binding protein and early growth response gene 1 (Egr-1) in 85% and 87% of patients with JMML respectively, both of which play a role in GM-CSF signaling and monocytic differentiation.75 Egr-1 protein is encoded by EGR1, a tumor suppressor gene that is downregulated in many cancers, and mIR-183 has been shown to decrease PTEN and Egr-1 levels in tumor cell lines.76 The authors hypothesize that mIR-183 overexpression plays a role in both Egr-1 deficiency and the monocytic predominance in JMML, although the mechanism and implications have yet to be elucidated.75
Genotype phenotype correlations in JMML
Despite the fact that all known driver mutations thus far converge on a single primary pathway, JMML patients exhibit substantial clinical heterogeneity, with some succumbing to aggressive disease and others experiencing a more indolent course with rare spontaneous resolution. Ras pathway alterations, although helpful diagnostically, have not been shown to be individually prognostic, and the determinants of therapeutic responses are unknown. Several published series have examined the hypothesis that mutational status may correlate with JMML clinical features and prognosis; however, this remains controversial. Yoshida et al77 evaluated the clinical course and laboratory findings of 49 JMML patients, 32 of whom harbored mutations in NF1, KRAS, NRAS, or PTPN11. In their series, the mutation of PTPN11 was associated with older age at diagnosis (>24 months), increased hemoglobin F (>10%), and reduced overall survival, and importantly, appeared to predict relapse after transplantation. A recent Korean study also found a trend toward an unfavorable prognostic implication with the PTPN11 mutation, with reduced 5-year EFS and 5-year overall survival, although the results were not statistically significant and the numbers were small.78
Although the current standard of care for JMML is allogeneic HSCT, continued controversy exists about identifying those patients who need to be moved quickly to HSCT versus those rare patients who might be observed. At this time, the worst prognostic factors remain primarily clinical variables4,77,79,80 and include poor prognosis in older age (>1.4-4years), increased fetal hemoglobin (hemoglobin F, >40%), reduced platelets (<33 000/μL), increased blasts (>20%), monosomy 7, and PTPN11 mutational status. Certainly, we now anticipate that most patients with NS who present with a JMML-like MPN in the neonatal period will spontaneously resolve during the first year of life, somewhat akin to the transient myeloproliferative disorder of Down syndrome. However, unlike patients with Down syndrome, NS/MPN patients do not appear to be at an increased risk for myeloid malignancies later in childhood. Rare NS patients may require some low- or intermediate-dose chemotherapy to improve splenomegaly and control uncomfortably high white blood cell counts.
For patients without syndromic JMML, there have been multiple case reports in the literature describing a phenomenon of “self-resolving” JMML. Matsuda et al6 observed that 3 children bearing NRAS or KRASG12S demonstrated a milder clinical course with spontaneous clinical improvement. Flotho et al7 were unable to confirm in the EWOG-MDS database that patients harboring NRAS or KRASG12S were associated with long-term survival in the absence of HSCT. However, among 12 patients observed without HSCT for more than 3 years from diagnosis, there were 5 patients experiencing long-term survival who harbored various RAS mutations. Of great interest in both series is that spontaneously resolving patients all presented with favorable clinical prognostic factors. Although patients with homozygous CBL mutations have a high rate of spontaneous resolution, we know this is not a uniform phenomenon, as some children with CBL lesions have progressed and relapsed after HSCT.
Other attempts to identify risk stratification markers for JMML have included gene expression and methylation signatures. Bresolin et al80 reported that gene expression signatures can segregate JMML patients into those who displayed an AML-type signature versus those who did not. These signatures were significantly associated with outcome, with those who displayed an AML-type signature experiencing a 10-year EFS of 6% in contrast to those without the AML-type signature, who experienced a 63% EFS. Of interest was the statistically significant correlation between an AML-type signature and known prognostic variables at diagnosis, including older age, lower platelet count, and higher hemoglobin F levels. Interestingly, genotype was not associated with an AML-type gene expression profile.
An additional report by Olk-Batz et al81 indicated that hypermethylation of 4 genes was associated with an inferior outcome for patients and raised the possibility of therapeutic interventions using DNA methyl transferase inhibitors, which is currently being tested in an EWOG trial of JMML and MDS.
Modeling JMML: insights from the bench
Defining the key mutated genes in JMML has allowed basic researchers to develop genetically accurate experimental models to improve our understanding of the processes implicated in disease pathogenesis. Preclinical studies in murine models that recapitulate the genetic, biochemical, and cell biologic features of JMML have identified promising therapeutic candidates that are now being translated back to the clinic. Recent advances in stem cell technology have also permitted development of a new tool in modeling JMML, the induced pluripotent stem cell (iPSC).
Murine models
Conditional inactivation of Nf1 in the hematopoietic compartment induces a moderate MPN at 6 months with 100% penetrance.82 The spleens of these Mx1-Cre, Nf1flox/flox mice are heavily infiltrated with myeloid lineage cells at all stages of differentiation, with the marrow demonstrating significant expansion of the LSK and pre-GM compartments compared with WT mice.83 In comparison, a conditional model that expresses oncogenic KrasG12D from its endogenous locus generates a much more aggressive MPN that develops at 2 months of age.45 The bone marrow of these Mx1-Cre, KrasG12D mice show expansion of the pre-GM population and a diminished number of premegakaryocyte erythroid progenitors, indicating a myeloid bias in lineage commitment.84 A heterozygous conditional knock-in model with endogenous NRASG12D expression demonstrated a more variable and indolent MPN,85 thus implying that biologic differences between loss of Nf1 vs expression of different oncogenic Ras isoforms can influence the severity of hematopoietic growth dysregulation. Furthermore, differences exist in how these mutations affect stem cell properties. Division of a HSC typically results in either proliferation of more differentiated cells or self-renewal of the HSC.86 Indeed, KrasG12D drives HSCs into cycle and reduces HSC frequency.87 Li et al88 recently described a bimodal response to expression of a single NRASG12D allele in murine HSCs that showed both enhanced proliferation and self-renewal, all before leukemia initiation.
This mechanism for promoting preleukemic clonal expansion could explain how germline NRAS mutations predispose to JMML89 or how patients with JMML who undergo remission continue to carry NRAS mutations in their hematopoietic cells.6,90 Intriguingly, signal transducer and activator of transcription 5 (STAT5) signaling is required for increased competitiveness by NRASG12D HSCs88 and identifies a potential therapeutic target for eradicating not only the rapidly proliferating HSCs but also those that are quiescent. This finding is of particular interest, as MEK inhibition alone did not reliably eliminate mutant HSCs in Mx1-Cre, KrasG12D or Mx1-Cre, Nf1flox/flox mice with MPN.83,84
The Shp-2 D61G alteration occurs in both NS and de novo JMML. Expression of D61G from the endogenous murine Ptpn11 locus yields mice with cardiac and skeletal defects that develop a mild MPN with a latency of 5 months.91 Studies of the somatic E76K protein reveal it displays the highest phosphatase activity, induces profound hypersensitivity to GM-CSF in fetal liver cells, and transforms transduced BaF3 cell lines to cytokine independence.29,39,40 Mice transplanted with retrovirally transduced murine HSCs with gain-of-function Ptpn11E76K and Ptpn11D61Y mutations developed an MPN, with hematopoietic cells exhibiting elevated levels of pERK, pAKT, and pSTAT5.40 A conditional knock-in model, LSL- Ptpn11D61Y mice, develops an aggressive MPN by 5 months and exhibits an expanded LSK stem cell compartment with increased ERK and Akt activation and colony formation.46
A conditional Cbl, Cbl-b double knockout mouse modeled an aggressive, fully penetrant MPN characterized by leukocytosis and splenomegaly with myeloid infiltration.92 Hematopoietic progenitors demonstrated GM-CSF hypersensitivity, and there was expansion of the LSK compartment in the bone marrow. However, Lin− cells did not show enhanced ERK or STAT5 activation with GM-CSF or SCF stimulation,92 which may reflect the absence of an oncogenic CblY371H allele. Further functional characterization of this model with comparisons with other JMML murine models may help elucidate a potentially unique role of Cbl-family proteins in MPN pathogenesis.
iPSCs
Genetically engineered murine models of JMML have proven thus far to be powerful tools, although differences in the biology of malignant hematopoiesis and/or Ras signaling may exist between mice and humans. Unfortunately, primary JMML samples have proven difficult to expand in culture, and there are no established human JMML cell lines. Access to primary patient samples is also limited by disease rarity and the young age of the patient population. iPSCs represent a renewable and innovative platform to investigate JMML pathogenesis on the basis of patient-derived samples.
Investigators have been able to create iPSCs from patients with myeloproliferative disease and leukemia.93 Most recently, Gandre-Babbe et al94 successfully generated iPSCs from 2 unrelated JMML patients harboring the somatic Shp-2 p.E76K mutation, the most common and most active PTPN11 mutation found in JMML. These iPSCs recapitulated key pathologic features of primary JMML cells, including cytokine-independent growth and GM-CSF hypersensitivity in methylcellulose assays. They also demonstrated increased p-ERK and p-STAT5 signaling in response to low concentrations of GM-CSF, similar to the phosphoflow signature in JMML patient samples described by Kotecha et al.90
Myeloid progeny of these mutant iPSCs were treated with a various pharmacologic agents suspected to be active in JMML and were selectively sensitive to MEK inhibition, validating findings from preclinical studies in Kras and Nf1 mutant mice with MPN.83,84 Interestingly, treatment with a JAK inhibitor had no effect. Nevertheless, the possibility that different JMML mutations could trigger distinct biochemical effects and drug sensitivity profiles must be considered. Creation of iPSCs from patients whose disease harbors other canonical JMML mutations may allow investigators to delineate genotype-phenotype correlations, which will ideally translate into refined stratification and treatment programs based on molecular genetics.
Translating discoveries: therapeutic implications
Advances in diagnosis and surveillance
JMML was historically difficult to diagnose clinically, as patients present with symptoms shared by other hematologic malignancies or viral infections. In 1991, Emanuel et al95 demonstrated that JMML cells selectively form abnormal numbers of CFU-GM colonies in methylcellulose cultures containing low concentrations of GM-CSF. Indeed, GM-CSF hypersensitivity remains an assay used to help aid in the diagnosis of JMML, but it, too, is neither sensitive nor specific and is performed differently across laboratories internationally. With the complete genomic landscape of JMML nearly defined, molecular testing has taken a fundamental role in establishing the diagnosis.
Detection of a mutation in NF1, NRAS, KRAS, PTPN11, or CBL strongly supports a diagnosis of JMML in patients with suggestive clinical findings,9 which led to a revision of the proposed JMML diagnostic criteria.96 As of 2013, the most commonly mutated exons in JMML (CBL exons 8 and 9, KRAS exons 2 and 3, NRAS exons 2 and 3, and PTPN11 exons 3, 4, and 13) make up a diagnostic JMML panel in a Clinical Laboratory Improvement Amendments-approved setting. Using the 454Jr Pyrosequencing platform, the sensitivity of detecting a point mutation is 1%. To address the need for a more refined surveillance method of minimal residual disease after HSCT, our laboratory developed an MRD assay referred to as TaqMAMA.97 This method allowed preferential amplification of the mutant allele compared with the WT allele. We demonstrated sensitivity and specificity in selectively identifying these point mutations and were able to detect molecular relapse well ahead of clinical relapse, and frequently ahead of falling donor chimerism measurements. Importantly, detection of the mutant allele from peripheral blood cells correlated with detection in bone marrow cells. Given the very recent developments of using sensitive 454 sequencing methodologies to detect JMML-specific mutations in our diagnostic panel, we are also comparing our allele-specific PCR assays against 454 sequencing results in the setting of a Children’s Oncology Group (COG)-sponsored JMML HSCT clinical trial ASCT1221.
Biochemical signatures can also be beneficial, given the heterogeneity of the disease for any given mutation. Kotecha et al90 reported that a subpopulation of CD33+CD14+CD38lo bone marrow cells from patients with JMML, CMML, and the M4 or M5 subtypes of an AML display a distinct pattern of STAT5 hyperphosphorylation in response to GM-CSF. This biochemical signature was observed in patients with mutations in multiple different JMML genes, including NF1. This phospho-STAT5 signature was recently validated as a diagnostic tool in 83 specimens (22 JMML, 47 controls, 7 cases with diseases other than JMML) and demonstrated a sensitivity of 91% (95% confidence interval [CI], 59%-100%) and a specificity of 87% (95% CI, 70%-96%), with positive and negative predictive values of 71% (95% CI, 42%-92%) and 96% (95% CI, 82%-100%), respectively.98
Approaches to HSCT
Pretransplant chemotherapy for JMML has not demonstrated any benefit on EFS or overall survival,4 although results are difficult to interpret, as patients with higher disease burdens were likely to have received more intense pretreatment. An alternative approach is to use the differentiating agent, 13-cis retinoic acid, which inhibits spontaneous proliferation of JMML myeloid progenitors in vitro. In a pilot study of 10 children with JMML, monotherapy with 13-cis retinoic acid reduced organomegaly and white blood cell counts in 50% of patients, although less than 10% achieved durable remissions.99 Given the heterogeneity of disease burden at diagnosis, it is not feasible to be prescriptive about specific pre-HSCT therapies for JMML, although guidelines are provided for physician consideration (Table 1).
Pre-HSCT treatment guidelines for JMML
| Asymptomatic . | High white blood cell count, pulmonary problems, and/or prominent organomegaly . | Moderately/Severely Ill . |
|---|---|---|
| No pre-HSCTtherapy | Oral 6-mercaptopurine (50 mg/m2 per day) | Low-dose intravenous cytarabine (40 mg/m2 per day × 5 days); if no improvement of symptoms: high-dose intravenous cytarabine (2 g/m2 per day × 5 days) + fludarabine (30 mg/m2 per day × 5 days) |
| Asymptomatic . | High white blood cell count, pulmonary problems, and/or prominent organomegaly . | Moderately/Severely Ill . |
|---|---|---|
| No pre-HSCTtherapy | Oral 6-mercaptopurine (50 mg/m2 per day) | Low-dose intravenous cytarabine (40 mg/m2 per day × 5 days); if no improvement of symptoms: high-dose intravenous cytarabine (2 g/m2 per day × 5 days) + fludarabine (30 mg/m2 per day × 5 days) |
These pre-HSCT guidelines are provided for physician consideration.
There is some controversy over current conditioning regimens before HSCT for JMML. Chemotherapy-based conditioning regimens, such as the one published by the EWOG-MDS, use busulfan, cyclophosphamide, and melphalan (Bu-Cy-Mel) with reasonable transplant-related mortality and morbidity.4,100 A total body irradiation-based approach was used in the COG trial AAML0122, but compliance with using total body irradiation was limited because of investigator concerns about the late effects of total body irradiation on young children. Although Bu-Cy-Mel has the most published experience,4 a survey of transplant physicians in the United States reveals that the majority are not using this regimen. Furthermore, survival rates are not improving over time, as evidenced by the poor EFS seen in patients undergoing umbilical cord blood transplant between 1995 and 2010.79 Small, nonrandomized studies suggest that elimination of Mel101 and/or substitution of Cy with fludarabine102 may decrease acute graft-versus-host disease and acute toxicities without affecting overall survival. The current COG ASCT1221 trial is testing the hypothesis that the main mechanism of disease elimination in patients with JMML is primarily a result of the alloreactive graft-versus-leukemia effect, and therefore that less toxic regimens could potentially be used to establish successful engraftment with similar rates of survival. Patients are randomly assigned on the basis of donor type and PTPN11 mutation status to receive either Bu-Cy-Mel or Bu-fludarabine to determine whether comparable survival rates result with decreased toxicity of 1 regimen over the other.
Once engraftment is established, the graft-versus-leukemia effect can potentially be harnessed via either rapid withdrawal of immunosuppression and/or donor lymphocyte infusions.103 Advances in lineage-specific chimerism104 and JMML mutation-specific MRD testing97,105 can help guide the aggressiveness of the immunomodulation strategies. Finally, for those JMML patients who relapse after HSCT, recent data indicate that up to 50% of patients can be salvaged using a second HSCT with either the original or an alternative donor. 4,103,106
Targeted therapy
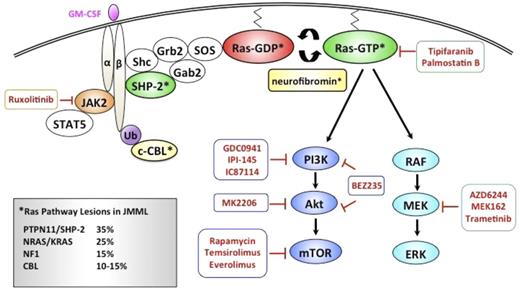
Although HSCT cures ∼50% of JMML patients, significant morbidities and late effects in this young population have driven the search for more effective and less toxic strategies. As aberrant Ras signaling plays a fundamental role leukemogenesis, molecular approaches of targeting this pathway are being evaluated (Figure 3).
Schematic diagram depicting pharmacologic agents currently in clinical development that target relevant nodes within the Ras signaling pathway in JMML.
Schematic diagram depicting pharmacologic agents currently in clinical development that target relevant nodes within the Ras signaling pathway in JMML.
Attempts to develop pharmacologic agents that target Ras proteins have, until very recently, been unsuccessful. Initial efforts focused on inhibiting posttranslational processing of Ras, which is necessary for plasma membrane translocation and activation. Specifically, the essential lipid modification at the C-terminal CAAX sequence catalyzed by farnesyl transferase was targeted with farnesyl transferase inhibitors (FTIs).107 Preclinical studies in a Nf−/− murine model of JMML with FTI L744 832 did not inhibit Nf−/− hematopoietic cell growth or splenic infiltration with myeloid progenitors.108 FTI treatment was associated with a decrease in farnesylation of Hras, but not Nras or Kras,108 likely because Kras and Nras (but not Hras) can undergo lipid modification via geranylgeranyl transferase when FTase is inhibited.107 Response rate and acute toxicities of the FTI Zarnesta were studied in the COG Phase II AAML0122 trial, and despite some responses in treated patients with decreased leukocytosis and organomegaly, EFS was not affected by the FTI.109
The palmitoylation/depalmitoylation cycle is another potential target for selectively inhibiting growth of NRAS mutant cancers. Xu et al110 demonstrated that mutating the palmitoylation site in NRASG12D inhibited plasma membrane localization and prevented myeloid disease in a retroviral transduction/transplantation model of MPN. When transduced hematopoietic cells from NRASG12D mice were exposed to palmostatin B, a novel acyl-protein thioesterase 1 inhibitor, NRASG12D was mislocalized away from the plasma membrane, and CFU-GM colony growth was reduced.110 Importantly, because the KRAS 4B isoform is not palmitoylated, acyl-protein thioesterase 1 inhibitors would only be applicable to NRAS mutants.
Most efforts to block activated Ras have focused on downstream effector pathways including the RAF-MEK-ERK and PI3K/AKT pathways. Preclinical studies in KrasG12D mutant and Nf1 mutant mice with MPN have provided a compelling biologic rationale for MEK inhibition in the treatment of JMML. In both studies, mutant mice treated with the allosteric MEK inhibitor PD0325901 (901) exhibited a rapid and sustained improvement of hematologic parameters (decreased leukocytosis, resolution of anemia, decreased reticulocytosis) and dramatic reduction in spleen size.83,84 Both models recapitulated the myeloid differentiation bias in JMML, with an expansion of the pre-GM population at the expense of the premegakaryocyte erythroid population in murine bone marrow. Importantly in both models, MEK inhibition worked by modifying the behavior of the mutant clone, restoring a normal program of hematopoietic output, rather than eliminating it. As a result of unacceptable ocular and neurologic toxicity, clinical development of 901 was halted. Current studies of MEK inhibitor AZD6244 are ongoing in pediatric gliomas and plexiform neurofibromas. Future trials of the US Food and Drug Administration-approved MEK inhibitor trametinib will investigate the effect of MEK inhibition on children with Ras-pathway-associated cancers, including JMML.
The PI3K/Akt/mammalian target of rapamycin pathway is activated in cancers with oncogenic Ras. Treatment of primary PTPN11 E76K JMML cells with low-dose rapamycin demonstrated antileukemic activity (M.L.L., unpublished data, June 2007). A preclinical trial in KrasG12D mutant mice with MPN using PI3K inhibitor GDC0941 demonstrated some effect with improvement of anemia in these mice, but abrogation of the MPN was not as significant as with MEK inhibition (Jon Akutagawa, Tannie Queen Huang, T.Y.C., Monique Dail, Lori S. Friedman, Deepak Sampath, and Benjamin S. Braun, manuscript submitted January 2014). Recent studies have begun to describe a key role of the PI3K/Akt pathway in PTPN11 and Cbl- mutated JMML. Chan et al111 reported that Shp2 E76K-expressing Pik3r1−/− cells displayed elevated protein levels of the PI3K catalytic subunit p110δ, suggesting it may be a mediator in downstream activation. Notably, treatment with the p110δ-specific inhibitor, IC87114, or GDC-0941 reduced GM-CSF hypersensitivity and hyperproliferation and hyperphosphorylation of Akt in gain-of-function Shp2 E76K-expressing cells.111 Bunda et al112 recently demonstrated that the JMML-associated Cbl(Y371H) mutant, in complex with the Src family kinase Lyn, promotes Cbl’s adapter function, leading to increased association to PI3K regulatory subunit p85 and Lyn-dependent AKT prosurvival signaling112,113 Pharmacologic inhibition of Src family kinases with PP2 or dasatinib reversed GM-CSF hypersensitivity of primary JMML cells and attenuated the hyperphosphorylation of Cbl(Y371H), p85 recruitment, and subsequent AKT phosphorylation. Furthermore, pharmacologic inhibition of the Lyn-PI3K/AKT pathway, but not the Ras/MAPK pathway, markedly increased the sensitivity of the otherwise chemoresistant Cbl mutant-JMML cells to chemotherapeutic agents.113
We and others described phosphorylation of STAT5 to low-dose GM-CSF in subsets of JMML cells.90,98,114 Whole-exome sequencing has also identified JAK3 mutations in patients with JMML.66 ADVL1011 was a phase 1 trial studying the JAK inhibitor Ruxolitinib in children with refractory solid tumors, leukemias, and MPN (NCT01164163). Given the findings from Li et al88 regarding the STAT5-dependent bimodal effect of NrasG12D expression in HSCs, it will be important to study the effects of downstream phosphorylated epitopes after exposure to JAK inhibitors.
A recent study evaluated the combination of JAK and MEK inhibition in a constitutively active oncogenic NrasG12D/G12D murine MPN and found that the combination abrogated murine MPN longer than the MEK inhibitor AZ6244 alone and suppressed transformation to T-ALL, although it is unclear how well AZ6244 achieved target inhibition.115
Finally, recent reports of aberrant DNA methylation contributing to increased risk for relapse81 has resulted in an EWOG/MDS and Innovative Therapies for Children with Cancer trial using the DNA hypomethylating agent azacitidine for treatment of pediatric patients with newly diagnosed or relapsed high-grade MDS or JMML (EUDRACT no. 2010-022235-10). Although azacitidine has been shown to improve survival in adults with MDS, to date, this strategy has only been published as a successful anecdote in a single JMML patient with monosomy 7 and KRAS mutation. This patient experienced resolution of organomegaly, monocytosis, thrombocytopenia, and monosomy 7 with molecular remission after an 18-month course, which likely facilitated successful HSCT.116
Summary and directions for future work
Similar to many pediatric cancers, JMML is an example of developmental hematopoiesis gone awry. From the bedside, astute observations in patients with developmental syndromes ultimately uncovered the genetic underpinnings of this disease. Functional in vitro and in vivo studies using patient samples and genetically accurate animal models confirmed that JMML is fundamentally a disease of hyperactive Ras. Despite our understanding of this disease, however, JMML remains one of the most difficult pediatric cancers to diagnose and treat. Recent advances at the bench have provided further insights to the molecular pathogenesis of JMML, improved means of diagnosis and surveillance, and importantly, generated the data to support our first clinical trials in patients using targeted agents.
Although rare, JMML is an excellent disease in which to study response and resistance to targeted therapy. First, the molecular genetics of this disease implicate hyperactive Ras as an essential initiating event for the majority of patients. Second, unlike adult cancers, the JMML genome is relatively quiet (few somatic mutations per patient have thus far been described) and may be more likely to be susceptible to specific therapeutic vulnerabilities, particularly in pathways reliant on Ras signaling. Finally, as work in AML has shown, hematopoietic malignancies lend themselves to studies on clonal evolution. Indeed, as clinical trials of MEK inhibitors in JMML begin, it will be critical to examine serial samples to understand why certain patients respond and others do not. Importantly, those who initially respond but ultimately relapse will provide insights to the potential mechanisms of resistance.
Authorship
Contribution: T.Y.C., C.C.D., and M.L.L. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Mignon Loh, Health Sciences East 302, 513 Parnassus Ave, San Francisco, CA 94143-0519; e-mail: lohm@peds.ucsf.edu.